MALDI MSI and Raman Spectroscopy Application in the Analysis of the Structural Components and Flavonoids in Brassica napus Stem
Abstract
1. Introduction
2. Materials and Methods
2.1. Biofertilizer Preparation
2.2. Rape Cultivation
2.3. Raman Sample Preparation and Data Acquisition
2.4. Raman Spectroscopic Imaging Data Curation
2.5. Sample Preparation for MALDI MS Imaging
2.6. MALDI Mass Spectrometry Imaging
3. Results
3.1. The Comparison of Different Groups in the Cortex Raman Spectra
3.2. The Comparison of Different Groups in the Pith Raman Spectra
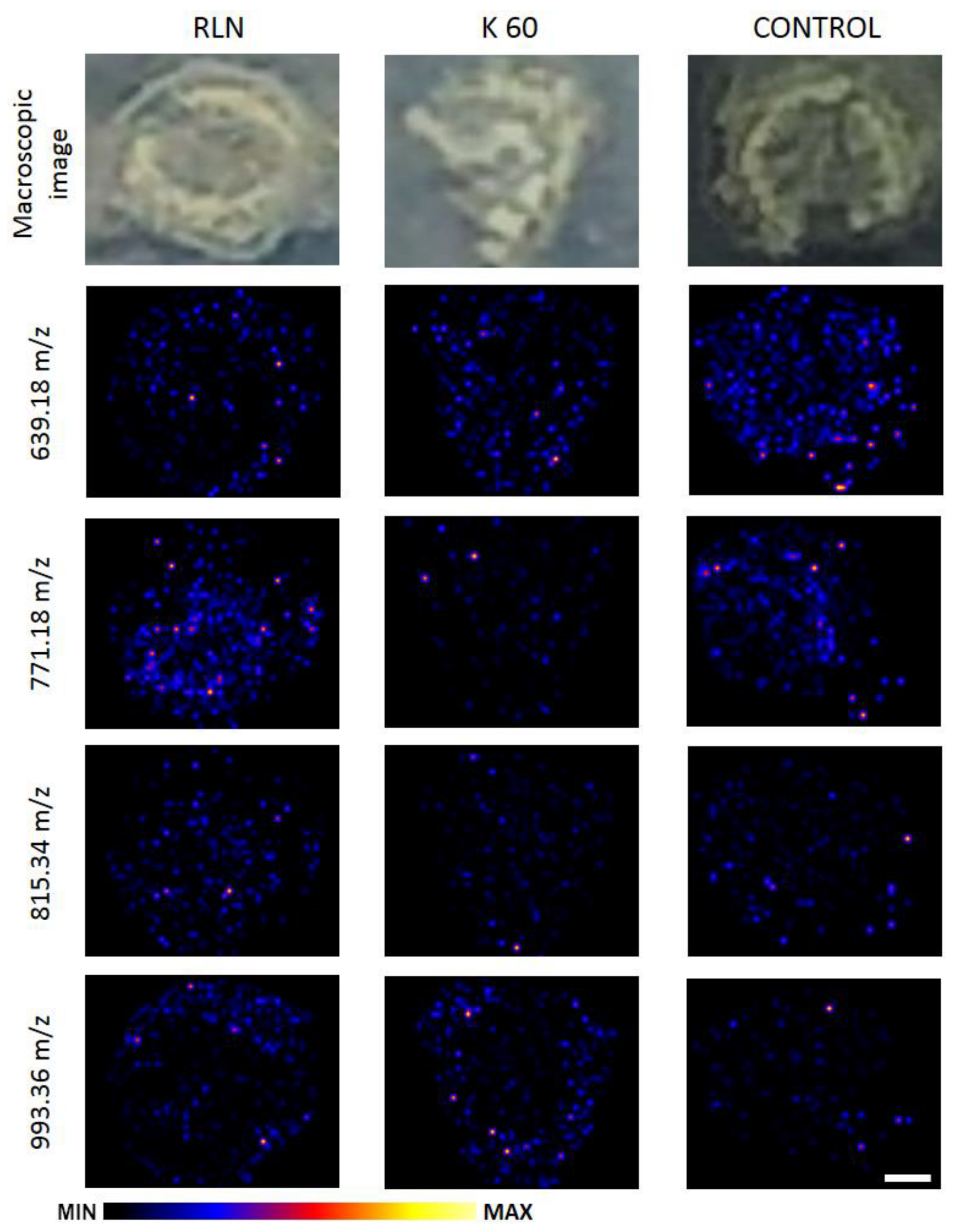
3.3. The Distribution of the Flavonoids in the Rapeseed Stem
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abiodun, O.A. The Role of Oilseed Crops in Human Diet and Industrial Use. In Oilseed Crops: Yield and Adaptations under Environmental Stress; Wiley-Blackwell: Hoboken, NJ, USA, 2017; pp. 249–263. [Google Scholar]
- Rashid, U.; Anwar, F. Production of Biodiesel through Optimized Alkaline-Catalyzed Transesterification of Rapeseed Oil. Fuel 2008, 87, 265–273. [Google Scholar] [CrossRef]
- Bell, J.M. Nutrients and Toxicants in Rapeseed Meal: A Review. J. Anim. Sci. 1984, 58, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-M.; Gu, W.-R.; Li, C.-F.; Li, J.; Wei, S. Effects of Nitrogen Fertilizer and Chemical Regulation on Spring Maize Lodging Characteristics, Grain Filling and Yield Formation under High Planting Density in Heilongjiang Province, China. J. Integr. Agric. 2021, 20, 511–526. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, J.; Zhang, G.; Chen, J.; Xie, R.; Ming, B.; Hou, P.; Wang, K.; Li, S. Nitrogen Split Application Can Improve the Stalk Lodging Resistance of Maize Planted at High Density. Agriculture 2020, 10, 364. [Google Scholar] [CrossRef]
- Raey, Y.; Ghassemi-Golezani, K. Yield-Density Relationship for Potato (Solarium tuberosum) and Common Bean (Phaseolus vulgaris) in Intercropping. N. Z. J. Crop Hortic. Sci. 2009, 37, 141–147. [Google Scholar] [CrossRef]
- Khan, S.; Anwar, S.; Kuai, J.; Ullah, S.; Fahad, S.; Zhou, G. Optimization of Nitrogen Rate and Planting Density for Improving Yield, Nitrogen Use Efficiency, and Lodging Resistance in Oilseed Rape. Front. Plant Sci. 2017, 8, 532. [Google Scholar] [CrossRef]
- Sieling, K.; Kage, H. Efficient N Management Using Winter Oilseed Rape. A Review. Agron. Sustain. Dev. 2010, 30, 271–279. [Google Scholar] [CrossRef]
- Zhang, W.-J.; Li, G.-H.; Yang, Y.-M.; Li, Q.; Zhang, J.; Liu, J.-Y.; Wang, S.; Tang, S.; Ding, Y.-F. Effects of Nitrogen Application Rate and Ratio on Lodging Resistance of Super Rice with Different Genotypes. J. Integr. Agric. 2014, 13, 63–72. [Google Scholar] [CrossRef]
- Armengot, L.; Ferrari, L.; Milz, J.; Velásquez, F.; Hohmann, P.; Schneider, M. Cacao Agroforestry Systems Do Not Increase Pest and Disease Incidence Compared with Monocultures under Good Cultural Management Practices. Crop. Prot. 2020, 130, 105047. [Google Scholar] [CrossRef]
- Deterioration, E.; Health, H. Environmental Deterioration and Human Health; Malik, A., Grohmann, E., Akhtar, R., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; ISBN 978-94-007-7889-4. [Google Scholar]
- Komárek, M.; Čadková, E.; Chrastný, V.; Bordas, F.; Bollinger, J.C. Contamination of Vineyard Soils with Fungicides: A Review of Environmental and Toxicological Aspects. Environ. Int. 2010, 36, 138–151. [Google Scholar] [CrossRef]
- Treutter, D. Significance of Flavonoids in Plant Resistance: A Review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Thoison, O.; Sévenet, T.; Niemeyer, H.M.; Russell, G.B. Insect Antifeedant Compounds from Nothofagus Dombeyi and N. Pumilio. Phytochemistry 2004, 65, 2173–2176. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Ohmura, W.; Doi, S.; Aoyama, M. Termite Feeding Deterrent from Japanese Larch Wood. Bioresour. Technol. 2004, 95, 129–134. [Google Scholar] [CrossRef]
- Widstrom, N.W.; Snook, M.E. Recurrent Selection for Maysin, a Compound in Maize Silks, Antibiotic to Earworm. Plant Breed. 2001, 120, 357–359. [Google Scholar] [CrossRef]
- Skadhauge, B.; Thomsen, K.K.; Von Wettstein, D. The Role of the Barley Testa Layer and Its Flavonoid Content in Resistance to Fusarium Infections. Hereditas 1997, 126, 147–160. [Google Scholar] [CrossRef]
- Kidaj, D.; Krysa, M.; Susniak, K.; Matys, J.; Komaniecka, I.; Sroka-Bartnicka, A. Biological Activity of Nod Factors. Acta Biochim. Pol. 2020, 67, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Susniak, K.; Krysa, M.; Kidaj, D.; Szymanska-Chargot, M.; Komaniecka, I.; Zamlynska, K.; Choma, A.; Wielbo, J.; Ilag, L.L.; Sroka-Bartnicka, A. Multimodal Spectroscopic Imaging of Pea Root Nodules to Assess the Nitrogen Fixation in the Presence of Biofertilizer Based on Nod-Factors. Int. J. Mol. Sci. 2021, 22, 12991. [Google Scholar] [CrossRef] [PubMed]
- Prithiviraj, B.; Zhou, X.; Souleimanov, A.; Kahn, W.M.; Smith, D.L. A Host-Specific Bacteria-to-Plant Signal Molecule (Nod Factor) Enhances Germination and Early Growth of Diverse Crop Plants. Planta 2003, 216, 437–445. [Google Scholar] [CrossRef]
- Souleimanov, A.; Prithiviraj, B.; Smith, L. The Major Nod Factor of Bradyrhizobium Japonicum Promotes Early Growth of Soybean and Corn. J. Exp. Bot. 2002, 53, 1929–1934. [Google Scholar] [CrossRef]
- Khan, W.; Prithiviraj, B.; Smith, D.L. Nod Factor [Nod Bj V (C18:1, MeFuc)] and Lumichrome Enhance Photosynthesis and Growth of Corn and Soybean. J. Plant Physiol. 2008, 165, 1342–1351. [Google Scholar] [CrossRef]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.H.; Qiu, J.; Stacey, G. Nonlegumes Respond to Rhizobial Nod Factors by Suppressing the Innate Immune Response. Science 2013, 341, 1384–1387. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Ye, S.; Xu, L.; Hu, R.; Jin, L.; Xu, H.; Liu, J.; Liu, W. Study on Baseline Correction Methods for the Fourier Transform Infrared Spectra with Different Signal-to-Noise Ratios. Appl. Opt. 2018, 57, 5794. [Google Scholar] [CrossRef] [PubMed]
- Toplak, M.; Read, S.T.; Sandt, C.; Borondics, F. Quasar: Easy Machine Learning for Biospectroscopy. Cells 2021, 10, 2300. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python Fabian. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Agarwal, U.P. An Overview of Raman Spectroscopy as Applied to Lignocellulosic Materials. In Advances in Lignocellulosics Characterization; TAPPI Press: Peachtree Corners, GA, USA, 1999; pp. 201–225. [Google Scholar]
- Agarwal, U.P. Raman Imaging to Investigate Ultrastructure and Composition of Plant Cell Walls: Distribution of Lignin and Cellulose in Black Spruce Wood (Picea mariana). Planta 2006, 224, 1141–1153. [Google Scholar] [CrossRef]
- Gierlinger, N.; Schwanninger, M. Chemical Imaging of Poplar Wood Cell Walls by Confocal Raman Microscopy. Plant Physiol. 2006, 140, 1246–1254. [Google Scholar] [CrossRef]
- Zeise, I.; Heiner, Z.; Holz, S.; Joester, M.; Büttner, C.; Kneipp, J. Raman Imaging of Plant Cell Walls in Sections of Cucumis Sativus. Plants 2018, 7, 7. [Google Scholar] [CrossRef]
- Gorzsás, A. Chemical Imaging of Xylem by Raman Microspectroscopy. In Xylem: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1544, ISBN 9781493967223. [Google Scholar]
- Edwards, H.G.M.; Farwell, D.W.; Webster, D. FT Raman Microscopy of Untreated Natural Plant Fibres. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 1997, 53, 2383–2392. [Google Scholar] [CrossRef]
- Del Río, J.C.; Prinsen, P.; Rencoret, J.; Nieto, L.; Jiménez-Barbero, J.; Ralph, J.; Martínez, Á.T.; Gutiérrez, A. Structural Characterization of the Lignin in the Cortex and Pith of Elephant Grass (Pennisetum Purpureum) Stems. J. Agric. Food Chem. 2012, 60, 3619–3634. [Google Scholar] [CrossRef]
- Tuan Rohadi, T.N.; Ridzuan, M.J.M.; Abdul Majid, M.S.; Cheng, E.M. Characterisation and Comparison of Pith and Cortex of Napier Grass Stem. IOP Conf. Ser. Mater. Sci. Eng. 2020, 864, 012138. [Google Scholar] [CrossRef]
- Niemann, G.J.; Baayen, R.P.; Boon, J.J. Differentiation between Tissues from Carnation (Dianthus Caryophyllus) Stems by Pyrolysis-Mass Spectrometry. Ann. Bot. 1990, 65, 461–472. [Google Scholar] [CrossRef]
- Gominho, J.; Fernandez, J.; Pereira, H. Cynara Cardunculus L.—A New Fibre Crop for Pulp and Paper Production. Ind. Crops Prod. 2001, 13, 1–10. [Google Scholar] [CrossRef]
- Farag, M.A.; Sharaf Eldin, M.G.; Kassem, H.; Abou El Fetouh, M. Metabolome Classification of Brassica napus L. Organs via UPLC-QTOF-PDA-MS and Their Anti-Oxidant Potential. Phytochem. Anal. 2013, 24, 277–287. [Google Scholar] [CrossRef]
- Rademacher, W. Growth Retardants: Effects on Gibberellin Biosynthesis and Other Metabolic Pathways. Annu. Rev. Plant Biol. 2000, 51, 501–531. [Google Scholar] [CrossRef] [PubMed]
- Berry, P.M.; Spink, J.H. Understanding the Effect of a Triazole with Anti-Gibberellin Activity on the Growth and Yield of Oilseed Rape (Brassica napus). J. Agric. Sci. 2009, 147, 273–285. [Google Scholar] [CrossRef]
- Wittkop, B.; Snowdon, R.; Friedt, W. Improvement of Rapeseed Meal Quality via Reduction of Seed Fibre-Fractions. In Quality, Nutrition, Processing and Trade, Proceedings of the 12th International Rapeseed Congress, Wuhan, China, 26–30 March 2007; Tingdong, F., Chunyun, G., Yongming, Z., Hanzhong, W., Dianrong, L., Eds.; Science Press USA Inc.: Monmouth Junction, NJ, USA, 2007; pp. 135–137. [Google Scholar]
- Mallikarjuna, N.; Kranthi, K.R.; Jadhav, D.R.; Kranthi, S.; Chandra, S. Influence of Foliar Chemical Compounds on the Development of Spodoptera Litura (Fab.) in Interspecific Derivatives of Groundnut. J. Appl. Entomol. 2004, 128, 321–328. [Google Scholar] [CrossRef]
- Pandino, G.; Lombardo, S.; Antonino, L.M.; Ruta, C.; Mauromicale, G. In Vitro Micropropagation and Mycorrhizal Treatment Influences the Polyphenols Content Profile of Globe Artichoke under Field Conditions. Food Res. Int. 2017, 99, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Tegelberg, R.; Julkunen-Tiitto, R.; Aphalo, P.J. Red:Far-Red Light Ratio and UV-B Radiation: Their Effects on Leaf Phenolics and Growth of Silver Birch Seedlings. Plant Cell Environ. 2004, 27, 1005–1013. [Google Scholar] [CrossRef]
- Kolb, C.A.; Käser, M.A.; Kopecký, J.; Zotz, G.; Riederer, M.; Pfündel, E.E. Effects of Natural Intensities of Visible and Ultraviolet Radiation on Epidermal Ultraviolet Screening and Photosynthesis in Grape Leaves. Plant Physiol. 2001, 127, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Auger, B.; Marnet, N.; Gautier, V.; Maia-Grondard, A.; Leprince, F.; Renard, M.; Guyot, S.; Nesi, N.; Routaboul, J.M. A Detailed Survey of Seed Coat Flavonoids in Developing Seeds of Brassica napus L. J. Agric. Food Chem. 2010, 58, 6246–6256. [Google Scholar] [CrossRef] [PubMed]

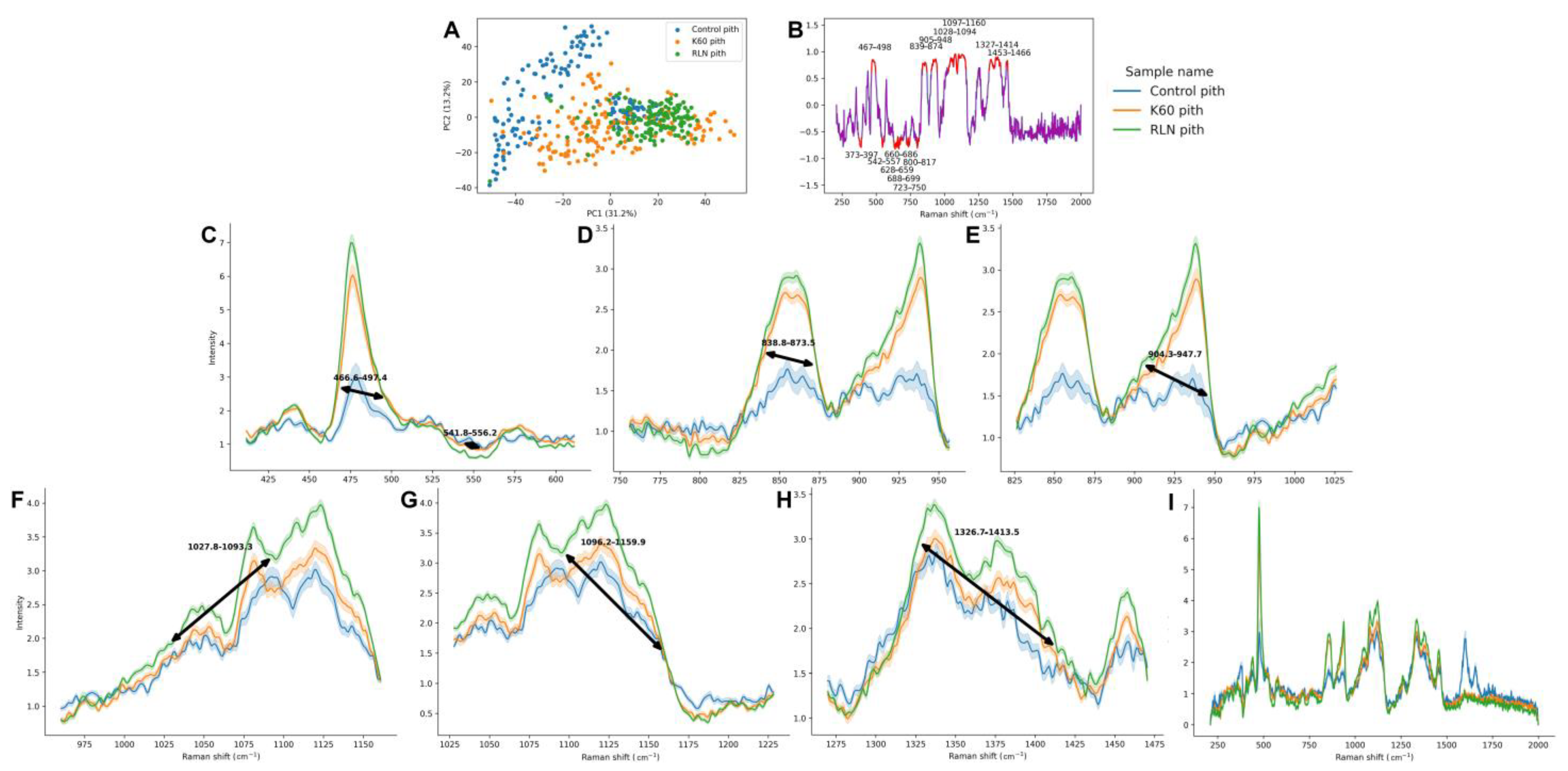
| Raman Shift [cm−1] | Source | Functional Group | Literature |
|---|---|---|---|
| ~1650 | Lignin | Ring C=C stretching of coniferyl alcohol, C=O stretching of coniferaldehyde | [29,30] |
| ~1600 | Lignin | Symmetric aryl ring stretching | [29,30] |
| ~1462 | Lignin and cellulose | HCH HOC bending | [31] |
| ~1393 | Lignin | OH bending | [29] |
| ~1376 | Cellulose | HCC, HOC, and HCO bending | [31] |
| ~1333 | Cellulose | HCC and HCO bending | [31] |
| ~1274 | Lignin | Aryl–O of aryl OH and aryl O–CH3; guaiacyl ring (with C=O group) mode | [31] |
| 1178–978 | Cellulose | CC and CO stretching mainly + HCC, HCO bending | [29,30] |
| ~927 | Lignin | CCH wagging | [32] |
| 860–825 | Pectin | COC skeletal vibrations | [33] |
| 515–475 | Hemicellulose | Composite bending vibrations involving the C6 position | [33] |
| ~436 | Cellulose | CCO ring stretching | [34] |
| 300–400 | Cellulose (crystallinity) | Out-of-plane bending of C–O–C glycosidic bonds and OH groups coupled with simultaneous expansion and contraction in all rings | [29,30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krysa, M.; Susniak, K.; Kubas, A.; Kidaj, D.; Sroka-Bartnicka, A. MALDI MSI and Raman Spectroscopy Application in the Analysis of the Structural Components and Flavonoids in Brassica napus Stem. Metabolites 2023, 13, 687. https://doi.org/10.3390/metabo13060687
Krysa M, Susniak K, Kubas A, Kidaj D, Sroka-Bartnicka A. MALDI MSI and Raman Spectroscopy Application in the Analysis of the Structural Components and Flavonoids in Brassica napus Stem. Metabolites. 2023; 13(6):687. https://doi.org/10.3390/metabo13060687
Chicago/Turabian StyleKrysa, Mikolaj, Katarzyna Susniak, Adrianna Kubas, Dominika Kidaj, and Anna Sroka-Bartnicka. 2023. "MALDI MSI and Raman Spectroscopy Application in the Analysis of the Structural Components and Flavonoids in Brassica napus Stem" Metabolites 13, no. 6: 687. https://doi.org/10.3390/metabo13060687
APA StyleKrysa, M., Susniak, K., Kubas, A., Kidaj, D., & Sroka-Bartnicka, A. (2023). MALDI MSI and Raman Spectroscopy Application in the Analysis of the Structural Components and Flavonoids in Brassica napus Stem. Metabolites, 13(6), 687. https://doi.org/10.3390/metabo13060687






