Metabolic Reprogramming of Barley in Response to Foliar Application of Dichlorinated Functional Analogues of Salicylic Acid as Priming Agents and Inducers of Plant Defence
Abstract
1. Introduction
2. Materials and Methods
2.1. Barley Plant Material and Growth Conditions
2.2. Barley Plant Treatment with Priming Inducers
2.3. Metabolite Extraction from Seedlings and Pre-Analytical Sample Preparation
2.4. Sample Analyses on Mass Spectrometry-Based Analytical Platforms (Ultra-High Performance Liquid Chromatography—High Definition Mass Spectrometry (UHPLC-HDMS))
2.5. Chemometrics: Data Mining
2.6. Metabolite Annotation
3. Results
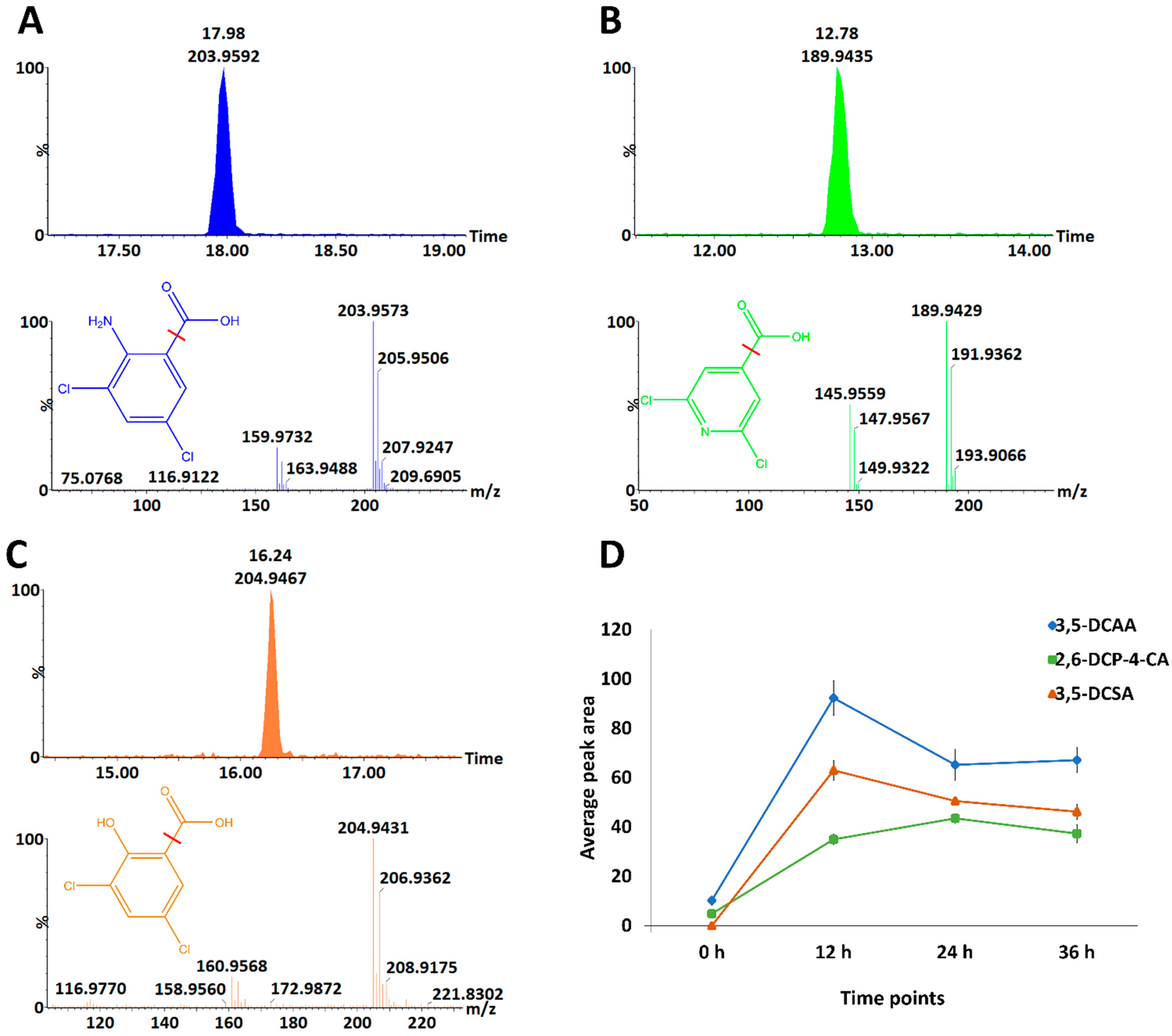
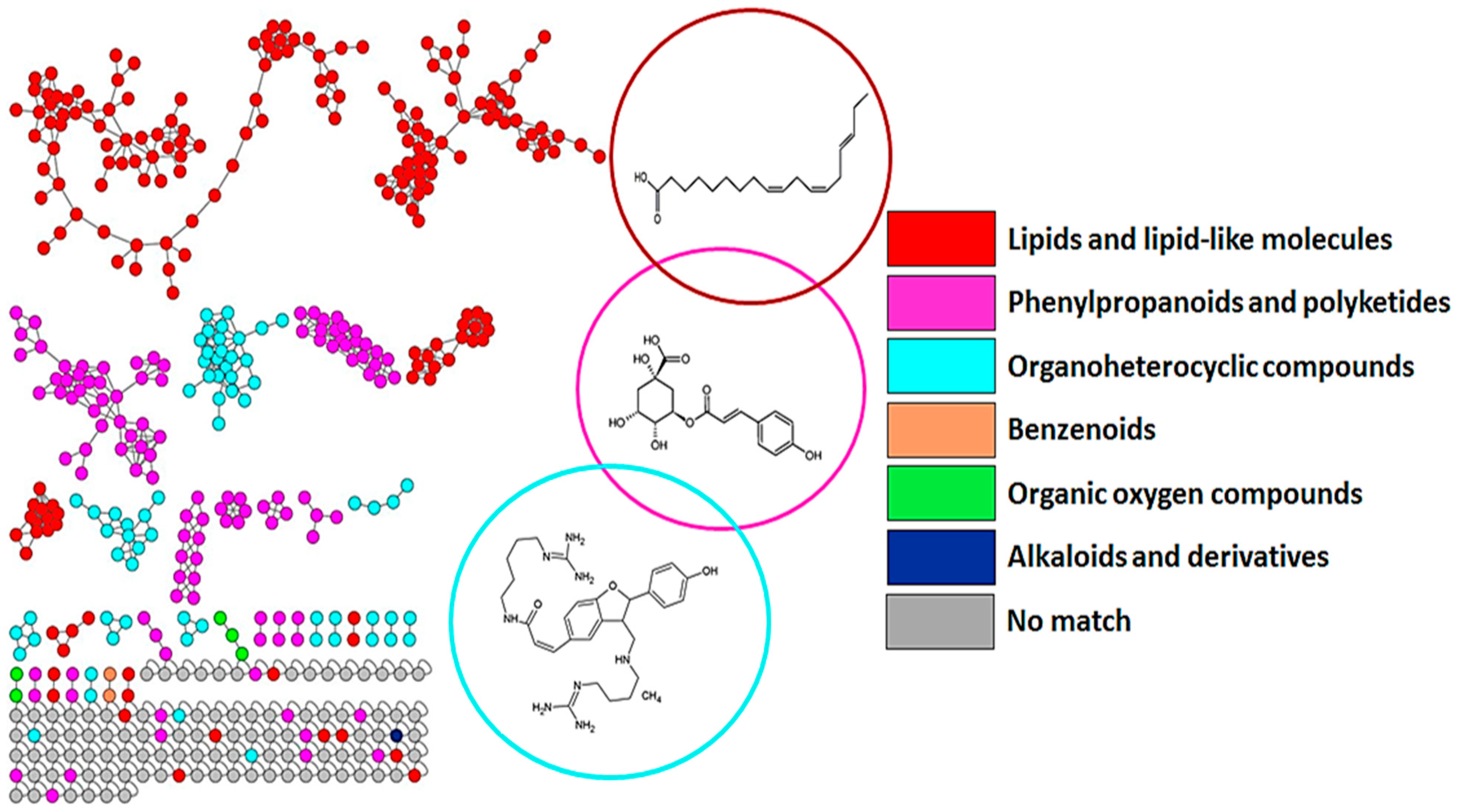
3.1. Chromatographic and Mass Spectrometric Analyses and Molecular Networking Approach to Uncover the Metabolic Space of Barley Leaves Treated with Inducers
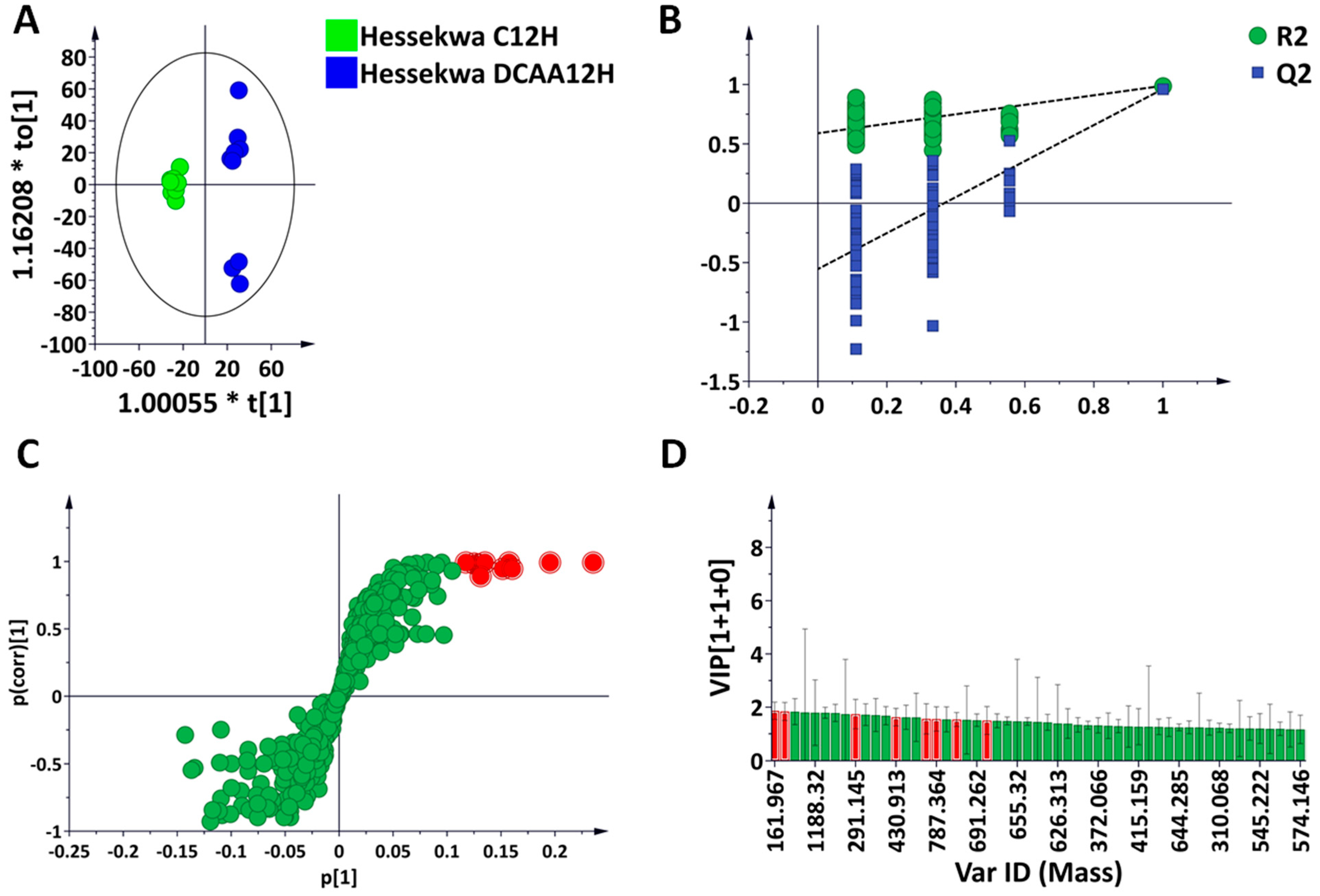
3.2. Multivariate Data Analyses: Statistical Description, Evaluation, and Exploration of Changes Observed in the UHPLC-MS Data
3.3. Discriminant Analyses: Treatment- and Time-Related Metabolites and Fold Changes
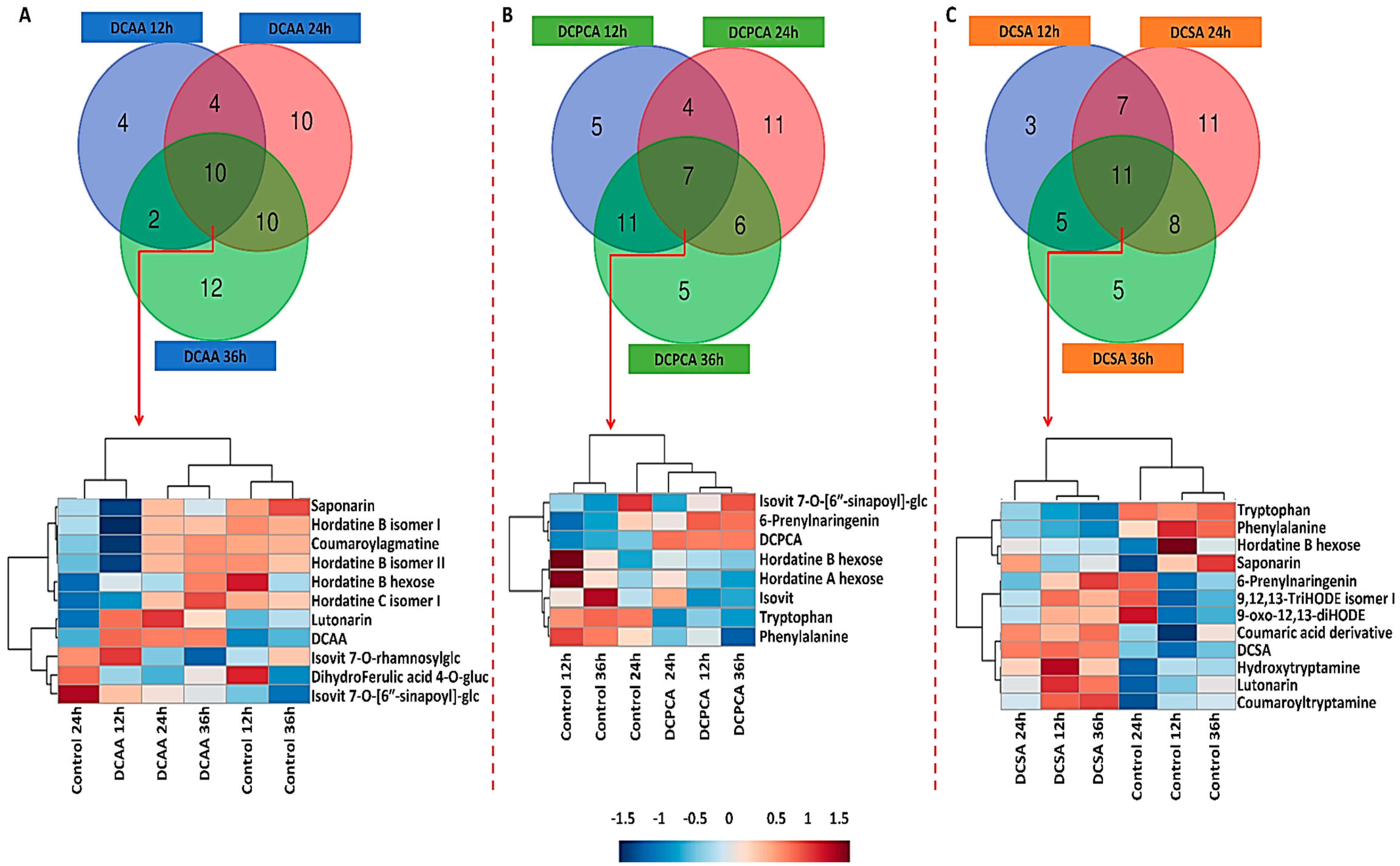
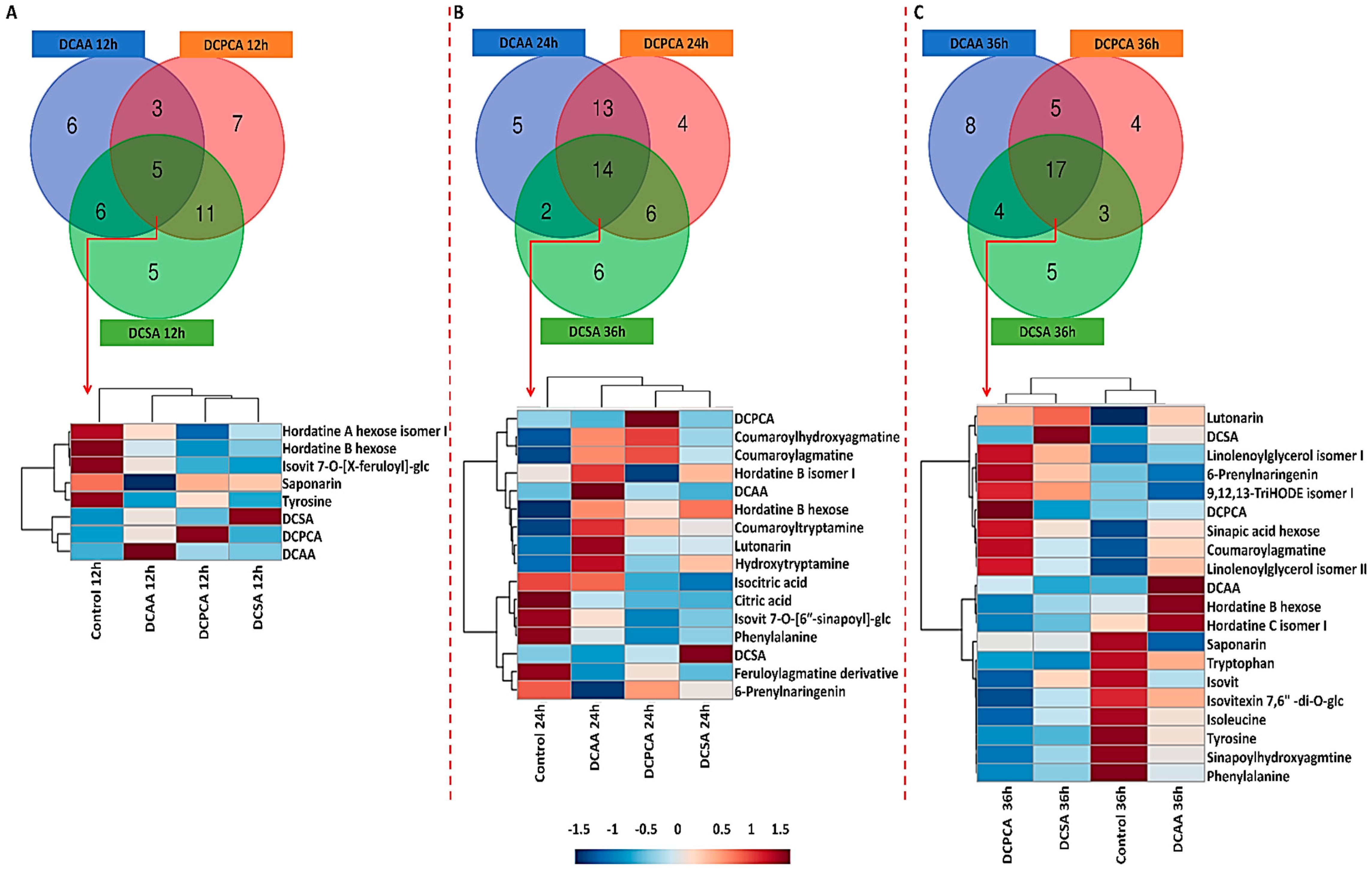
3.4. Time-Related Differences and Similarities in the Chemical Profiles of Barley Leaves, following Foliar Application of 3,5-DCAA, 2,6-DCP-4-CA and 3,5-DCSA
3.5. Treatment-Related Similarities and Differences in the Chemical Profile of Barley Leaves, following Foliar Application of 3,5-DCAA, 2,6-DCP-4-CA, and 3,5-DCSA
4. Discussion
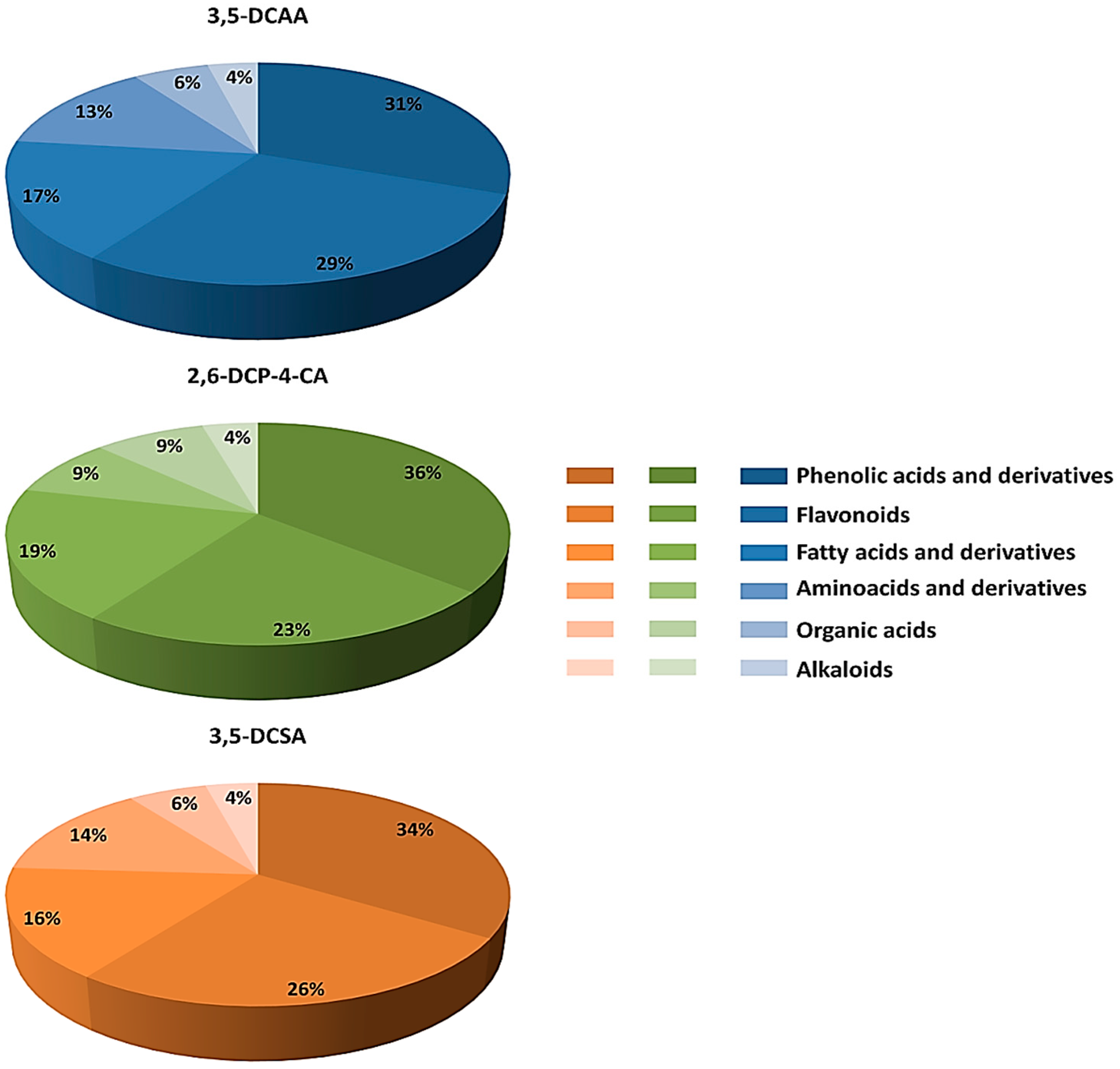
4.1. Distribution of Metabolite Classes and Metabolic Pathways Analyses for Biological Interpretation
4.2. Biological Implication with Regard to Plant Protection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, D.Z.; Wang, G.X.; Zhou, J.M. Receptor Kinases in Plant-Pathogen Interactions: More than Pattern Recognition. Plant Cell 2018, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant–Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wei, T.; Lou, H.; Shu, X.; Chen, Q. A critical review on communication mechanism within plant-endophytic fungi interactions to cope with biotic and abiotic stresses. J. Fungi 2021, 7, 719. [Google Scholar] [CrossRef] [PubMed]
- Prasch, C.M.; Sonnewald, U. Signaling events in plants: Stress factors in combination change the picture. Environ. Exp. Bot. 2015, 114, 4–14. [Google Scholar] [CrossRef]
- Llorens, E.; González-Hernández, A.I.; Scalschi, L.; Fernández-Crespo, E.; Camañes, G.; Vicedo, B.; García-Agustín, P. Priming mediated stress and cross-stress tolerance in plants: Concepts and opportunities. In Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants; Academic Press: Cambridge, MA, USA, 2020; pp. 1–20. [Google Scholar]
- Dempsey, D.A.; Klessig, D.F. SOS-too many signals for systemic acquired resistance? Trends Plant Sci. 2012, 17, 538–545. [Google Scholar] [CrossRef]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef]
- Shine, M.B.; Xiao, X.; Kachroo, P.; Kachroo, A. Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 2019, 279, 81–86. [Google Scholar] [CrossRef]
- Conrath, U.; Pieterse, C.M.; Mauch-Mani, B. Priming in plant–pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef]
- Pieterse, C.M.J. Prime time for transgenerational defense. Plant Physiol. 2012, 158, 545. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Gao, Q.M.; Kachroo, A.; Kachroo, P. Chemical inducers of systemic immunity in plants. J. Exp. Bot. 2014, 65, 1849–1855. [Google Scholar] [CrossRef]
- Baccelli, I.; Mauch-Mani, B. Beta aminobutyric acid priming of plant defense: The role of ABA and other hormones. Plant Mol. Biol. 2016, 91, 703–711. [Google Scholar] [CrossRef]
- Henry, E.; Yadeta, K.A.; Coaker, G. Recognition of bacterial plant pathogens: Local, systemic and transgenerational immunity. New Phytol. 2013, 199, 908–915. [Google Scholar] [CrossRef]
- Bektas, Y.; Eulgem, T. Synthetic plant defense elicitors. Front. Plant Sci. 2015, 5, 804. [Google Scholar] [CrossRef]
- Merewitz, E. Chemical priming-induced drought stress tolerance in plants. In Drought Stress Tolerance in Plants; Springer: Cham, Switzerland, 2016; Volume 1, pp. 77–103. [Google Scholar]
- Yi, H.S.; Yang, J.W.; Ryu, C.M. ISR meets SAR outside: Additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front. Plant Sci. 2013, 4, 122. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.A.; Pieterse, C.M.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef]
- Buswell, W.; Schwarzenbacher, R.E.; Luna, E.; Sellwood, M.; Chen, B.; Flors, V.; Petriacq, P.; Ton, J. Chemical priming of immunity without costs to plant growth. New Phytol. 2018, 218, 1205–1216. [Google Scholar] [CrossRef]
- Walters, D.R.; Avrova, A.; Bingham, I.J.; Burnett, F.J.; Fountaine, J.; Havis, N.D.; Hoad, S.P.; Hughes, G.; Looseley, M.; Oxley, S.J.; et al. Control of foliar diseases in barley: Towards an integrated approach. Eur. J. Plant Pathol. 2012, 133, 33–73. [Google Scholar] [CrossRef]
- Beckers, G.J.M.; Conrath, U. Priming for stress resistance: From the lab to the field. Curr. Opin. Plant Biol. 2007, 10, 425–431. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Klessig, D.F. Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 1996, 271, 28492–28501. [Google Scholar] [CrossRef] [PubMed]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef] [PubMed]
- Maruri-López, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and extracellular journey of the phytohormone salicylic acid. Front. Plant Sci. 2019, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Bacelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Knoth, C.; Salus, M.S.; Girke, T.; Eulgem, T. The synthetic elicitor 3,5-dichloroanthranilic acid induces NPR1-dependent and NPR1-independent mechanisms of disease resistance in Arabidopsis. Plant Physiol. 2009, 150, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Knoth, C.; Eulgem, T. High-throughput screening of small-molecule libraries for inducers of plant defense responses. In Plant Chemical Genomics; Humana Press: Totowa, NJ, USA, 2014; pp. 45–49. [Google Scholar]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, W. Recent advances in synthetic chemical inducers of plant immunity. Front. Plant Sci. 2018, 9, 1613. [Google Scholar] [CrossRef]
- Faize, L.; Faize, M. Functional analogues of salicylic acid and their use in crop protection. Agronomy 2018, 8, 5. [Google Scholar] [CrossRef]
- Silverman, F.P.; Petracek, P.D.; Heiman, D.F.; Fledderman, C.M.; Warrior, P. Salicylate activity. 3. Structure relationship to systemic acquired resistance. J. Agri. Food Chem. 2005, 53, 9775–9780. [Google Scholar] [CrossRef]
- Conrath, U.; Chen, Z.; Ricigliano, J.R.; Klessig, D.F. Two inducers of plant defense responses, 2, 6-dichloroisonicotinec acid and salicylic acid, inhibit catalase activity in tobacco. Proc. Natl. Acad. Sci. USA 1995, 92, 7143–7147. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/ (accessed on 29 March 2022).
- Giraldo, P.; Benavente, E.; Manzano-Agugliaro, F.; Gimenez, E. Worldwide Research Trends on Wheat and Barley: A Bibliometric Comparative Analysis. Agronomy 2019, 9, 352. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Piater, L.A.; Steenkamp, P.A.; Tugizimana, F.; Dubery, I.A. A metabolomics approach and chemometric tools for differentiation of barley cultivars and biomarker discovery. Metabolites 2021, 11, 578. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Steenkamp, P.A.; Piater, L.A.; Tugizimana, F.; Dubery, I.A. Hordatines and associated precursors dominate metabolite profiles of barley (Hordeum vulgare L.) seedlings: A metabolomics study of five cultivars. Metabolites 2022, 12, 310. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 4, W388–W396. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, 353–361. [Google Scholar] [CrossRef]
- Abf Converter. Available online: https://www.reifycs.com/AbfConverter/ (accessed on 28 January 2022).
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Tinte, M.M.; Masike, K.; Steenkamp, P.A.; Huyser, J.; van der Hooft, J.J.; Tugizimana, F. Computational Metabolomics Tools Reveal Metabolic Reconfigurations Underlying the Effects of Biostimulant Seaweed Extracts on Maize Plants under Drought Stress Conditions. Metabolites 2022, 12, 487. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Gozzo, F. Systemic acquired resistance in crop protection: From nature to a chemical approach. J. Agric. Food Chem. 2003, 51, 4487–4503. [Google Scholar] [CrossRef] [PubMed]
- White, P.S. Pattern, process, and natural disturbance in vegetation. Bot. Rev. 1979, 45, 229–299. [Google Scholar] [CrossRef]
- Stadnik, M.J.; Buchenauer, H. Inhibition of phenylalanine ammonia-lyase suppresses the resistance induced by benzothiadiazole in wheat to Blumeria graminis f. sp. tritici. Physiol. Mol. Plant Path. 2000, 57, 25–34. [Google Scholar] [CrossRef]
- Ramos, O.F.; Smith, C.M.; Fritz, A.K.; Madl, R.L. Salicylic Acid-Mediated Synthetic Elicitors of Systemic Acquired Resistance Administered to Wheat Plants at Jointing Stage Induced Phenolics in Mature Grains. Crop. Sci. 2017, 57, 3122–3128. [Google Scholar] [CrossRef]
- Pastor, V.; Balmer, A.; Gamir, J.; Flors, V.; Mauch-Mani, B. Preparing to fight back: Generation and storage of priming compounds. Front. Plant Sci. 2014, 5, 295. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef]
- Häusler, R.E.; Ludewig, F.; Krueger, S. Amino acids–a life between metabolism and signaling. Plant Sci. 2014, 229, 225–237. [Google Scholar] [CrossRef]
- Pratelli, R.; Pilot, G. Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 2014, 65, 5535–5556. [Google Scholar] [CrossRef]
- Balmer, D.; de Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013, 74, 213–225. [Google Scholar] [CrossRef]
- Killiny, N.; Hijaz, F. Amino acids implicated in plant defense are higher in Candidatus liberibacter asiaticus-tolerant citrus varieties. Plant Signal. Behav. 2016, 11, e1171449. [Google Scholar] [CrossRef]
- Kirma, M.; Araújo, W.L.; Fernie, A.R.; Galili, G. The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 2012, 63, 4995–5001. [Google Scholar] [CrossRef]
- Hildebrandt, T.M.; Nesi, A.N.; Araújo, W.L.; Braun, H.P. Amino acid catabolism in plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef]
- Ogura, Y.; Ishihara, A.; Iwamura, H. Induction of hydroxycinnamic acid amides and tryptophan by jasmonic acid, abscisic acid and osmotic stress in barley leaves. Z. Naturforsch. C 2001, 56, 193–202. [Google Scholar] [CrossRef]
- Ishihara, A.; Kumeda, R.; Hayashi, N.; Yagi, Y.; Sakaguchi, N.; Kokubo, Y.; Ube, N.; Tebayashi, S.I.; Ueno, K. Induced accumulation of tyramine, serotonin, and related amines in response to Bipolaris sorokiniana infection in barley. Biosci. Biotechnol. Biochem. 2017, 81, 1090–1098. [Google Scholar] [CrossRef]
- Lopez-Bucio, J.; Nieto-Jacobo, M.F.; Ramırez-Rodrıguez, V.; Herrera-Estrella, L. Organic acid metabolism in plants: From adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 2000, 160, 1–13. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Eprintsev, A.T. Organic acids: The pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front. Plant Sci. 2016, 7, 1042. [Google Scholar] [CrossRef]
- Gidda, S.K.; Miersch, O.; Levitin, A.; Schmidt, J.; Wasternack, C.; Varin, L. Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 17895–17900. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Madala, N.E.; Dubery, I.A. Mass spectrometric approaches to study the metabolism of jasmonates: Biotransformation of exogenously supplemented methyl jasmonate by cell suspension cultures of Moringa oleifera. In Jasmonate in Plant Biology; Humana: New York, NY, USA, 2020; pp. 211–226. [Google Scholar]
- Pretorius, C.J.; Zeiss, D.R.; Dubery, I.A. The presence of oxygenated lipids in plant defense in response to biotic stress: A metabolomics appraisal. Plant Signal. Behav. 2021, 16, 1989215. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Pena-Cortés, H.; Albrecht, T.; Prat, S.; Weiler, E.W.; Willmitzer, L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 1993, 191, 123–128. [Google Scholar] [CrossRef]
- Tamaoki, D.; Seo, S.; Yamada, S.; Kano, A.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 2013, 8, e24260. [Google Scholar] [CrossRef]
- Marti, G.; Erb, M.; Boccard, J.; Glauser, G.; Doyen, G.R.; Villard, N.; Robert, C.A.M.; Turlings, T.C.; Rudaz, S.; Wolfender, J.L. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant Cell Environ. 2013, 36, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Zeiss, D.R.; Piater, L.A.; Dubery, I.A. Hydroxycinnamate amides: Intriguing conjugates of plant protective metabolites. Trends Plant Sci. 2021, 26, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Ramaroson, M.L.; Koutouan, C.; Helesbeux, J.J.; Le Clerc, V.; Hamama, L.; Geoffriau, E.; Briard, M. Role of Phenylpropanoids and Flavonoids in Plant Resistance to Pests and Diseases. Molecules 2022, 27, 8371. [Google Scholar] [CrossRef]
- Mathesius, U. Flavonoid functions in plants and their interactions with other organisms. Plants 2018, 7, 30. [Google Scholar] [CrossRef]
- Benedet, J.A.; Umeda, H.; Shibamoto, T. Antioxidant activity of flavonoids isolated from young green barley leaves toward biological lipid samples. J. Agric. Food Chem. 2007, 55, 5499–5504. [Google Scholar] [CrossRef]
- Markham, K.R.; Mitchell, K.A. The misidentification of the major antioxidant flavonoids in young barley (Hordeum vulgare) leaves. Z. Naturforsch. C 2003, 58, 53–56. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Adamczak, A.; Ożarowski, M. Antibacterial activity of apigenin, luteolin, and their C-glucosides. In Proceedings of the 5th International Electronic Conference on Medicinal Chemistry, Online, 1–30 November 2019. [Google Scholar] [CrossRef]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.P.; Ryals, J.A. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef]
- Kessmann, H.; Staub, T.; Hofmann, C.; Maetzke, T.; Herzog, J.; Ward, E.; Uknes, S.; Ryals, J. Induction of systemic acquired disease resistance in plants by chemicals. Ann. Rev. Phytopathol. 1994, 32, 439–459. [Google Scholar] [CrossRef]
- Schwachtje, J.; Whitcomb, S.; Firmino, A.A.P.; Zuther, E.; Hincha, D.K.; Kopka, J. Induced, Imprinted, and Primed Responses to Changing Environments: Does Metabolism Store and Process Information? Front. Plant Sci. 2019, 10, 106. [Google Scholar] [CrossRef]


| ESI Mode | Compounds | Rt (min) | m/z | DCAA Fold Change | DCPCA Fold Change | DCSA Fold Change | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 36 h | 12 h | 24 h | 36 h | 12 h | 24 h | 36 h | |||||
| 1 | – | p-Coumaric acid derivative | 0.86 | 404.103 | ● | ● | ● | ● | ● | ● | 1.48 | 1.18 | 1.16 |
| 2 | – | 4-O-p-Coumaroylquinic acid | 1.15 | 337.084 | ● | ● | 5.58 | ● | ● | ● | ● | ● | ● |
| 3 | – | 3-Hydroxycoumarin | 1.25 | 161.043 | ● | ● | ● | ● | 0.58 | ● | ● | ● | ● |
| 4 | – | 3-O-p-Coumaroylquinic acid | 3.06 | 337.112 | ● | ● | ● | ● | 0.83 | ● | ● | 0.78 | |
| 5 | – | Sinapic acid hexose | 5.14 | 385.113 | ● | 0.65 | 2.18 | 2.21 | ● | 4.59 | 1.86 | ● | 2.12 |
| 6 | – | Dihydroferulic acid 4-O-glucuronide | 7.01 | 371.096 | 0.87 | 0.84 | 1.087 | ● | ● | ● | 0.82 | 0.84 | ● |
| 7 | + | Coumaroylputrescine | 2.39 | 235.145 | ● | ● | ● | ● | 1.68 | ● | ● | ● | ● |
| 8 | + | Coumaroylhydroxyagmatine | 2.57 | 293.157 | ● | 1.37 | 1.27 | ● | 1.45 | ● | ● | 1.15 | 1.09 |
| 9 | + | Coumaroylagmatine | 4.06 | 277.161 | 0.35 | 1.33 | 1.09 | ● | 1.62 | 1.17 | ● | 1.34 | 1.07 |
| 10 | – | Feruloylhydroxyagmatine | 4.49 | 323.133 | ● | 0.73 | ● | ● | 0.84 | ● | ● | 0.74 | ● |
| 11 | + | Feruloylagmatine | 5.30 | 307.172 | 0.612 | ● | ● | ● | ● | 1.14 | ● | ● | ● |
| 12 | + | Sinapoylagmatine | 6.17 | 337.186 | ● | ● | 3.08 | ● | 0.32 | ● | ● | ● | ● |
| 13 | – | Sinapoylhydroxyagmatine | 6.30 | 351.126 | ● | 0.93 | 0.93 | 0.88 | ● | 0.89 | ● | 0.92 | 0.92 |
| 14 | – | Hordatine B hexose | 3.81 | 787.364 | 0.818 | 1.14 | 1.17 | 0.74 | 1.10 | 0.91 | 0.78 | 1.16 | 1.22 |
| 15 | – | Hordatine A hexose | 4.14 | 757.353 | 0.80 | ● | ● | 0.63 | 1.12 | 0.80 | 0.75 | ● | ● |
| 16 | – | Hordatine C hexose isomer I | 4.63 | 817.376 | ● | ● | ● | 0.68 | 1.24 | ● | 0.72 | 1.31 | ● |
| 17 | – | Hordatine C hexose isomer II | 6.26 | 771.200 | ● | 1.92 | ● | ● | ● | ● | ● | ● | ● |
| 18 | – | Hordatine B isomer I | 7.28 | 579.304 | 0.63 | 1.13 | 1.11 | 0.86 | 0.67 | ● | 1.14 | ● | |
| 19 | + | Hordatine A Isomer I | 7.72 | 551.304 | 0.01 | 1.34 | ● | 0.85 | ● | ● | ● | 1.32 | 0.92 |
| 20 | – | Hordatine A isomer II | 7.76 | 549.294 | ● | ● | ● | ● | ● | 2.61 | ● | ● | 3.49 |
| 21 | + | Hordatine B isomer II | 7.97 | 581.319 | 0.06 | 1.41 | 0.82 | ● | ● | ● | ● | 1.50 | ● |
| 22 | + | Hordatine C + 46 isomer I | 7.99 | 655.320 | 0.83 | 1.09 | 1.12 | 0.87 | ● | 0.88 | ● | 1.17 | 0.92 |
| 23 | + | Hordatine C isomer II | 8.67 | 611.330 | ● | ● | ● | ● | ● | ● | ● | 1.38 | ● |
| 24 | – | Isoorientin 7-O-glucoside(Lutonarin) | 6.43 | 609.144 | 1.45 | 1.81 | 1.11 | ● | 1.22 | 1.13 | 1.35 | 1.24 | 1.15 |
| 25 | – | Isoorientin 7-O-[6″-sinapoyl]-glucoside | 10.53 | 815.205 | 1.40 | 1.26 | ● | ● | 0.73 | ● | ● | ● | ● |
| 26 | – | Isovitexin 7,6″-di-O-glucoside | 8.15 | 755.205 | ● | ● | 0.85 | 0.53 | ● | 0.55 | ● | ● | 0.73 |
| 27 | + | Isovitexin -7-O-glucoside (Saponarin) | 8.39 | 595.166 | 0.56 | 1.21 | 0.78 | 0.93 | ● | 0.87 | 1.06 | 1.31 | 0.87 |
| 28 | – | Isovitexin 7-O-rhamnosylglucoside | 8.81 | 739.208 | 1.09 | 0.93 | 0.89 | 0.91 | ● | 0.86 | ● | ● | ● |
| 29 | – | Isovitexin 2″-O-glucoside | 9.79 | 593.150 | ● | ● | ● | ● | ● | ● | ● | 0.01 | 10.97 |
| 30 | – | Isovitexin 2″-O-arabinoside | 9.93 | 563.139 | ● | 0.01 | 6.34 | ● | ● | ● | ● | 0.01 | 11.45 |
| 31 | – | Isovitexin | 10.59 | 431.097 | 0.66 | ● | 0.29 | 0.40 | 2.09 | 0.13 | ● | 2.02 | 0.43 |
| 32 | – | Isovitexin 7-O-[6″-sinapoyl]-glucoside | 11.42 | 799.210 | 1.16 | 0.77 | 1.18 | 1.09 | 0.65 | 1.47 | ● | 0.70 | ● |
| 33 | – | Isovitexin 7-O-[X″-feruloyl]-glucoside (Feruloylsaponarin) | 11.82 | 769.200 | 0.89 | 0.93 | ● | 0.85 | ● | ● | 0.84 | ● | ● |
| 34 | – | Apigenin 7-O-arabinosylglucoside | 11.90 | 563.140 | ● | ● | 0.82 | ● | ● | ● | ● | ● | 0.85 |
| 35 | – | Apigenin 6-C-arabinoside 8-C-glucoside | 8.97 | 563.140 | 1.52 | ● | ● | ● | ● | ● | ● | ● | ● |
| 36 | – | Isoscoparin 7-O-glucoside | 8.99 | 623.160 | ● | 1.31 | 0.85 | ● | 1.29 | 0.78 | 1.10 | 1.26 | ● |
| 37 | + | Isoscoparin 2″,6”-di-O-glucoside | 10.89 | 787.209 | ● | 1.53 | ● | ● | 1.29 | ● | ● | ● | ● |
| 38 | – | Isoscoparin 7-O-[6″-sinapoyl]-glucoside | 11.53 | 829.221 | ● | ● | ● | 0.72 | ● | ● | ● | ● | ● |
| 39 | – | Isoscoparin 7-O-[6″-feruloyl]-glucoside | 11.95 | 799.211 | ● | ● | ● | ● | 1.32 | 0.72 | ● | 1.25 | ● |
| 40 | – | Isorhamnetin-3-O-glucoside | 9.74 | 477.107 | ● | 3.49 | ● | ● | ● | ● | ● | 2.43 | ● |
| 41 | – | 6-Prenylnaringenin | 19.02 | 339.215 | ● | 0.19 | 0.68 | 5.42 | 0.82 | 3.60 | 3.07 | 0.53 | 1.80 |
| 42 | + | Hydroxytryptamine | 1.67 | 177.102 | ● | 2.47 | 2.50 | ● | 1.31 | ● | 1.84 | 1.76 | 1.26 |
| 43 | – | Coumaroyltryptamine | 2.60 | 289.129 | ● | 1.29 | 1.55 | 0.76 | 1.19 | ● | 1.15 | 1.14 | 1.15 |
| 44 | + | Valine | 0.88 | 118.086 | ● | ● | 0.77 | ● | ● | ● | 1.26 | ● | ● |
| 45 | + | Tyrosine | 1.14 | 182.081 | 0.50 | ● | 0.72 | 0.70 | ● | 0.58 | 0.63 | ● | 0.61 |
| 46 | + | Tyrosine derivatives | 1.14 | 276.107 | 0.33 | 1.60 | ● | ● | ● | ● | ● | ● | ● |
| 47 | + | Isoleucine | 1.31 | 132.102 | ● | ● | 0.74 | ● | 0.72 | 0.55 | 0.61 | ● | 0.69 |
| 48 | + | Phenylalanine | 1.65 | 166.087 | ● | 0.82 | 0.58 | 0.64 | 0.743 | 0.44 | 0.51 | 0.76 | 0.52 |
| 49 | + | Proline betaine | 2.05 | 144.139 | ● | ● | ● | ● | ● | ● | ● | ● | 15.26 |
| 50 | – | Tryptophan | 2.43 | 203.080 | ● | ● | 0.84 | 0.76 | 0.65 | 0.65 | 0.71 | 0.74 | 0.63 |
| 51 | – | Asparaginylglucose | 4.24 | 293.122 | ● | 0.79 | ● | ● | ● | ● | ● | 0.81 | ● |
| 52 | – | Citric acid | 1.14 | 191.017 | ● | 0.55 | ● | ● | 0.50 | ● | ● | 0.49 | ● |
| 53 | – | Isocitric acid | 0.93 | 191.017 | ● | 0.69 | ● | 1.29 | 0.42 | ● | 0.41 | ● | |
| 54 | – | Citric acid derivative | 1.39 | 306.117 | ● | ● | ● | ● | 0.85 | ● | ● | ● | ● |
| 55 | – | Malic acid | 0.95 | 133.012 | ● | ● | 0.74 | ● | 0.74 | 1.22 | 1.27 | 0.73 | ● |
| 56 | – | 12-Hydroxyjasmonate sulfate | 16.61 | 305.129 | ● | ● | 2.93 | ● | ● | ● | ● | ● | ● |
| 57 | – | Dihydrojasmonic acid (H2JA) conjugate | 16.80 | 419.173 | ● | ● | ● | ● | ● | 2.14 | ● | ● | 1.34 |
| 58 | – | 9,12,13,TriHODE isomer I | 16.53 | 327.216 | ● | 0.39 | 0.70 | 3.52 | ● | 2.23 | 1.67 | 0.61 | 1.69 |
| 59 | – | 9,12,13,TriHODE Isomer II | 16.64 | 327.216 | ● | ● | ● | 10.73 | ● | ● | ● | ● | ● |
| 60 | – | TriHOME | 17.25 | 329.232 | ● | 0.31 | ● | 4.59 | ● | ● | 2.13 | 0.46 | ● |
| 61 | – | 9-Oxo-12,13-diHODE | 17.45 | 325.200 | ● | 0.14 | ● | 6.31 | ● | 3.51 | 2.41 | 0.43 | 2.27 |
| 62 | – | OPDA conjugate | 19.55 | 309.205 | ● | 0.23 | ● | 3.90 | ● | 2.47 | 3.73 | 0.31 | ● |
| 63 | + | Linolenoylglycerol isomer I | 20.69 | 353.267 | ● | ● | 2.12 | 1.93 | ● | 3.08 | 1.71 | ● | 1.71 |
| 64 | + | Linolenoylglycerol isomer II | 20.98 | 353.265 | ● | ● | 1.70 | ● | ● | 2.71 | 1.64 | ● | 1.70 |
| 65 | + | Linolenoylglycerol isomer III | 21.80 | 353.263 | ● | ● | 1.72 | ● | ● | ● | ● | ● | ● |
| 66 | + | Linolenoylglycerol isomer VI | 22.07 | 353.263 | 2.57 | ● | ● | ● | ● | 1.63 | 2.17 | 0.70 | ● |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamany Djande, C.Y.; Steenkamp, P.A.; Piater, L.A.; Tugizimana, F.; Dubery, I.A. Metabolic Reprogramming of Barley in Response to Foliar Application of Dichlorinated Functional Analogues of Salicylic Acid as Priming Agents and Inducers of Plant Defence. Metabolites 2023, 13, 666. https://doi.org/10.3390/metabo13050666
Hamany Djande CY, Steenkamp PA, Piater LA, Tugizimana F, Dubery IA. Metabolic Reprogramming of Barley in Response to Foliar Application of Dichlorinated Functional Analogues of Salicylic Acid as Priming Agents and Inducers of Plant Defence. Metabolites. 2023; 13(5):666. https://doi.org/10.3390/metabo13050666
Chicago/Turabian StyleHamany Djande, Claude Y., Paul A. Steenkamp, Lizelle A. Piater, Fidele Tugizimana, and Ian A. Dubery. 2023. "Metabolic Reprogramming of Barley in Response to Foliar Application of Dichlorinated Functional Analogues of Salicylic Acid as Priming Agents and Inducers of Plant Defence" Metabolites 13, no. 5: 666. https://doi.org/10.3390/metabo13050666
APA StyleHamany Djande, C. Y., Steenkamp, P. A., Piater, L. A., Tugizimana, F., & Dubery, I. A. (2023). Metabolic Reprogramming of Barley in Response to Foliar Application of Dichlorinated Functional Analogues of Salicylic Acid as Priming Agents and Inducers of Plant Defence. Metabolites, 13(5), 666. https://doi.org/10.3390/metabo13050666








