Quantitative Analytical and Computational Workflow for Large-Scale Targeted Plasma Metabolomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Chemical, Reagent, and Standards
2.2. Preparation of Standards
2.3. Methods
2.3.1. Plasma Sample Preparation
2.3.2. LC-MS Method
2.3.3. AutoQC
2.4. Data Analysis
2.4.1. Peak Extraction and Processing with Skyline
2.4.2. Statistical Analysis
2.4.3. Method Validation Parameter Calculations
2.4.4. Quantification Using Relative Response Factors (RRF)
2.4.5. Targeted Identification and Fragmentation (MS/MS) Data
2.4.6. Data Availability
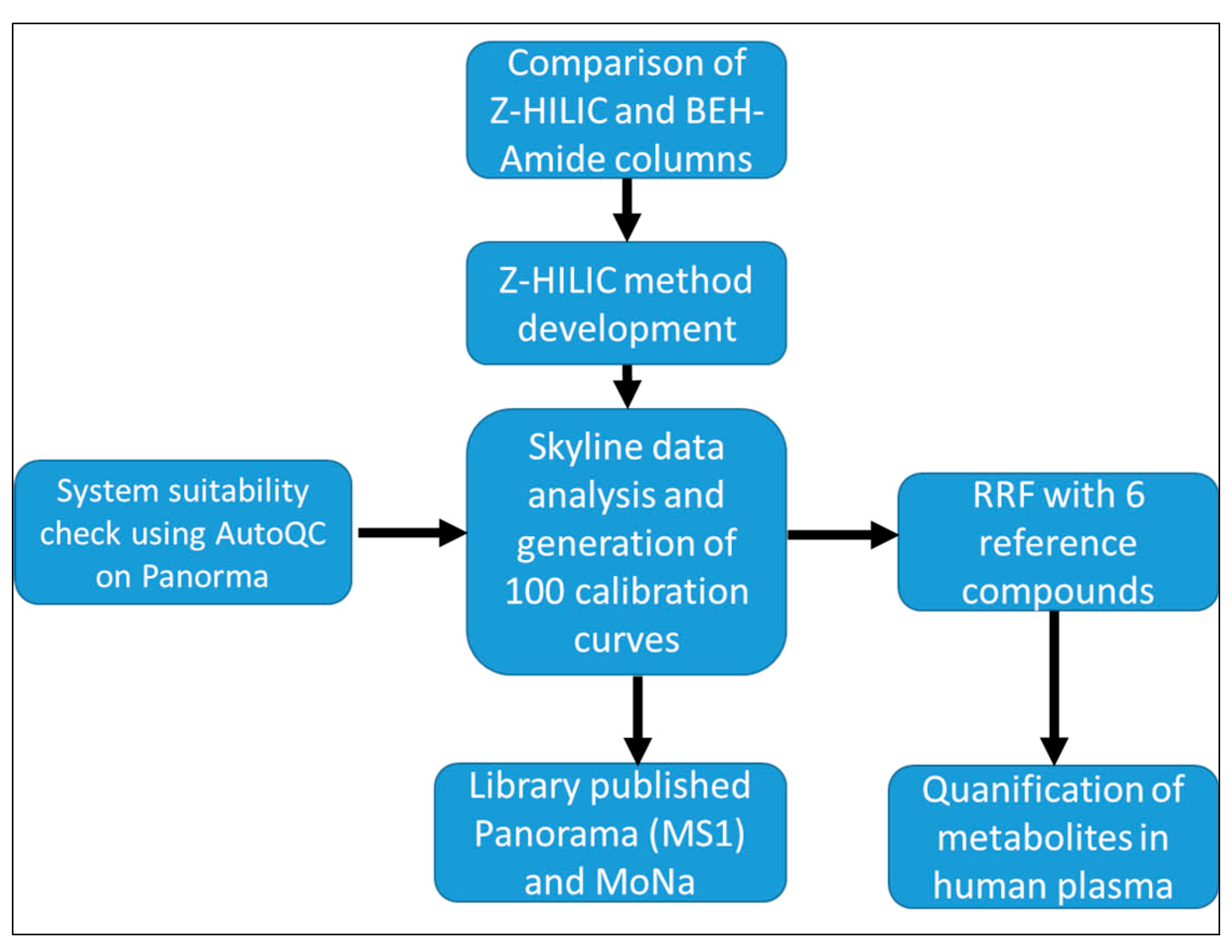
3. Results and Discussion
3.1. LC-MS Method Development
3.2. Overview of ISAS Quant Metabolomics Library (IQML)

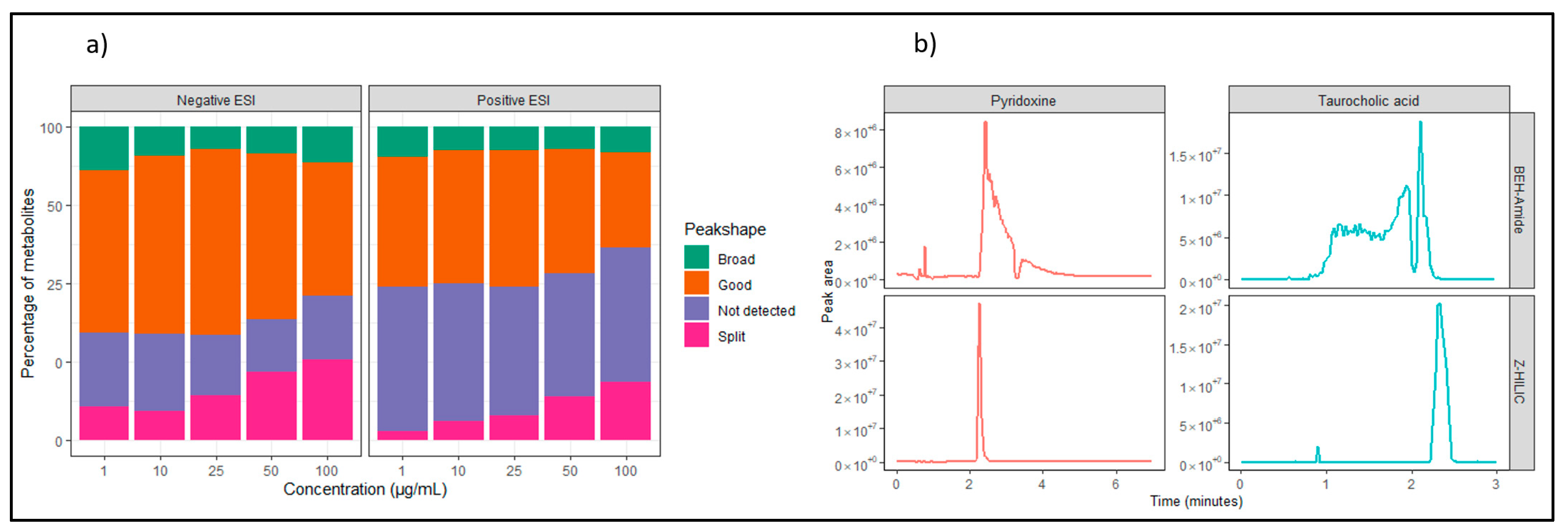
Quality Control, System Suitability, and Reproducibility
3.3. Quantitative Approach and Method Validation
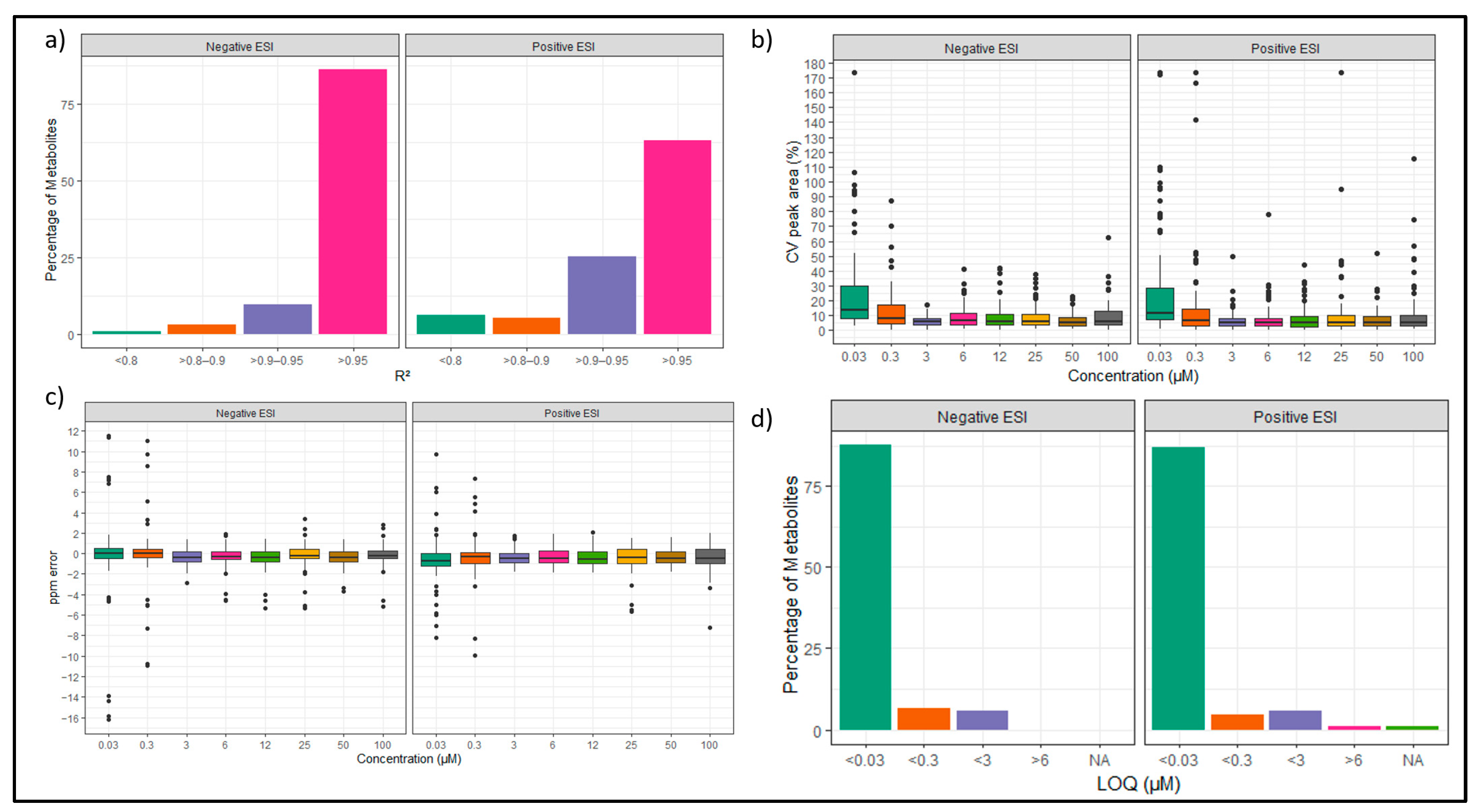
3.3.1. External Calibration Curve Evaluation for Linearity, Limit of Quantification (LOQ), and Dynamic Range
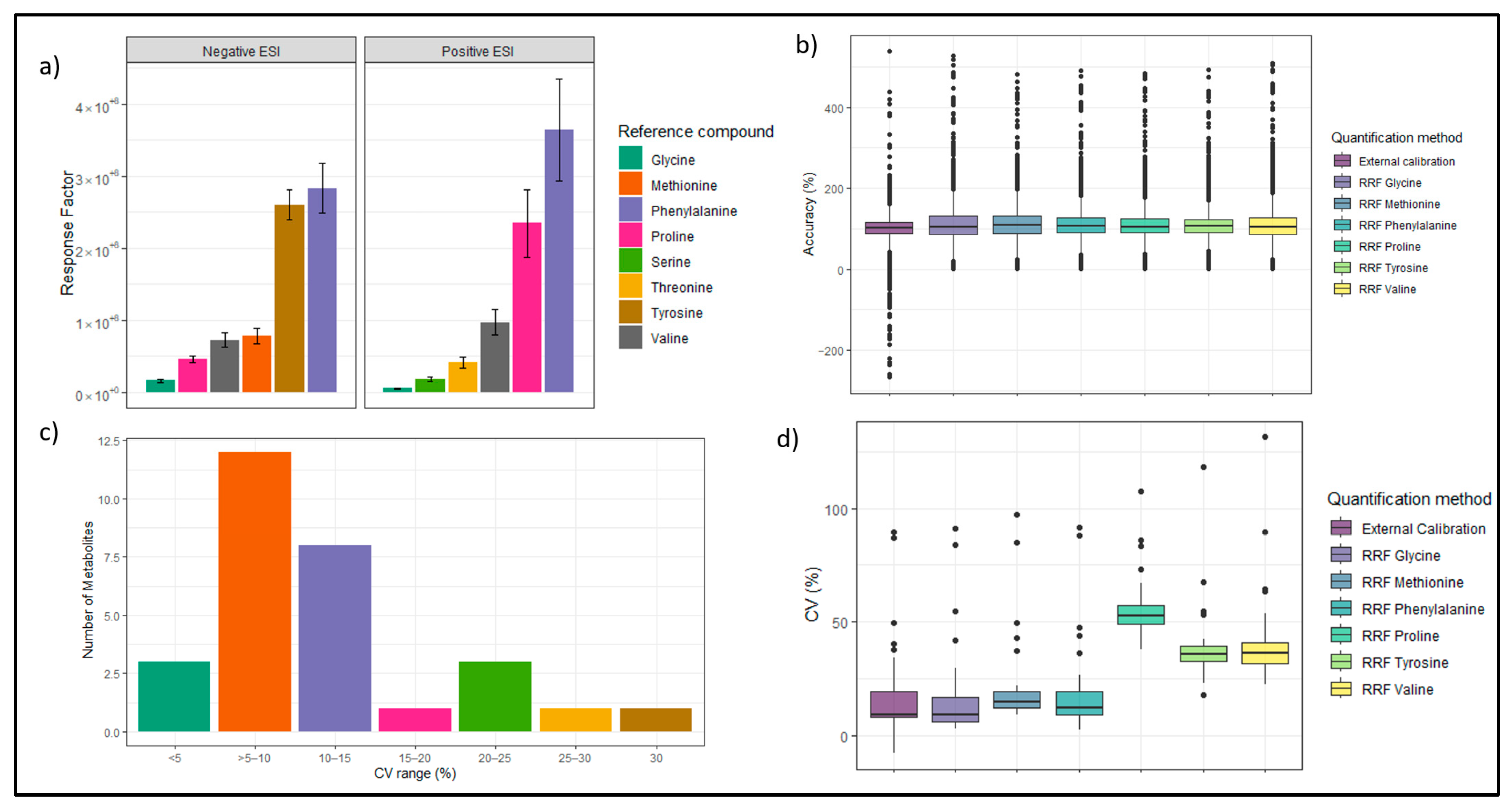
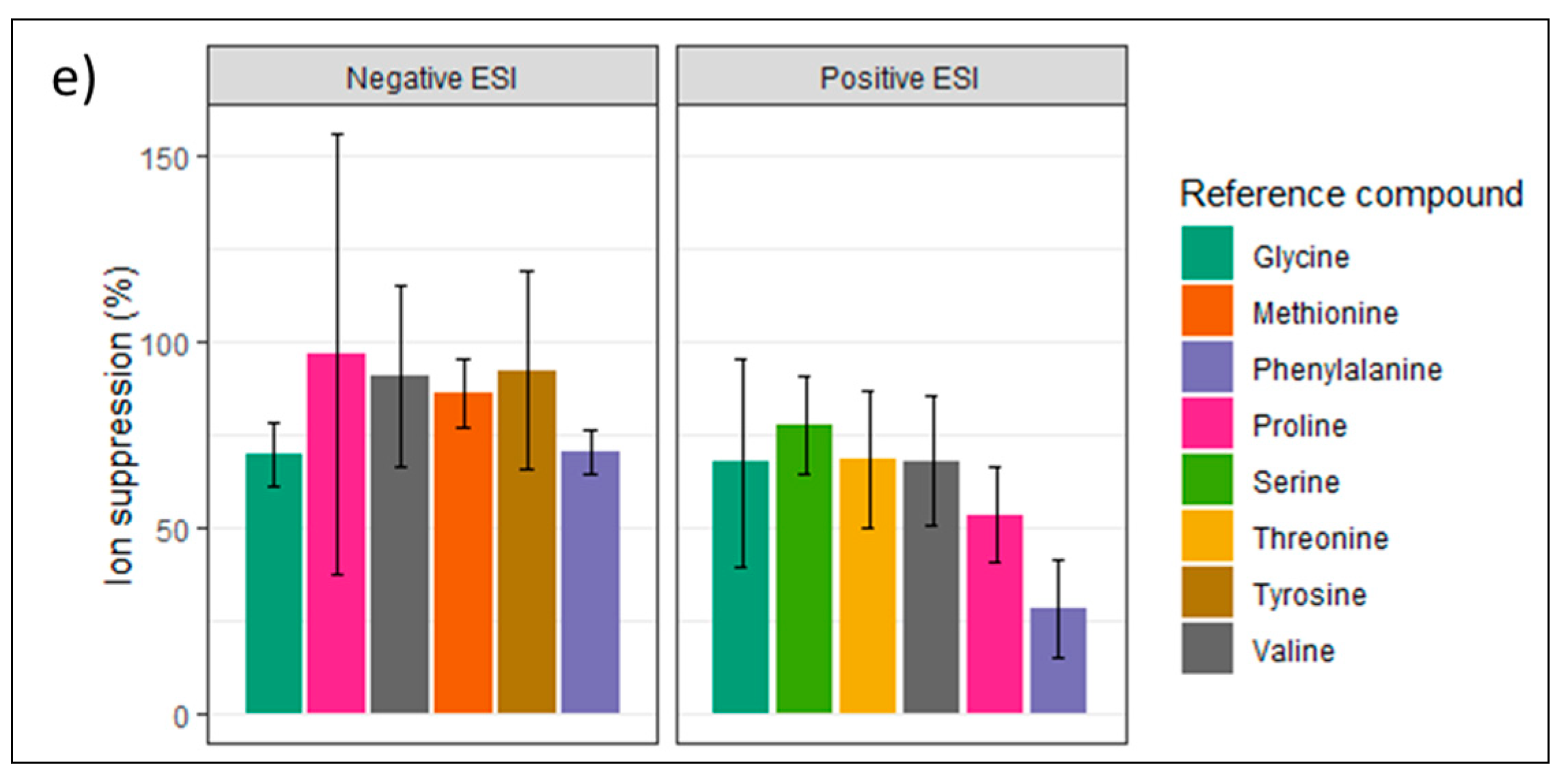
3.3.2. Relative Response Factor Evaluation in Standards
3.3.3. RRF-Based Quantification of Plasma Compared to External Calibration
3.3.4. Benchmarking with Reference Plasma Material (NIST 1950 SRM)
3.4. Limitations and Outlook of the Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Phapale, P. Pharmaco-metabolomics opportunities in drug development and clinical research. Anal. Sci. Adv. 2021, 2, 611–616. [Google Scholar] [CrossRef]
- Phapale, P.; Kim, S.-D.; Lee, H.W.; Lim, M.; Kale, D.; Kim, Y.-L.; Cho, J.-H.; Hwang, D.; Yoon, Y.-R. An Integrative Approach for Identifying a Metabolic Phenotype Predictive of Individualized Pharmacokinetics of Tacrolimus. Clin. Pharmacol. Ther. 2010, 87, 426–436. [Google Scholar] [CrossRef]
- Ussher, J.R.; Elmariah, S.; Gerszten, R.E.; Dyck, J.R. The Emerging Role of Metabolomics in the Diagnosis and Prognosis of Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2850–2870. [Google Scholar] [CrossRef]
- Kruve, A. Strategies for Drawing Quantitative Conclusions from Nontargeted Liquid Chromatography–High-Resolution Mass Spectrometry Analysis. Anal. Chem. 2020, 92, 4691–4699. [Google Scholar] [CrossRef]
- Groff, L.C., 2nd; Grossman, J.N.; Kruve, A.; Minucci, J.M.; Lowe, C.N.; McCord, J.P.; Kapraun, D.F.; Phillips, K.A.; Purucker, S.T.; Chao, A.; et al. Uncertainty Estimation Strategies for Quantitative Non-Targeted Analysis. Anal. Bioanal. Chem. 2022, 414, 4919–4933. [Google Scholar] [CrossRef] [PubMed]
- Sindelar, M.; Patti, G.J. Chemical Discovery in the Era of Metabolomics. J. Am. Chem. Soc. 2020, 142, 9097–9105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yin, Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2015, 88, 524–545. [Google Scholar] [CrossRef]
- Wood, P.L. Mass Spectrometry Strategies for Clinical Metabolomics and Lipidomics in Psychiatry, Neurology, and Neuro-Oncology. Neuropsychopharmacology 2013, 39, 24–33. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, F.; Zhu, J. Bridging Targeted and Untargeted Mass Spectrometry-Based Metabolomics via Hybrid Approaches. Metabolites 2020, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Phapale, P.B.; Blasche, S.; Patil, K.R.; Alexandrov, T. Quantification of Duloxetine in the Bacterial Culture and Medium to Study Drug-gut Microbiome Interactions. Bio-Protocol 2021, 11, e4214. [Google Scholar] [CrossRef] [PubMed]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Strategies and challenges in method development and validation for the absolute quantification of endogenous biomarker metabolites using liquid chromatography-tandem mass spectrometry. Mass Spectrom. Rev. 2019, 40, 31–52. [Google Scholar] [CrossRef]
- Thakare, R.; Chhonker, Y.S.; Gautam, N.; Alamoudi, J.A.; Alnouti, Y. Quantitative analysis of endogenous compounds. J. Pharm. Biomed. Anal. 2016, 128, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Nellis, M.; Uppal, K.; Ma, C.; Tran, V.; Liang, Y.; Walker, D.I.; Jones, D.P. Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem. 2020, 92, 8836–8844. [Google Scholar] [CrossRef]
- Gu, H.; Zhao, Y.; DeMichele, M.; Zheng, N.; Zhang, Y.J.; Pillutla, R.; Zeng, J. In-Sample Calibration Curve Using Multiple Isotopologue Reaction Monitoring of a Stable Isotopically Labeled Analyte for Instant LC-MS/MS Bioanalysis and Quantitative Proteomics. Anal. Chem. 2019, 91, 2536–2543. [Google Scholar] [CrossRef]
- Sun, F.; Wu, P.; Abdallah, M.F.; Tan, H.; Li, Y.; Yang, S. One sample multi-point calibration curve as a novel approach for quantitative LC-MS analysis: The quantitation of six aflatoxins in milk and oat-based milk as an example. Food Chem. 2023, 420, 135593. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Abdallah, M.F.; Tan, H.; Li, J.; Liu, S.; Zhang, R.; Sun, F.; Li, Y.; Yang, S. Novel one-point calibration strategy for high-throughput quantitation of microcystins in freshwater using LC-MS/MS. Sci. Total. Environ. 2023, 858, 159345. [Google Scholar] [CrossRef]
- Rule, G.S.; Rockwood, A.L. Improving quantitative precision and throughput by reducing calibrator use in liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2016, 919, 55–61. [Google Scholar] [CrossRef]
- Visconti, G.; Olesti, E.; González-Ruiz, V.; Glauser, G.; Tonoli, D.; Lescuyer, P.; Vuilleumier, N.; Rudaz, S. Internal calibration as an emerging approach for endogenous analyte quantification: Application to steroids. Talanta 2022, 240, 123149. [Google Scholar] [CrossRef]
- Khamis, M.M.; Klemm, N.; Adamko, D.J.; El-Aneed, A. Comparison of accuracy and precision between multipoint calibration, single point calibration, and relative quantification for targeted metabolomic analysis. Anal. Bioanal. Chem. 2018, 410, 5899–5913. [Google Scholar] [CrossRef] [PubMed]
- van der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical Error Reduction Using Single Point Calibration for Accurate and Precise Metabolomic Phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef] [PubMed]
- Le-Trilling, V.T.K.; Mennerich, D.; Schuler, C.; Sakson, R.; Lill, J.K.; Kasarla, S.S.; Kopczynski, D.; Loroch, S.; Flores-Martinez, Y.; Katschinski, B.; et al. Identification of Herbal Teas and Their Compounds Eliciting Antiviral Activity against SARS-CoV-2 in Vitro. BMC Biol. 2022, 20, 264. [Google Scholar] [CrossRef]
- Coglianese, A.; Charlier, B.; Mensitieri, F.; Filippelli, A.; Izzo, V.; Piaz, F.D. Standard addition method (SAM) in LC-MS/MS to quantify gluten-derived metabolites in urine samples. J. Pharm. Biomed. Anal. 2023, 232, 115416. [Google Scholar] [CrossRef] [PubMed]
- Liigand, J.; Kruve, A.; Liigand, P.; Laaniste, A.; Girod, M.; Antoine, R.; Leito, I. Transferability of the Electrospray Ionization Efficiency Scale between Different Instruments. J. Am. Soc. Mass Spectrom. 1923, 26, 5079–5086. [Google Scholar] [CrossRef]
- Liigand, P.; Liigand, J.; Kaupmees, K.; Kruve, A. 30 Years of research on ESI/MS response: Trends, contradictions and applications. Anal. Chim. Acta 2020, 1152, 238117. [Google Scholar] [CrossRef]
- Liigand, J.; Laaniste, A.; Kruve, A. pH Effects on Electrospray Ionization Efficiency. J. Am. Soc. Mass Spectrom. 2016, 28, 461–469. [Google Scholar] [CrossRef]
- Sonnenberg, R.A.; Naz, S.; Cougnaud, L.; Vuckovic, D. Comparison of underivatized silica and zwitterionic sulfobetaine hydrophilic interaction liquid chromatography stationary phases for global metabolomics of human plasma. J. Chromatogr. A 2019, 1608, 460419. [Google Scholar] [CrossRef]
- Serafimov, K.; Lämmerhofer, M. Metabolic profiling workflow for cell extracts by targeted hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2022, 1684, 463556. [Google Scholar] [CrossRef]
- Sharma, V.; Eckels, J.; Schilling, B.; Ludwig, C.; Jaffe, J.D.; MacCoss, M.J.; MacLean, B. Panorama Public: A Public Repository for Quantitative Data Sets Processed in Skyline. Mol. Cell. Proteom. 2018, 17, 1239–1244. [Google Scholar] [CrossRef]
- Adams, K.J.; Pratt, B.; Bose, N.; Dubois, L.G.; St. John-Williams, L.; Perrott, K.M.; Ky, K.; Kapahi, P.; Sharma, V.; MacCoss, M.J.; et al. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res. 2020, 19, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Rardin, M.J. Rapid Assessment of Contaminants and Interferences in Mass Spectrometry Data Using Skyline. J. Am. Soc. Mass Spectrom. 2018, 29, 1327–1330. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Phapale, P.; Palmer, A.; Gathungu, R.M.; Kale, D.; Brügger, B.; Alexandrov, T. Public LC-Orbitrap Tandem Mass Spectral Library for Metabolite Identification. J. Proteome Res. 2021, 20, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- MassBank of North America. Available online: https://mona.fiehnlab.ucdavis.edu/ (accessed on 5 June 2023).
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A Positive/Negative Ion–Switching, Targeted Mass Spectrometry–Based Metabolomics Platform for Bodily Fluids, Cells, and Fresh and Fixed Tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef]
- Bereman, M.S.; Beri, J.; Sharma, V.; Nathe, C.; Eckels, J.; MacLean, B.; MacCoss, M.J. An Automated Pipeline to Monitor System Performance in Liquid Chromatography–Tandem Mass Spectrometry Proteomic Experiments. J. Proteome Res. 2016, 15, 4763–4769. [Google Scholar] [CrossRef]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef]
- Wang, J.; Peake, D.A.; Mistrik, R.; Huang, Y. A Platform to Identify Endogenous Metabolites Using a Novel High Performance Orbitrap MS and the MzCloud Library. Blood 2013, 4, 2–8. [Google Scholar]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Simón-Manso, Y.; Lowenthal, M.S.; Kilpatrick, L.E.; Sampson, M.L.; Telu, K.H.; Rudnick, P.A.; Mallard, W.G.; Bearden, D.W.; Schock, T.B.; Tchekhovskoi, D.V.; et al. Metabolite Profiling of a NIST Standard Reference Material for Human Plasma (SRM 1950): GC-MS, LC-MS, NMR, and Clinical Laboratory Analyses, Libraries, and Web-Based Resources. Anal. Chem. 2013, 85, 11725–11731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tong, M.; Xu, H.; Yin, Y.; Gao, P.; Bi, K.; Zhang, Y.; Li, Q. Accurate determination for lipidomics based on LC-tandem-MS parameters modeling, prediction, and database: Monitoring the progression of hepatocellular carcinoma. J. Pharm. Biomed. Anal. 2023, 223, 115126. [Google Scholar] [CrossRef]
- Lam, S.M.; Tian, H.; Shui, G. Lipidomics, en route to accurate quantitation. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2017, 1862, 752–761. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Han, X. Tutorial on lipidomics. Anal. Chim. Acta 2019, 1061, 28–41. [Google Scholar] [CrossRef]
- Musmade, B.D.; Sawant, A.V.; Kulkarni, S.V.; Nage, S.D.; Bhope, S.G.; Padmanabhan, S.; Lohar, K.S. Method Development, Validation and Estimation of Relative Response Factor for the Quantitation of Known Impurities in Mometasone Furoate Nasal Spray Dosage form by RP-HPLC with UV/PDA Detector. Pharm. Chem. J. 2022, 56, 538–544. [Google Scholar] [CrossRef]
- Blanz, J.; Williams, G.; Dayer, J.; Délémonté, T.; Gertsch, W.; Ramstein, P.; Aichholz, R.; Trunzer, M.; Pearson, D. Evaluation of relative MS response factors of drug metabolites for semi-quantitative assessment of chemical liabilities in drug discovery. J. Mass Spectrom. 2017, 52, 210–217. [Google Scholar] [CrossRef]
- Liu, L.; Mouallem, A.; Xiao, K.P.; Meisel, J. Assay of active pharmaceutical ingredients in drug products based on relative response factors: Instrumentation insights and practical considerations. J. Pharm. Biomed. Anal. 2020, 194, 113760. [Google Scholar] [CrossRef]
| Metabolite Super Class | Use | Number of Metabolites |
|---|---|---|
| Organic acids and derivatives | Standard | 74 |
| Lipids and lipid-like molecules | Standard | 51 |
| Organoheterocyclic compounds | Standard | 42 |
| Organic oxygen compounds | Standard | 40 |
| Nucleosides, nucleotides, and analogues | Standard | 33 |
| Benzenoids | Standard | 19 |
| Organic nitrogen compounds | Standard | 11 |
| Phenylpropanoids and polyketides | Standard | 8 |
| Alkaloids and derivatives | Standard | 1 |
| Organosulfur compounds | Standard | 1 |
| 13C Amino acids | Internal standard/Reference compound | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fecke, A.; Saw, N.M.M.T.; Kale, D.; Kasarla, S.S.; Sickmann, A.; Phapale, P. Quantitative Analytical and Computational Workflow for Large-Scale Targeted Plasma Metabolomics. Metabolites 2023, 13, 844. https://doi.org/10.3390/metabo13070844
Fecke A, Saw NMMT, Kale D, Kasarla SS, Sickmann A, Phapale P. Quantitative Analytical and Computational Workflow for Large-Scale Targeted Plasma Metabolomics. Metabolites. 2023; 13(7):844. https://doi.org/10.3390/metabo13070844
Chicago/Turabian StyleFecke, Antonia, Nay Min Min Thaw Saw, Dipali Kale, Siva Swapna Kasarla, Albert Sickmann, and Prasad Phapale. 2023. "Quantitative Analytical and Computational Workflow for Large-Scale Targeted Plasma Metabolomics" Metabolites 13, no. 7: 844. https://doi.org/10.3390/metabo13070844
APA StyleFecke, A., Saw, N. M. M. T., Kale, D., Kasarla, S. S., Sickmann, A., & Phapale, P. (2023). Quantitative Analytical and Computational Workflow for Large-Scale Targeted Plasma Metabolomics. Metabolites, 13(7), 844. https://doi.org/10.3390/metabo13070844









