Differential Inhibition of Anaplerotic Pyruvate Carboxylation and Glutaminolysis-Fueled Anabolism Underlies Distinct Toxicity of Selenium Agents in Human Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culturing, Se Treatments, and Metabolite Extraction
2.2. Ex Vivo Organotypic Tissue Culture (OTC) Experiments
2.3. NMR Spectroscopy
2.4. GC-MS Analysis
2.5. Ion Chromatography-Ultrahigh Resolution Fourier Transform Mass Spectrometry (IC-UHR-FTMS1)
2.6. FT-ICR-MS and FT-MS Analysis
2.7. Cell Proliferation Assay
2.8. Apoptosis/Necrosis, ROS, and Cell Cycle Analyses
2.9. Cell Cycle Analysis
2.10. Transwell Migration Assay
2.11. Enzyme Assays
2.12. Oxygen Consumption Analysis
2.13. shRNA Knockdown Experiments
2.14. Reverse Phase Protein Array (RPPA) and Western Blot Analysis
2.15. Immunohistofluorescence Analysis
2.16. Statistical Analyses
3. Results
3.1. Lung Cancer Cell Proliferation Is Inhibited by Selenite, MSeA, and SeM but Their EC50 Differs Widely and Is Cell Line-Dependent
3.2. Selenite, MSeA, and SeM Differentially Induce ROS/Apoptosis and Inhibit Metastatic Potential in Lung Cancer Cells
3.3. Selenite and MSeA Perturb Glycolysis and Major Nutrient Consumption in Lung Cancer Cells but These Effects Cannot Account for the Altered Phenotypes
3.4. MSeA and Selenite Block the Krebs Cycle Activity in Lung Cancer Cells While SeM Does Not, Which May Underlie Their Differential Effect on Proliferation
3.5. Selenite, MSeA, and SeM Differentially Block O2 Consumption and Mitochondrial Respiratory Capacity in Lung Cancer Cells
3.6. Selenite, MSeA, and SeM Differentially Block Glutaminolysis in Lung Cancer Cells
3.7. Inhibition of PC Activity and Suppression of GLS1 Expression Respectively Contribute to Reduced Krebs Cycle Activity Induced by MSeA and Selenite
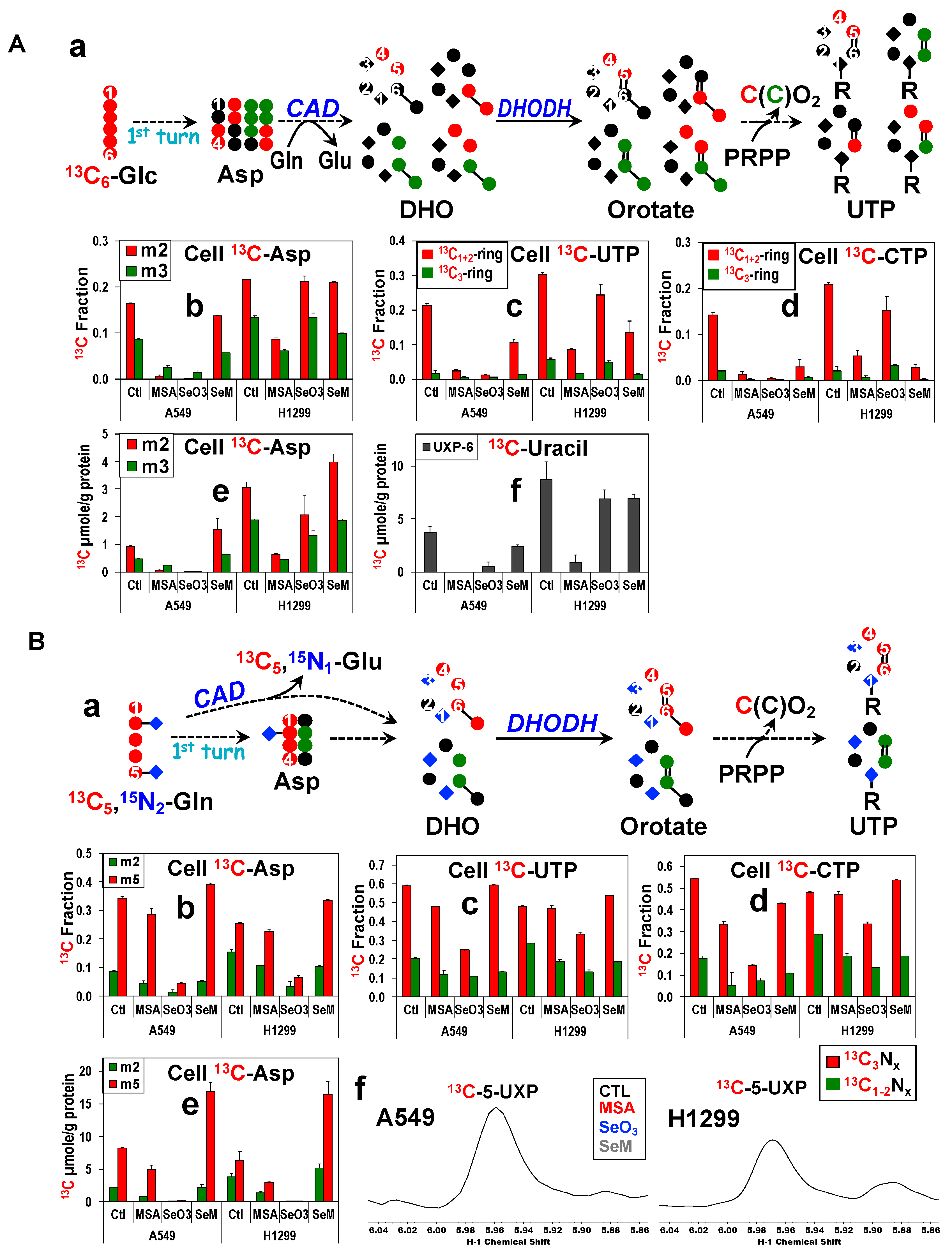
3.8. MSeA Inhibition of PC and Selenite Suppression of GAC/PC/CAD Lead to Reduced Pyrimidine Nucleotide Synthesis
3.9. MseA and Selenite Block Purine Ring Synthesis in A549 Cells via Reduced Gly and Ribose Synthesis
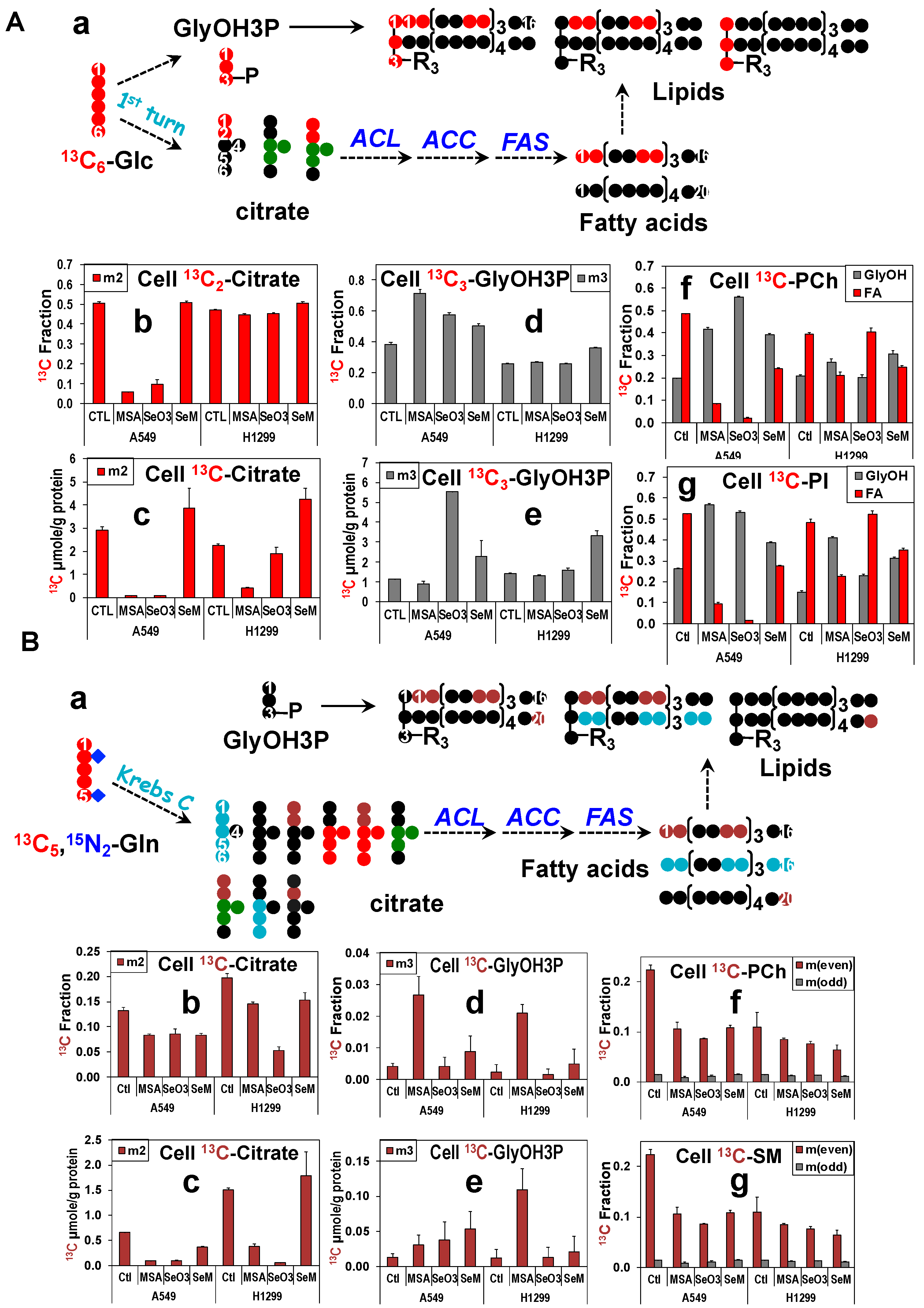
3.10. MSeA, Selenite, and SeM Differentially Block Glucose or Gln-Fueled Fatty Acyl Synthesis but Not Glycerol Backbone Incorporation into Lipids
3.11. PC or GLS1 Knockdown Inhibits Lung Cancer Cell Proliferation but Their Effects on ROS Production, Cell Cycle Arrest, and Cell Death Are Cell Type-Dependent
3.12. MSeA or Selenite Block Pyruvate Carboxylation and/or Glutaminolysis, Which Was Accompanied by Necrosis in Ex Vivo Organotypic Cultures of Human NSCLC Tissues
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayat, M.J.; Howlader, N.; Reichman, M.E.; Edwards, B.K. Cancer statistics, trends, and multiple primary cancer analyses from the surveillance, epidemiology, and end results (SEER) program. Oncologist 2007, 12, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Liu, L.Z.; Dong, M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol. Cancer 2021, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Azim, H.A.; Ganti, A.K. Targeted therapy in advanced non-small cell lung cancer (NSCLC): Where do we stand? Cancer Treat. Rev. 2006, 32, 630–636. [Google Scholar] [CrossRef]
- Pirozynski, M. 100 years of lung cancer. Respir. Med. 2006, 100, 2073–2084. [Google Scholar] [CrossRef]
- Clark, L.C.; Dalkin, B.; Krongrad, A.C., Jr.; Turnbull, B.W.; Slate, E.H.; Witherington, R.; Herlong, J.H.; Janosko, E.; Carpenter, D.; Borosso, C.; et al. Decreased incidence of prostate cancer with selenium supplementation: Results of a double-blind cancer prevention trial. Br. J. Urol. 1998, 81, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.C.; Combs, G.F., Jr.; Turnbull, B.W.; Slate, E.H.; Chalker, D.K.; Chow, J.; Davis, L.S.; Glover, R.A.; Graham, G.F.; Gross, E.G.; et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA 1996, 276, 1957–1963. [Google Scholar] [CrossRef]
- Zu, K.; Bihani, T.; Lin, A.; Park, Y.-M.; Mori, K.; Ip, C. Enhanced selenium effect on growth arrest by BiP/GRP78 knockdown in p53-null human prostate cancer cells. Oncogene 2006, 25, 546–554. [Google Scholar] [CrossRef]
- Kaeck, M.; Lu, J.; Strange, R.; Ip, C.; Ganther, H.E.; Thompson, H.J. Differential induction of growth arrest inducible genes by selenium compounds. Biochem. Pharmacol. 1997, 53, 921–926. [Google Scholar] [CrossRef]
- Jönsson-Videsäter, K.; Björkhem-Bergman, L.; Hossain, A.; Söderberg, A.; Eriksson, L.C.; Paul, C.; Rosén, A.; Björnstedt, M. Selenite-induced apoptosis in doxorubicin-resistant cells and effects on the thioredoxin system. Biochem. Pharmacol. 2004, 67, 513–522. [Google Scholar] [CrossRef]
- Li, L.; Xie, Y.; El-Sayed, W.M.; Szakacs, J.G.; Roberts, J.C. Characteristics of selenazolidine prodrugs of selenocysteine: Toxicity, selenium levels, and glutathione peroxidase induction in A/J mice. Life Sci. 2004, 75, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Swede, H.; Dong, Y.; Reid, M.; Marshall, J.; Ip, C. Cell cycle arrest biomarkers in human lung cancer cells after treatment with selenium in culture. Cancer Epidemiol. Biomark. Prev. 2003, 12, 1248–1252. [Google Scholar]
- El-Bayoumy, K.; Sinha, R. Molecular chemoprevention by selenium: A genomic approach. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2005, 591, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. Selenium in cancer prevention: A review of the evidence and mechanism of action. Proc. Nutr. Soc. 2005, 64, 527–542. [Google Scholar] [CrossRef]
- Hu, H.; Jiang, C.; Ip, C.; Rustum, Y.M.; Lu, J. Methylseleninic acid potentiates apoptosis induced by chemotherapeutic drugs in androgen-independent prostate cancer cells. Clin. Cancer Res. 2005, 11, 2379–2388. [Google Scholar] [CrossRef]
- Shah, Y.M.; Al-Dhaheri, M.; Dong, Y.; Ip, C.; Jones, F.E.; Rowan, B.G. Selenium disrupts estrogen receptor (alpha) signaling and potentiates tamoxifen antagonism in endometrial cancer cells and tamoxifen-resistant breast cancer cells. Mol. Cancer Ther. 2005, 4, 1239–1249. [Google Scholar] [CrossRef]
- Cao, S.; Durrani, F.; Rustum, Y. Selective modulation of the therapeutic efficacy of anticancer drugs by selenium containing compounds against human tumor xenografts. Clin. Cancer Res. 2004, 10, 2561–2569. [Google Scholar] [CrossRef]
- Selenius, M.; Rundlöf, A.-K.; Olm, E.; Fernandes, A.P.; Björnstedt, M. Selenium and the selenoprotein thioredoxin reductase in the prevention, treatment and diagnostics of cancer. Antioxid. Redox Signal. 2010, 12, 867–880. [Google Scholar] [CrossRef]
- Kandas, N.O.; Randolph, C.; Bosland, M.C. Differential effects of selenium on benign and malignant prostate epithelial cells: Stimulation of LNCaP cell growth by noncytotoxic, low selenite concentrations. Nutr. Cancer 2009, 61, 251–264. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Tang, Y.-C.; Chen, Y.-H.; Wong, C.-M.; Tsou, A.-P. Selenite-induced survival of HuH7 hepatoma cells involves activation of focal adhesion kinase-phosphatidylinositol 3-kinase-Akt pathway and Rac1. J. Biol. Chem. 2003, 278, 39615–39624. [Google Scholar] [CrossRef]
- Kim, A.; Jung, J.-Y.; Son, M.; Lee, S.-H.; Lim, J.-S.; Chung, A.-S.; Kim, Y. Long exposure of non-cytotoxic concentrations of methylselenol suppresses the invasive potential of B16F10 melanoma. Oncol. Rep. 2008, 20, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Endo, M.; Shinohara, F.; Echigo, S.; Rikiishi, H. Differential apoptotic response of human cancer cells to organoselenium compounds. Cancer Chemother. Pharmacol. 2010, 66, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Demars, L.C. Dietary supplementation with methylseleninic acid, but not selenomethionine, reduces spontaneous metastasis of Lewis lung carcinoma in mice. Int. J. Cancer J. Int. Cancer 2011, 131, 1260–1266. [Google Scholar] [CrossRef]
- Liu, C.; Liu, H.; Li, Y.; Wu, Z.; Zhu, Y.; Wang, T.; Gao, A.C.; Chen, J.; Zhou, Q. Intracellular glutathione content influences the sensitivity of lung cancer cell lines to methylseleninic acid. Mol. Carcinog. 2012, 51, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Sinha, I.; Null, K.; Wolter, W.; Suckow, M.A.; King, T.; Pinto, J.T.; Sinha, R. Methylseleninic acid downregulates hypoxia-inducible factor-1alpha in invasive prostate cancer. Int. J. Cancer J. Int. Cancer 2012, 130, 1430–1439. [Google Scholar] [CrossRef]
- Poerschke, R.L.; Franklin, M.R.; Moos, P.J. Modulation of redox status in human lung cell lines by organoselenocompounds: Selenazolidines, selenomethionine, and methylseleninic acid. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2008, 22, 1761–1767. [Google Scholar] [CrossRef]
- Olm, E.; Fernandes, A.P.; Hebert, C.; Rundlöf, A.-K.; Larsen, E.H.; Danielsson, O.; Björnstedt, M. Extracellular thiol-assisted selenium uptake dependent on the x(c)- cystine transporter explains the cancer-specific cytotoxicity of selenite. Proc. Natl. Acad. Sci. USA 2009, 106, 11400–11405. [Google Scholar] [CrossRef]
- Selenius, M.; Fernandes, A.P.; Brodin, O.; Björnstedt, M.; Rundlöf, A.-K. Treatment of lung cancer cells with cytotoxic levels of sodium selenite: Effects on the thioredoxin system. Biochem. Pharmacol. 2008, 75, 2092–2099. [Google Scholar] [CrossRef]
- Suzuki, M.; Endo, M.; Shinohara, F.; Echigo, S.; Rikiishi, H. Rapamycin suppresses ROS-dependent apoptosis caused by selenomethionine in A549 lung carcinoma cells. Cancer Chemother. Pharmacol. 2011, 67, 1129–1136. [Google Scholar] [CrossRef]
- Li, S.; Zhou, Y.; Wang, R.; Zhang, H.; Dong, Y.; Ip, C. Selenium sensitizes MCF-7 breast cancer cells to doxorubicin-induced apoptosis through modulation of phospho-Akt and its downstream substrates. Mol. Cancer Ther. 2007, 6, 1031–1038. [Google Scholar] [CrossRef]
- Combs, G.F.; Clark, L.C.; Turnbull, B.W. An analysis of cancer prevention by selenium. Biofactors 2001, 14, 153–159. [Google Scholar] [CrossRef]
- Reid, M.E.; Duffield-Lillico, A.J.; Garland, L.; Turnbull, B.W.; Clark, L.C.; Marshall, J.R. Selenium supplementation and lung cancer incidence: An update of the nutritional prevention of cancer trial. Cancer Epidemiol. Biomark. Prev. 2002, V11, 1285–1291. [Google Scholar]
- Reid, M.E.; Stratton, M.; Lillico, A.J.; Fakih, M.; Natarajan, R.; Clark, L.C.; Marshall, J.R. A report of high-dose selenium supplementation: Response and toxicities. J. Trace Elem. Med. Biol. 2004, 18, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kamat, A.; Svatek, R.; Sm, L.; Ea, K.; Pj, G.; Lucia; Im, T.; Lg, F.; Hl, P.; Lm, M. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: The Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 2009, 301, 39–51. [Google Scholar]
- Karp, D.; Lee, S.; Keller, S.; Wright, G.S.; Aisner, S.; Belinsky, S.A.; Johnson, D.; Johnston, M.; Goodman, G.; Clamon, G.; et al. Randomized, Double-Blind, Placebo-Controlled, Phase III Chemoprevention Trial of Selenium Supplementation in Patients With Resected Stage I Non-Small-Cell Lung Cancer: ECOG 5597. J. Clin. Oncol. 2013, 31, 4179–4187. [Google Scholar] [CrossRef]
- Algotar, A.M.; Stratton, M.S.; Ahmann, F.R.; Ranger-Moore, J.; Nagle, R.B.; Thompson, P.A.; Slate, E.; Hsu, C.H.; Dalkin, B.L.; Sindhwani, P.; et al. Phase 3 clinical trial investigating the effect of selenium supplementation in men at high-risk for prostate cancer. Prostate 2013, 73, 328–335. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Del Giovane, C.; Dennert, G.; Zwahlen, M.; Brinkman, M.; Zeegers, M.P.; Horneber, M.; Roberto D’Amico; Crespi, C. Selenium for preventing cancer. Cochrane Database Syst. Rev. 2018, 2018, CD005195. [Google Scholar] [CrossRef]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef]
- Hwang, J.-T.; Kim, Y.M.; Surh, Y.-J.; Baik, H.W.; Lee, S.-K.; Ha, J.; Park, O.J. Selenium Regulates Cyclooxygenase-2 and Extracellular Signal-Regulated Kinase Signaling Pathways by Activating AMP-Activated Protein Kinase in Colon Cancer Cells. Cancer Res. 2006, 66, 10057–10063. [Google Scholar] [CrossRef]
- Lee, Y.-K.; Park, S.Y.; Kim, Y.-M.; Kim, D.C.; Lee, W.S.; Surh, Y.-J.; Park, O.J. Suppression of mTOR via Akt dependent and independent mechanisms in selenium treated colon cancer cells: Involvement of AMPK [10]1. Carcinogenesis 2010, 31, 1092–1099. [Google Scholar] [CrossRef]
- Huang, F.; Nie, C.; Yang, Y.; Yue, W.; Ren, Y.; Shang, Y.; Wang, X.; Jin, H.; Xu, C.; Chen, Q. Selenite induces redox-dependent Bax activation and apoptosis in colorectal cancer cells. Free Radic. Biol. Med. 2009, 46, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Jiang, W.; Ip, C.; Ganther, H.; Lu, J. Selenium-induced inhibition of angiogenesis in mammary cancer at chemopreventive levels of intake. Mol. Carcinog. 1999, 26, 213–225. [Google Scholar] [CrossRef]
- Ip, C.; Thompson, H.J.; Zhu, Z.; Ganther, H.E. In vitro and in vivo studies of methylseleninic acid: Evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 2000, 60, 2882–2886. [Google Scholar] [PubMed]
- Brozmanová, J.; Mániková, D.; Vlčková, V.; Chovanec, M. Selenium: A double-edged sword for defense and offence in cancer. Arch. Toxicol. 2010, 84, 919–938. [Google Scholar] [CrossRef] [PubMed]
- Prokopczyk, B.; Amin, S.; Desai, D.H.; Kurtzke, C.; Upadhyaya, P.; El-Bayoumy, K. Effects of 1,4-phenylenebis(methylene)selenocyanate and selenomethionine on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced tumorigenesis in A/J mouse lung. Carcinogenesis 1997, 18, 1855–1857. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-X.; Lee, H.-J.; Wang, Z.; Hu, H.; Liao, J.D.; Watts, J.C.; Combs, G.F., Jr.; Lü, J. Superior in vivo inhibitory efficacy of methylseleninic acid against human prostate cancer over selenomethionine or selenite. Carcinogenesis 2008, 29, 1005–1012. [Google Scholar] [CrossRef]
- Hu, H.; Li, G.-X.; Wang, L.; Watts, J.; Combs, G.F.; Lu, J. Methylseleninic acid enhances taxane drug efficacy against human prostate cancer and down-regulates antiapoptotic proteins Bcl-XL and survivin. Clin. Cancer Res. 2008, 14, 1150–1158. [Google Scholar] [CrossRef]
- Poerschke, R.L.; Moos, P.J. Thioredoxin reductase 1 knockdown enhances selenazolidine cytotoxicity in human lung cancer cells via mitochondrial dysfunction. Biochem. Pharmacol. 2011, 81, 211–221. [Google Scholar] [CrossRef]
- Yan, L.; Spallholz, J.E. Generation of Reactive Oxygen Species from the Reaction of Selenium Compounds with Thiols and Mammary Tumor Cells. Biochem. Pharmacol. 1993, 45, 429–437. [Google Scholar]
- Li, G.-X.; Hu, H.; Jiang, C.; Schuster, T.; Lü, J. Differential involvement of reactive oxygen species in apoptosis induced by two classes of selenium compounds in human prostate cancer cells. Int. J. Cancer 2007, 120, 2034–2043. [Google Scholar] [CrossRef]
- Zhao, R.; Xiang, N.; Domann, F.E.; Zhong, W. Expression of p53 enhances selenite-induced superoxide production and apoptosis in human prostate cancer cells. Cancer Res. 2006, 66, 2296–2304. [Google Scholar] [CrossRef] [PubMed]
- Xiang, N.; Zhao, R.; Zhong, W. Sodium selenite induces apoptosis by generation of superoxide via the mitochondrial-dependent pathway in human prostate cancer cells. Cancer Chemother. Pharm. 2009, 63, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Shilo, S.; Aronis, A.; Komarnitsky, R.; Tirosh, O. Selenite sensitizes mitochondrial permeability transition pore opening in vitro and in vivo: A possible mechanism for chemo-protection. Biochem. J. 2003, 370, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Domann, F.E.; Zhong, W. Apoptosis induced by selenomethionine and methioninase is superoxide mediated and p53 dependent in human prostate cancer cells. Mol. Cancer Ther. 2006, 5, 3275–3284. [Google Scholar] [CrossRef]
- Chiang, E.C.; Bostwick, D.G.; Waters, D.J. Homeostatic housecleaning effect of selenium: Evidence that noncytotoxic oxidant-induced damage sensitizes prostate cancer cells to organic selenium-triggered apoptosis. Biofactors 2013, 39, 575–588. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Bandura, L.L.; Higashi, R.M.; Lane, A.N. Metabolomics-edited transcriptomics analysis of Se anticancer action in human lung cancer cells. Metabolomics 2005, 1, 325–339. [Google Scholar] [CrossRef]
- Nelson, D.L.; Cox, M. Lehninger Principles of Biochemistry, 5th ed.; W. H. Freeman: New York, NY, USA, 2008. [Google Scholar]
- Abdullaev, F.I.; Frenkel, G.D. Time-course of inhibition of cellular nucleic acid synthesis by selenite. J. Inorg. Biochem. 1994, 55, 113–121. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; Tan, J.; McKinney, M.M.; Lane, A.N. Stable Isotope Resolved Metabolomics Analysis of Ribonucleotide and RNA Metabolism in Human Lung Cancer Cells. Metabolomics 2012, 8, 517–527. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Lane, A.N.; Higashi, R.M.; Farag, M.A.; Gao, H.; Bousamra, M.; Miller, D.M. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 2009, 8, 41. [Google Scholar] [CrossRef]
- Sellers, K.; Fox, M.P.; Bousamra, M., 2nd; Slone, S.P.; Higashi, R.M.; Miller, D.M.; Wang, Y.; Yan, J.; Yuneva, M.O.; Deshpande, R.; et al. Pyruvate carboxylase is critical for non–small-cell lung cancer proliferation. J. Clin. Investig. 2015, 125, 687–698. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.-C.; Lee, Y.-S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Yuneva, M.; Fan, T.; Allen, T.; Higashi, R.; Ferraris, D.; Tsukamoto, T.; Matés, J.; Alonso, F.; Wang, C.; Seo, Y.; et al. The Metabolic Profile of Tumors Depends on Both the Responsible Genetic Lesion and Tissue Type. Cell Metab. 2012, 15, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.-Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; Le, A.; Gao, P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 2009, 15, 6479–6483. [Google Scholar] [CrossRef]
- Wang, J.-B.; Erickson, J.W.; Fuji, R.; Ramachandran, S.; Gao, P.; Dinavahi, R.; Wilson, K.F.; Ambrosio, A.L.; Dias, S.M.; Dang, C.V.; et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 2010, 18, 207–219. [Google Scholar] [CrossRef]
- Lorkiewicz, P.; Higashi, R.M.; Lane, A.N.; Fan, T.W.-M. High information throughput analysis of nucleotides and their isotopically enriched isotopologues by direct-infusion FTICR-MS. Metabolomics 2012, 8, 930–939. [Google Scholar] [CrossRef]
- Mattingly, S.J.; Xu, T.; Nantz, M.H.; Higashi, R.M.; Fan, T.W.-M. A Carbonyl Capture Approach for Profiling Oxidized Metabolites in Cell Extracts. Metabolomics 2012, 8, 989–996. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; Lorkiewicz, P.K.; Sellers, K.; Moseley, H.N.; Higashi, R.M.; Lane, A.N. Stable isotope-resolved metabolomics and applications for drug development. Pharmacol. Ther. 2012, 133, 366–391. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; Yuan, P.; Lane, A.N.; Higashi, R.M.; Wang, Y.; Hamidi, A.B.; Zhou, R.; Guitart, X.; Chen, G.; Manji, H.K.; et al. Stable Isotope-Resolved Metabolomic Analysis of Lithium Effects on Glial-Neuronal Metabolism and Interactions. Metabolomics 2010, 6, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.-M.; Lane, A.N.; Higashi, R.M.; Yan, J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics 2011, 7, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan TW, M.; Bousamra, M.; Higashi, R.M.; Yan, J.; Miller, D.M. Clinical Applications of Stable Isotope-Resolved Metabolomics (SIRM) in Non-Small Cell Lung Cancer. OMICS 2011, 15, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan, T.W.; Higashi, R.M.; Deleeuw, L.; Yang, T.H. Stable Isotope Tracing in Metabolic Pathways. Modelling Complex Biological Systems. In The Context of the Genome; Patrick Amar, F.K., Norris, V., Vandenbunder, B., Eds.; RTI Press: Research Triangle Park, NC, USA, 2008; pp. 69–78. [Google Scholar]
- Moseley, H.N.; Lane, A.N.; Belshoff, A.C.; Higashi, R.M.; Fan, T.W. A novel deconvolution method for modeling UDP-GlcNAc biosynthetic pathways based on 13C mass isotopologue profiles under non steady-state conditions. BMC Biol. 2011, 9, 37. [Google Scholar] [CrossRef]
- Guo, W.; Wu, S.; Liu, J.; Fang, B. Identification of a Small Molecule with Synthetic Lethality for K-Ras and Protein Kinase C Iota. Cancer Res. 2008, 68, 7403–7408. [Google Scholar] [CrossRef]
- Yoon, Y.K.; Kim, H.P.; Han, S.W.; Oh, D.Y.; Im, S.A.; Bang, Y.J.; Kim, T.Y. KRAS mutant lung cancer cells are differentially responsive to MEK inhibitor due to AKT or STAT3 activation: Implication for combinatorial approach. Mol. Carcinog. 2010, 49, 353–362. [Google Scholar] [CrossRef]
- Lu, W.; Lin, J.; Chen, J. Expression of p14ARF overcomes tumor resistance to p53. Cancer Res. 2002, 62, 1305–1310. [Google Scholar]
- Fan, T.W.-M.; Lane, A.N.; Higashi, R.M. Stable Isotope Resolved Metabolomics Studies in ex vivo Tissue Slices. Bio-Protocol 2016, 6, e1730. [Google Scholar] [CrossRef]
- El Harane, S.; Zidi, B.; El Harane, N.; Krause, K.H.; Matthes, T.; Preynat-Seauve, O. Variable metabolic and immune action of checkpoint inhibition in ex vivo patient-derived lung cancer tissue cultures. eLife 2021, 10, e69578. [Google Scholar]
- Waksman, B.H.; Porter, H.; Lees, M.D.; Adams, R.D.; Folch, J. A study of the chemical nature of components of bovine white matter effective in producing allergic encephalomyelitis in the rabbit. J. Exp. Med. 1954, 100, 451–471. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; Lane, A.N. Structure-based profiling of Metabolites and Isotopomers by NMR. Prog. NMR Spectrosc. 2008, 52, 69–117. [Google Scholar] [CrossRef]
- Lane, A.N.; Fan, T.; Higashi, R. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Biophys. Tools Biol. 2008, 84, 541–588. [Google Scholar]
- Sun, Q.; Fan, T.W.-M.; Lane, A.N.; Higashi, R.M. Ion Chromatography-Ultra High-resolution MS1/MS2 Method for Stable Isotope-Resolved Metabolomics (SIRM) Reconstruction of Metabolic Networks. Anal. Chem. 2021, 93, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan, T.W.-M.; Xie, Z.; Moseley, H.N.; Higashi, R.M. Isotopomer analysis of lipid biosynthesis by high resolution mass spectrometry and NMR. Anal. Chim. Acta 2009, 651, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Pike Winer, L.S.; Wu, M. Rapid analysis of glycolytic and oxidative substrate flux of cancer cells in a microplate. PLoS ONE 2014, 9, e109916. [Google Scholar] [CrossRef] [PubMed]
- Cassago, A.; Ferreira, A.P.S.; Ferreira, I.M.; Fornezari, C.; Gomes, E.R.M.; Greene, K.S.; Pereira, H.M.; Garratt, R.C.; Dias, S.M.G.; Ambrosio, A.L.B. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. USA 2012, 109, 1092–1109. [Google Scholar] [CrossRef]
- Sablina, A.A.; Budanov, A.V.; Ilyinskaya, G.V.; Agapova, L.S.; Kravchenko, J.E.; Chumakov, P. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005, 11, 1306–1313. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Bruntz, R.C.; Yang, Y.; Song, H.; Chernyavskaya, Y.; Deng, P.; Zhang, Y.; Shah, P.P.; Beverly, L.J.; Qi, Z.; et al. De novo synthesis of serine and glycine fuels purine nucleotide biosynthesis in human lung cancer tissues. J. Biol. Chem. 2019, 294, 13464–13477. [Google Scholar] [CrossRef]
- Bruntz, R.C.; Belshoff, A.C.; Zhang, Y.; Macedo, J.K.A.; Higashi, R.M.; Lane, A.N.; Fan, T.W. Inhibition of Anaplerotic Glutaminolysis Underlies Selenite Toxicity in Human Lung Cancer. Proteomics 2019, 19, e1800486. [Google Scholar] [CrossRef]
- Ghosh, J.; Sarveswaran, S.; Liroff, J.; Zhou, Z.; Nikitin, A.Y. Selenite triggers rapid transcriptional activation of p53, and p53-mediated apoptosis in prostate cancer cells: Implication for the treatment of early-stage prostate cancer. Int. J. Oncol. 2010, 36, 1419–1428. [Google Scholar] [CrossRef]
- Zhang, X.; Su, Y.; Lane, A.N.; Stromberg, A.J.; Fan, T.W.M.; Wang, C. Bayesian Kinetic Modeling for Tracer-Based Metabolomic Data. BMC Bioinform. 2023, 24, 108. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Zeller, K.I.; Potter, J.J.; Wonsey, D.R.; O’Donnell, K.A.; Kim, J.-W.; Yustein, J.T.; Lee, L.A.; Dang, C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell Biol. 2005, 25, 6225–6234. [Google Scholar] [CrossRef]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012, 481, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.-H.; Sullivan, L.B.; Cheng, T.; Yang, Y.; Linehan, W.M.; Chandel, N.S.; DeBerardinis, R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2012, 481, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Jitrapakdee, S.; St Maurice, M.; Rayment, I.; Cleland, W.W.; Wallace, J.C.; Attwood, P.V. Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 2008, 413, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishna, R.; Gundimeda, U. Antioxidant regulation of protein kinase C in cancer prevention. J. Nutr. 2002, 132, 3819S–3823S. [Google Scholar] [CrossRef]
- Park, E.-M.; Choi, K.-S.; Park, S.-Y.; Kong, E.-S.; Zu, K.; Wu, Y.; Zhang, H.; Ip, C.; Park, Y.-M. A Display Thiol-Proteomics Approach to Characterize Global Redox Modification of Proteins by Selenium: Implications for the Anticancer Action of Selenium. Cancer Genom. Proteom. 2005, 2, 25–35. [Google Scholar]
- Fan, T.W.-M.; Warmoes, M.; Sun, Q.; Song, H.; Turchan-Cholewo, J.; Martin, J.; Mahan, A.; Higashi, R.; Lane, A. Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator beta-glucan in a two-case ex vivo non-small-cell lung cancer study. Cold Spring Harb. Mol. Case Stud. 2016, 2, a000893. [Google Scholar] [CrossRef]
- Xie, H.; Hanai, J.-I.; Ren, J.-G.; Kats, L.; Burgess, K.; Bhargava, P.; Signoretti, S.; Billiard, J.; Duffy, K.J.; Grant, A.; et al. Targeting Lactate Dehydrogenase-A Inhibits Tumorigenesis and Tumor Progression in Mouse Models of Lung Cancer and Impacts Tumor-Initiating Cells. Cell Metab. 2014, 19, 795–809. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Lane, A.N.; Higashi, R.M.; Fan, T.W.-M. Metabolic reprogramming in tumors: Contributions of the tumor microenvironment. Genes Dis. 2019, 7, 85–198. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Mishra, A.; Gautam, P.; Feroz, Z.; Vijayaraghavalu, S.; Likos, E.M.; Shukla, G.C.; Kumar, M. Metabolic Pathways, Enzymes, and Metabolites: Opportunities in Cancer Therapy. Cancers 2022, 14, 5268. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Ortega, A.; Santeliz, R.; Garrido, B.; Chacín, M.; Galban, N.; Vera, I.; De Sanctis, J.B.; Bermúdez, V. Metabolic Reprogramming in Cancer Cells: Emerging Molecular Mechanisms and Novel Therapeutic Approaches. Pharmaceutics 2022, 14, 1303. [Google Scholar] [CrossRef]
- Nisar, H.; González, P.M.S.; Brauny, M.; Labonté, F.M.; Schmitz, C.; Roggan, M.D.; Konda, B.; Hellweg, C.E. Hypoxia Changes Energy Metabolism and Growth Rate in Non-Small Cell Lung Cancer Cells. Cancers 2023, 15, 2472. [Google Scholar] [CrossRef]
- Nong, S.; Han, X.; Xiang, Y.; Qian, Y.; Wei, Y.; Zhang, T.; Tian, K.; Shen, K.; Yang, J.; Ma, X. Metabolic reprogramming in cancer: Mechanisms and therapeutics. MedComm 2023, 4, e218. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Pan, Y.; Chen, F. The Metabolic Landscape of Breast Cancer and Its Therapeutic Implications. Mol. Diagn. Ther. 2023, 27, 349–369. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]




| Protein Targets | Vendor | Catalogue Number | Dilution |
|---|---|---|---|
| ACLY | Proteintech Group | 15421-1-AP | 1:100 |
| CAD | Proteintech Group | 16617-1-AP | 1:100 |
| CCND1 | Proteintech Group | 60186-1-Ig | 1:100 |
| FASN | Proteintech Group | 10624-2-AP | 1:100 |
| GAC a | Gift of Dr. S. Dias b | 1:3000 | |
| GLDC | Proteintech Group | 24827-1-AP | 1:100 |
| GLS2 | Invitrogen | PA5-72963 | 1:100 |
| KGA/GAC | Proteintech Group | 12855-1-AP | 1:100 |
| ME1 | Proteintech Group | 16619-1-AP | 1:100 |
| ME2 | Proteintech Group | 24944-1-AP | 1:100 |
| MTATP8 | Proteintech Group | 26723-1-AP | 1:100 |
| NDUFS1 | Proteintech Group | 12444-1-AP | 1:100 |
| PC | Proteintech group | 16588-1-AP | 1:100 |
| PHGDH | Proteintech Group | 14719-1-AP | 1:100 |
| PSAT1 | Proteintech group | 10501-1-AP | 1:100 |
| SHMT1 | Proteintech Group | 14149-1-AP | 1:100 |
| SHMT2 | Proteintech Group | 11099-1-AP | 1:100 |
| TFAM | Proteintech Group | 19998-1-AP | 1:100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, T.W.-M.; Winnike, J.; Al-Attar, A.; Belshoff, A.C.; Lorkiewicz, P.K.; Tan, J.L.; Wu, M.; Higashi, R.M.; Lane, A.N. Differential Inhibition of Anaplerotic Pyruvate Carboxylation and Glutaminolysis-Fueled Anabolism Underlies Distinct Toxicity of Selenium Agents in Human Lung Cancer. Metabolites 2023, 13, 774. https://doi.org/10.3390/metabo13070774
Fan TW-M, Winnike J, Al-Attar A, Belshoff AC, Lorkiewicz PK, Tan JL, Wu M, Higashi RM, Lane AN. Differential Inhibition of Anaplerotic Pyruvate Carboxylation and Glutaminolysis-Fueled Anabolism Underlies Distinct Toxicity of Selenium Agents in Human Lung Cancer. Metabolites. 2023; 13(7):774. https://doi.org/10.3390/metabo13070774
Chicago/Turabian StyleFan, Teresa W.-M., Jason Winnike, Ahmad Al-Attar, Alexander C. Belshoff, Pawel K. Lorkiewicz, Jin Lian Tan, Min Wu, Richard M. Higashi, and Andrew N. Lane. 2023. "Differential Inhibition of Anaplerotic Pyruvate Carboxylation and Glutaminolysis-Fueled Anabolism Underlies Distinct Toxicity of Selenium Agents in Human Lung Cancer" Metabolites 13, no. 7: 774. https://doi.org/10.3390/metabo13070774
APA StyleFan, T. W.-M., Winnike, J., Al-Attar, A., Belshoff, A. C., Lorkiewicz, P. K., Tan, J. L., Wu, M., Higashi, R. M., & Lane, A. N. (2023). Differential Inhibition of Anaplerotic Pyruvate Carboxylation and Glutaminolysis-Fueled Anabolism Underlies Distinct Toxicity of Selenium Agents in Human Lung Cancer. Metabolites, 13(7), 774. https://doi.org/10.3390/metabo13070774








