Bone Metabolite Profile Differs between Normal and Femur Head Necrosis (FHN/BCO)-Affected Broilers: Implications for Dysregulated Metabolic Cascades in FHN Pathophysiology
Abstract
1. Introduction
2. Materials and Methods
2.1. Birds and FHN Scoring
2.2. Sample Collection and Preparation
2.3. Ultra-High-Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC–HRMS) Metabolomics Analysis
2.4. Ingenuity Pathway Analysis (IPA)
2.5. RNA Isolation, Reverse Transcription, and Real-Time Quantitative PCR
2.6. Data Processing and Statistical Analysis
3. Results
3.1. Multivariate Analysis and Comparative Metabolomics Profile in FHN-Affected and Unaffected Bone
3.2. Metabolic Pathway and Network Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wideman, R.F.; Hamal, K.R.; Stark, J.M.; Blankenship, J.; Lester, H.; Mitchell, K.N.; Lorenzoni, G.; Pevzner, I. A wire-flooring model for inducing lameness in broilers: Evaluation of probiotics as a prophylactic treatment. Poult. Sci. 2012, 91, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Wideman, R.F. Bacterial chondronecrosis with osteomyelitis and lameness in broilers: A review. Poult. Sci. 2016, 95, 325–344. [Google Scholar] [CrossRef] [PubMed]
- Wideman, R.F.; Prisby, R.D. Bone circulatory disturbances in the development of spontaneous bacterial chondronecrosis with osteomyelitis: A translational model for the pathogenesis of femoral head necrosis. Front. Endocrinol. 2012, 3, 183. [Google Scholar] [CrossRef] [PubMed]
- Granquist, E.G.; Vasdal, G.; de Jong, I.C.; Moe, R.O. Lameness and its relationship with health and production measures in broiler chickens. Animal 2019, 13, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Bassler, A.W.; Arnould, C.; Butterworth, A.; Colin, L.; De Jong, I.C.; Ferrante, V.; Ferrari, P.; Haslam, S.; Wemelsfelder, F.; Blokhuis, H.J. Potential risk factors associated with contact dermatitis, lameness, negative emotional state, and fear of humans in broiler chicken flocks. Poult. Sci. 2013, 92, 2811–2826. [Google Scholar] [CrossRef]
- Kittelsen, K.E.; David, B.; Moe, R.O.; Poulsen, H.D.; Young, J.F.; Granquist, E.G. Associations among gait score, production data, abattoir registrations, and postmortem tibia measurements in broiler chickens. Poult. Sci. 2017, 96, 1033–1040. [Google Scholar] [CrossRef]
- McGeown, D.; Danbury, T.C.; Waterman-Pearson, A.E.; Kestin, S.C. Effect of carprofen on lameness in broiler chickens. Vet. Rec. 1999, 144, 668–671. [Google Scholar] [CrossRef]
- Caplen, G.; Baker, L.; Hothersall, B.; McKeegan, D.E.; Sandilands, V.; Sparks, N.H.; Waterman-Pearson, A.E.; Murrell, J.C. Thermal nociception as a measure of non-steroidal anti-inflammatory drug effectiveness in broiler chickens with articular pain. Vet. J. 2013, 198, 616–619. [Google Scholar] [CrossRef]
- Wijesurendra, D.S.; Chamings, A.N.; Bushell, R.N.; Rourke, D.O.; Stevenson, M.; Marenda, M.S.; Noormohammadi, A.H.; Stent, A. Pathological and microbiological investigations into cases of bacterial chondronecrosis and osteomyelitis in broiler poultry. Avian Pathol. 2017, 46, 683–694. [Google Scholar] [CrossRef]
- Ramser, A.; Greene, E.; Wideman, R.; Dridi, S. Local and Systemic Cytokine, Chemokine, and FGF Profile in Bacterial Chondronecrosis with Osteomyelitis (BCO)-Affected Broilers. Cells 2021, 10, 3174. [Google Scholar] [CrossRef]
- de Oliveira Peixoto, J.; Savoldi, I.R.; Ibelli, A.M.G.; Cantão, M.E.; Jaenisch, F.R.F.; Giachetto, P.F.; Settles, M.L.; Zanella, R.; Marchesi, J.A.P.; Pandolfi, J.R.; et al. Proximal femoral head transcriptome reveals novel candidate genes related to epiphysiolysis in broiler chickens. BMC Genom. 2019, 20, 1031. [Google Scholar] [CrossRef] [PubMed]
- Ibelli, A.M.G.; Peixoto, J.d.O.; Zanella, R.; Gouveia, J.J.d.S.; Cantão, M.E.; Coutinho, L.L.; Marchesi, J.A.P.; Pizzol, M.S.d.; Marcelino, D.E.P.; Ledur, M.C. Downregulation of growth plate genes involved with the onset of femoral head separation in young broilers. Front. Physiol. 2022, 13, 941134. [Google Scholar] [CrossRef] [PubMed]
- Florencio-Silva, R.; Sasso, G.R.; Sasso-Cerri, E.; Simões, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. BioMed Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef]
- Shen, F.; Ruddy, M.J.; Plamondon, P.; Gaffen, S.L. Cytokines link osteoblasts and inflammation: Microarray analysis of interleukin-17- and TNF-α-induced genes in bone cells. J. Leukoc. Biol. 2005, 77, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.M. An overview of bone cells and their regulating factors of differentiation. Malays. J. Med. Sci. 2008, 15, 4–12. [Google Scholar] [PubMed]
- Greene, E.; Cauble, R.; Dhamad, A.E.; Kidd, M.T.; Kong, B.; Howard, S.M.; Castro, H.F.; Campagna, S.R.; Bedford, M.; Dridi, S. Muscle Metabolome Profiles in Woody Breast-(un)Affected Broilers: Effects of Quantum Blue Phytase-Enriched Diet. Front. Vet. Sci. 2020, 7, 458. [Google Scholar] [CrossRef] [PubMed]
- Dridi, J.S.; Greene, E.S.; Maynard, C.W.; Brugaletta, G.; Ramser, A.; Christopher, C.J.; Campagna, S.R.; Castro, H.F.; Dridi, S. Duodenal Metabolic Profile Changes in Heat-Stressed Broilers. Animals 2022, 12, 1337. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Martino, C.; Powers, J.B.; Campagna, S.R.; Voy, B.H.; Donohoe, D.R.; Gaffney, J.; Embree, M.M.; Myer, P.R. Rumen Bacteria and Serum Metabolites Predictive of Feed Efficiency Phenotypes in Beef Cattle. Sci. Rep. 2019, 9, 19265. [Google Scholar] [CrossRef]
- Lu, W.; Clasquin, M.F.; Melamud, E.; Amador-Noguez, D.; Caudy, A.A.; Rabinowitz, J.D. Metabolomic Analysis via Re-versed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer. Anal. Chem. 2010, 82, 3212–3221. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Clasquin, M.F.; Melamud, E.; Rabinowitz, J.D. LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Curr. Protoc. Bioinform. 2012, 37, 14.11.1–14.11.23. [Google Scholar]
- Bazurto, J.V.; Dearth, S.P.; Tague, E.D.; Campagna, S.R.; Downs, D.M. Untargeted metabolomics confirms and extends the understanding of the impact of aminoimidazole carboxamide ribotide (AICAR) in the metabolic network of Salmonella enterica. Microb. Cell 2017, 5, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef]
- Nguyen, P.; Greene, E.; Ishola, P.; Huff, G.; Donoghue, A.; Bottje, W.; Dridi, S. Chronic Mild Cold Conditioning Modulates the Expression of Hypothalamic Neuropeptide and Intermediary Metabolic-Related Genes and Improves Growth Performances in Young Chicks. PLoS ONE 2015, 10, e0142319. [Google Scholar] [CrossRef]
- Greene, E.S.; Zampiga, M.; Sirri, F.; Ohkubo, T.; Dridi, S. Orexin system is expressed in avian liver and regulates hepatic lipogenesis via ERK1/2 activation. Sci. Rep. 2020, 10, 19191. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Alrubaye, A.A.K.; Ekesi, N.S.; Hasan, A.; Elkins, E.; Ojha, S.; Zaki, S.; Dridi, S.; Wideman, R.F.; Rebollo, M.A.; Rhoads, D.D. Chondronecrosis with osteomyelitis in broilers: Further defining lameness-inducing models with wire or litter flooring to evaluate protection with organic trace minerals. Poult. Sci. 2020, 99, 5422–5429. [Google Scholar] [CrossRef] [PubMed]
- Mandal, R.K.; Jiang, T.; Al-Rubaye, A.A.; Rhoads, D.D.; Wideman, R.F.; Zhao, J.; Pevzner, I.; Kwon, Y.M. An investigation into blood microbiota and its potential association with Bacterial Chondronecrosis with Osteomyelitis (BCO) in Broilers. Sci. Rep. 2016, 6, 25882. [Google Scholar] [CrossRef] [PubMed]
- Ferver, A.; Greene, E.; Wideman, R.; Dridi, S. Evidence of Mitochondrial Dysfunction in Bacterial Chondronecrosis with Osteomyelitis-Affected Broilers. Front. Vet. Sci. 2021, 8, 640901. [Google Scholar] [CrossRef] [PubMed]
- Ramser, A.; Greene, E.; Alrubaye, A.A.K.; Wideman, R.; Dridi, S. Role of autophagy machinery dysregulation in bacterial chondronecrosis with osteomyelitis. Poult. Sci. 2022, 101, 101750. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, H.C.; Ibelli, A.M.G.; Guimarães, S.E.F.; Cantão, M.E.; Peixoto, J.d.O.; Coutinho, L.L.; Ledur, M.C. RNA-seq reveals downregulated osteochondral genes potentially related to tibia bacterial chondronecrosis with osteomyelitis in broilers. BMC Genet. 2020, 21, 58. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.M.; Mahbub, S.; Habibalahi, A.; Paton, S.; Gronthos, S.; Goldys, E. Ageing human bone marrow mesenchymal stem cells have depleted NAD(P)H and distinct multispectral autofluorescence. GeroScience 2021, 43, 859–868. [Google Scholar] [CrossRef]
- Kim, H.-N.; Ponte, F.; Warren, A.; Ring, R.; Iyer, S.; Han, L.; Almeida, M. A decrease in NAD(+) contributes to the loss of osteoprogenitors and bone mass with aging. NPJ Aging Mech. Dis. 2021, 7, 8. [Google Scholar] [CrossRef]
- Li, B.; Shi, Y.; Liu, M.; Wu, F.; Hu, X.; Yu, F.; Wang, C.; Ye, L. Attenuates of NAD + impair BMSC osteogenesis and fracture repair through OXPHOS. Stem Cell Res. Ther. 2022, 13, 77. [Google Scholar] [CrossRef]
- Wang, P.; Du, H.; Zhou, C.-C.; Song, J.; Liu, X.; Cao, X.; Mehta, J.L.; Shi, Y.; Su, D.-F.; Miao, C.-Y. Intracellular NAMPT-NAD+-SIRT1 cascade improves post-ischaemic vascular repair by modulating Notch signalling in endothelial progenitors. Cardiovasc. Res. 2014, 104, 477–488. [Google Scholar] [CrossRef]
- Hong, L.; Ai, J.; Ma, D. Activation of Dusp14 protects against osteoclast generation and bone loss by regulating AMPKα-dependent manner. Biochem. Biophys. Res. Commun. 2019, 519, 445–452. [Google Scholar] [CrossRef]
- Mao, Z.; Zhu, Y.; Hao, W.; Chu, C.; Su, H. MicroRNA-155 inhibition up-regulates LEPR to inhibit osteoclast activation and bone resorption via activation of AMPK in alendronate-treated osteoporotic mice. IUBMB Life 2019, 71, 1916–1928. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.J.; Cho, G.W. Metformin promotes neuronal differentiation and neurite outgrowth through AMPK activation in human bone marrow–mesenchymal stem cells. Biotechnol. Appl. Biochem. 2017, 64, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, B.-J.; Choi, H.J.; Cho, S.W.; Shin, C.S.; Park, S.-Y.; Lee, Y.-S.; Lee, S.-Y.; Kim, H.-H.; Kim, G.S.; et al. (–)-Epigallocathechin-3-Gallate, an AMPK Activator, Decreases Ovariectomy-Induced Bone Loss by Suppression of Bone Resorption. Calcif. Tissue Int. 2012, 90, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.-E.; Shin, J.H.; Jang, E.S.; Park, S.J.; Park, D.R.; Ko, R.; Seo, D.-H.; Kim, H.-S.; Lee, S.H.; Choi, Y.; et al. Sirtuin 3 (SIRT3) maintains bone homeostasis by regulating AMPK-PGC-1β axis in mice. Sci. Rep. 2016, 6, 22511. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.L.; Bucholtz, K.M.; Kacsoh, B. Selective inhibition of human 3β-hydroxysteroid dehydrogenase type 1 as a potential treatment for breast cancer. J. Steroid Biochem. Mol. Biol. 2011, 125, 57–65. [Google Scholar] [CrossRef]
- Labrie, F.; Archer, D.F.; Portman, D. Efficacy of intravaginal dehydroepiandrosterone (DHEA) on moderate to severe dyspareunia and vaginal dryness, symptoms of vulvovaginal atrophy. Maturitas 2015, 82, 315–316. [Google Scholar] [CrossRef]
- Yanase, T.; Suzuki, S.; Goto, K.; Nomura, M.; Okabe, T.; Takayanagi, R.; Nawata, H. Aromatase in bone: Roles of vitamin D3 and androgens. J. Steroid Biochem. Mol. Biol. 2003, 86, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, J.; Tsuji, T.; Imazeki, I.; Ikeda, H.; Nishimura, T. Immunosteroid as a regulator for Th1/Th2 balance: Its possible role in autoimmune diseases. Autoimmunity 2005, 38, 369–375. [Google Scholar] [CrossRef]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.M.W.; Tam, S.; Sims, N.A.; Saleh, H.; McGregor, N.E.; Poulton, I.J.; Walker, E.C.; Scott, J.; Kemp, B.E.; Gillespie, M.T.; et al. Mice lacking AMP-activated kinase (AMPK) subunits β1 or β2 have low bone mass, while AICAR acts AMPK-independently to increase osteoclast formation. Bone 2009, 44, S136. [Google Scholar] [CrossRef]
- Czerner, C.P.; Klos, A.; Seifert, R.; Neumann, D. Histamine induces chemotaxis and phagocytosis in murine bone marrow-derived macrophages and RAW 264.7 macrophage-like cells via histamine H4-receptor. Inflamm. Res. 2014, 63, 239–247. [Google Scholar] [CrossRef]
- Wiercigroch, M.; Folwarczna, J. Histamine in regulation of bone remodeling processes. Postȩpy Hig. I Med. Doświadczalnej 2013, 67, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, Y.; Yonekawa, T.; Ohkuni, Y.; Kuribayashi, M.; Fukino, K.; Ueno, K. A comparative study of histamine activities on differentiation of osteoblasts and osteoclasts. J. Toxicol. Sci. 2007, 32, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Graham, L.S.; Tintut, Y.; Parhami, F.; Kitchen, C.M.R.; Ivanov, Y.; Tetradis, S.; Effros, R.B. Bone density and hyperlipidemia: The T-lymphocyte connection. J. Bone Miner. Res. 2010, 25, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Alekos, N.S.; Moorer, M.C.; Riddle, R.C. Dual Effects of Lipid Metabolism on Osteoblast Function. Front. Endocrinol. 2020, 11, 578194. [Google Scholar] [CrossRef]
- Wang, B.; Wang, H.; Li, Y.; Song, L. Lipid metabolism within the bone micro-environment is closely associated with bone metabolism in physiological and pathophysiological stages. Lipids Health Dis. 2022, 21, 5. [Google Scholar] [CrossRef]
- Durairaj, V.; Okimoto, R.; Rasaputra, K.; Clark, F.D.; Rath, N.C. Histopathology and Serum Clinical Chemistry Evaluation of Broilers with Femoral Head Separation Disorder. Avian Dis. 2009, 53, 21–25. [Google Scholar] [CrossRef]
- Liu, K.; Wang, K.; Wang, L.; Zhou, Z. Changes of lipid and bone metabolism in broilers with spontaneous femoral head necrosis. Poult. Sci. 2021, 100, 100808. [Google Scholar] [CrossRef]
- Packialakshmi, B.; Liyanage, R.; Lay, J.O.; Okimoto, R.; Rath, N.C. Proteomic Changes in the Plasma of Broiler Chickens with Femoral Head Necrosis. Biomark. Insights 2016, 11, BMI-S38291. [Google Scholar] [CrossRef]
- Wang, Y.; Russo, T.A.; Kwon, O.; Chanock, S.; Rumsey, S.C.; Levine, M. Ascorbate Recycling in Human Neutrophils: Induction by Bacteria. Proc. Natl. Acad. Sci. USA 1997, 94, 13816–13819. [Google Scholar] [CrossRef]
- Aghajanian, P.; Hall, S.; Wongworawat, M.D.; Mohan, S. The Roles and Mechanisms of Actions of Vitamin C in Bone: New Developments. J. Bone Miner. Res. 2015, 30, 1945–1955. [Google Scholar] [CrossRef] [PubMed]
- Qutob, S.; Dixon, S.J.; Wilson, J.X. Insulin stimulates vitamin C recycling and ascorbate accumulation in osteoblastic cells. Endocrinology 1998, 139, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Drury, A.N.; Szent-Györgyi, A. The physiological activity of adenine compounds with especial reference to their action upon the mammalian heart. J. Physiol. 1929, 68, 213–237. [Google Scholar] [CrossRef] [PubMed]
- Hoebertz, A.; Arnett, T.R.; Burnstock, G. Regulation of bone resorption and formation by purines and pyrimidines. Trends Pharmacol. Sci. 2003, 24, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Jørgensen, N.R. Extracellular purines and bone homeostasis. Biochem. Pharmacol. 2021, 187, 114425. [Google Scholar] [CrossRef] [PubMed]
- Elgueta, R.; Benson, M.J.; de Vries, V.C.; Wasiuk, A.; Guo, Y.; Noelle, R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 2009, 229, 152–172. [Google Scholar] [CrossRef] [PubMed]
- Pineda, B.; Tarín, J.J.; Hermenegildo, C.; Laporta, P.; Cano, A.; García-Pérez, M.Á. Gene–gene interaction between CD40 and CD40L reduces bone mineral density and increases osteoporosis risk in women. Osteoporos. Int. 2011, 22, 1451–1458. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Cahill, M.P.; Hostager, B.S.; Brosnahan, A.J.; Klingelhutz, A.J.; Gourronc, F.A.; Bishop, G.A.; Leung, D.Y.M. Staphylococcal Superantigens Stimulate Epithelial Cells through CD40 To Produce Chemokines. mBio 2019, 10, e00214-19. [Google Scholar] [CrossRef]
- Iena, F.M.; Lebeck, J. Implications of Aquaglyceroporin 7 in Energy Metabolism. Int. J. Mol. Sci. 2018, 19, 154. [Google Scholar] [CrossRef]
- Liu, Y.; Song, L.; Wang, Y.; Rojek, A.; Nielsen, S.; Agre, P.; Carbrey, J.M. Osteoclast differentiation and function in aquaglyceroporin AQP9-null mice. Biol. Cell 2009, 101, 133–140. [Google Scholar] [CrossRef]
- Graziano, A.C.E.; Avola, R.; Pannuzzo, G.; Cardile, V. Aquaporin1 and 3 modification as a result of chondrogenic differentiation of human mesenchymal stem cell. J. Cell. Physiol. 2018, 233, 2279–2291. [Google Scholar] [CrossRef] [PubMed]
- Pelagalli, A.; Nardelli, A.; Lucarelli, E.; Zannetti, A.; Brunetti, A. Autocrine signals increase ovine mesenchymal stem cells mi-gration through Aquaporin-1 and CXCR4 overexpression. J. Cell. Physiol. 2018, 233, 6241–6249. [Google Scholar] [CrossRef] [PubMed]
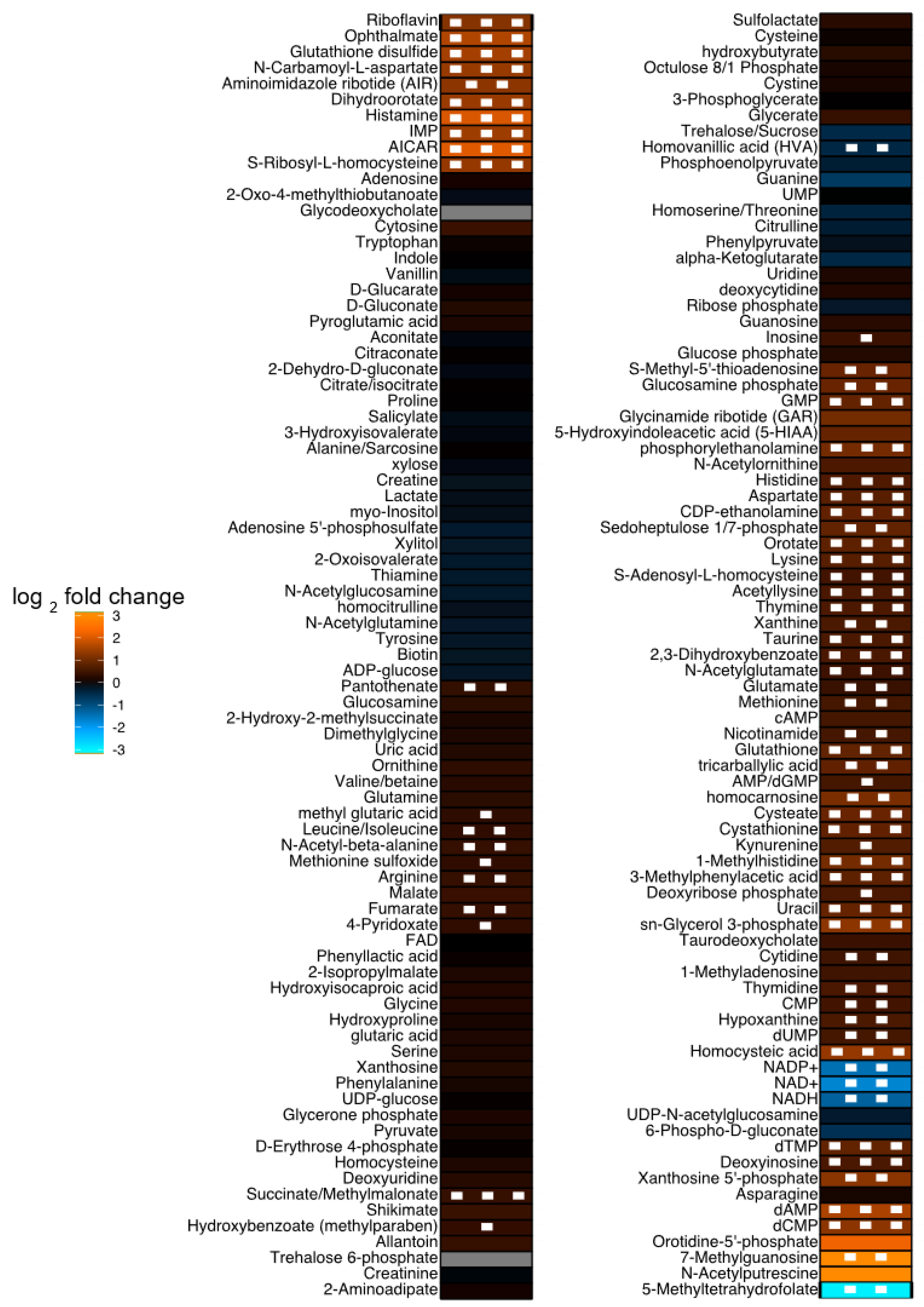




| HMDB ID | Metabolite Name | Fold Change | p-Value |
|---|---|---|---|
| Increased abundance | |||
| HMDB0001517 | AICAR | 4.166 | 0.00014 |
| HMDB0000870 | Histamine | 4.023 | 0.00077 |
| HMDB0005765 | Ophthalmate | 3.103 | 0.00062 |
| HMDB0000905 | dAMP | 2.981 | 0.00033 |
| HMDB0003337 | Glutathione disulfide | 2.841 | 0.00002 |
| HMDB0015536 | IMP | 2.764 | 0.00083 |
| HMDB0003349 | Dihydroorotate | 2.652 | 0.00086 |
| HMDB0000828 | N-Carbamoyl-l-aspartate | 2.621 | 0.00031 |
| HMDB0000676 | S-Ribosyl-l-homocysteine | 2.590 | 0.00167 |
| HMDB0002205 | Homocysteic acid | 2.558 | 0.00010 |
| HMDB0000126 | sn-Glycerol-3-phosphate | 2.450 | 0.00182 |
| HMDB0001202 | dCMP | 2.446 | 0.00110 |
| HMDB0001235 | Aminoimidazole ribotide | 2.438 | 0.01686 |
| HMDB0001554 | Xanthosine-5′-phosphate | 2.402 | 0.01264 |
| HMDB0000244 | Riboflavin | 2.340 | 0.00031 |
| HMDB0000745 | Homocarnosine | 2.163 | 0.03150 |
| HMDB0000224 | Phosphorylethanolamine | 2.108 | 0.00485 |
| HMDB0000001 | 1-Methylhistidine | 2.107 | 0.00337 |
| HMDB0002757 | Cysteate | 1.909 | 0.00011 |
| HMDB0001173 | S-Methyl-5′-thioadenosine | 1.904 | 0.01450 |
| HMDB0001254 | Glucosamine phosphate | 1.903 | 0.01654 |
| HMDB0000300 | Uracil | 1.866 | 0.00735 |
| HMDB0001397 | GMP | 1.844 | 0.00194 |
| HMDB0060509 | Sedoheptulose-1/7-phosphate | 1.830 | 0.03038 |
| HMDB0000125 | Glutathione | 1.826 | 0.00034 |
| HMDB0001227 | dTMP | 1.826 | 0.00054 |
| HMDB0001564 | CDP-ethanolamine | 1.818 | 0.00109 |
| HMDB0031193 | Tricarballylic acid | 1.805 | 0.02514 |
| HMDB0000099 | Cystathionine | 1.803 | 0.00095 |
| HMDB0000226 | Orotate | 1.785 | 0.00104 |
| HMDB0002222 | 3-Methylphenylacetic acid | 1.784 | 0.00025 |
| HMDB0003405 | Lysine | 1.696 | 0.00116 |
| HMDB0000191 | Aspartate | 1.678 | 0.00020 |
| HMDB0000177 | Histidine | 1.594 | 0.00093 |
| HMDB0000262 | Thymine | 1.562 | 0.00219 |
| HMDB0000251 | Taurine | 1.545 | 0.00111 |
| HMDB0000292 | Xanthine | 1.540 | 0.02493 |
| HMDB0000157 | Hypoxanthine | 1.534 | 0.01077 |
| HMDB0000397 | 2,3-Dihydroxybenzoate | 1.524 | 0.00089 |
| HMDB0000273 | Thymidine | 1.514 | 0.03461 |
| HMDB0000206 | Acetyllysine | 1.514 | 0.00196 |
| HMDB0000071 | Deoxyinosine | 1.511 | 0.00786 |
| HMDB0001409 | dUMP | 1.471 | 0.03402 |
| HMDB0000696 | Methionine | 1.463 | 0.02533 |
| HMDB0001406 | Nicotinamide | 1.459 | 0.02024 |
| HMDB0000095 | CMP | 1.452 | 0.01468 |
| HMDB0000939 | I-Adenosyl-l-homocysteine | 1.449 | 0.00441 |
| HMDB0000089 | Cytidine | 1.432 | 0.01883 |
| HMDB0001138 | N-Acetylglutamate | 1.395 | 0.00092 |
| HMDB0060475 | Glutamate | 1.344 | 0.01180 |
| HMDB0000202 | Succinate/Methylmalonate | 1.325 | 0.00358 |
| HMDB0000134 | Fumarate | 1.324 | 0.02559 |
| HMDB0000517 | Arginine | 1.314 | 0.03810 |
| HMDB0061880 | N-Acetyl-beta-alanine | 1.297 | 0.01459 |
| HMDB0000210 | Pantothenate | 1.290 | 0.03354 |
| HMDB0028932 | Leucine/Isoleucine | 1.255 | 0.03115 |
| HMDB0000118 | Homovanillic acid | 0.721 | 0.02502 |
| Decreased abundance | |||
| HMDB0001487 | NADH | 0.417 | 0.04303 |
| HMDB0000217 | NADP+ | 0.360 | 0.02809 |
| HMDB0000902 | NAD+ | 0.300 | 0.01608 |
| Canonical Pathway | Molecules | −log (p-Value) | Ratio |
|---|---|---|---|
| Ascorbate Recycling (Cytosolic) | glutathione, glutathione disulfide, NAD+, NADH, NADP+ | 6.99 | 0.625 |
| Purine Nucleotides Degradation II (Aerobic) | GMP, hypoxanthine, NAD+, NADH, xanthine, xanthosine monophosphate | 6.47 | 0.353 |
| Purine Nucleotides De Novo Biosynthesis II | AICAR, aminoimidazole ribotide, GMP, l-aspartic acid, NAD+, NADH, xanthosine monophosphate | 6.12 | 0.233 |
| Urate Biosynthesis/Inosine 5′-phosphate Degradation | NAD+, NADH, xanthine, xanthosine monophosphate | 4.88 | 0.444 |
| Guanosine Nucleotides Degradation III | GMP, NAD+, NADH, xanthine | 4.66 | 0.4 |
| Diseases and Functions | p-Value | # Molecules * |
|---|---|---|
| Cancer | 4.94 × 10−2–8.20 × 10−6 | 19 |
| Organismal Injury and Abnormalities | 4.94 × 10−2–8.20 × 10−6 | 23 |
| Dermatological Disease and Conditions | 3.78 × 10−2–1.73 × 10−4 | 6 |
| Hematological Disease | 1.36 × 10−2–4.95 × 10−4 | 4 |
| Gastrointestinal Disease | 3.78 × 10−2–7.36 × 10−4 | 14 |
| Molecular and Cellular Functions | p-Value | # Molecules * |
|---|---|---|
| Lipid Metabolism | 4.00 × 10−2–1.23 × 10−4 | 12 |
| Small Molecule Biochemistry | 4.93 × 10−2–1.23 × 10−4 | 17 |
| Cellular Development | 4.66 × 10−2–2.12 × 10−4 | 15 |
| Cellular Growth and Proliferation | 4.66 × 10−2–2.12 × 10−4 | 14 |
| Nucleic Acid Metabolism | 4.14 × 10−2–3.54 × 10−4 | 12 |
| Upstream Regulators * | p-Value | Z Score |
|---|---|---|
| ATM | 6.20 × 10−7 | −1.732 |
| CPT1B | 8.78 × 10−6 | −1.633 |
| CD40 | 1.20 × 10−4 | 2.236 |
| CD274 | 6.16 × 10−7 | 1 |
| CHKA | 9.55 × 10−7 | −0.577 |
| AQP7 | 1.86 × 10−4 | 1.566 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramser, A.; Hawken, R.; Greene, E.; Okimoto, R.; Flack, B.; Christopher, C.J.; Campagna, S.R.; Dridi, S. Bone Metabolite Profile Differs between Normal and Femur Head Necrosis (FHN/BCO)-Affected Broilers: Implications for Dysregulated Metabolic Cascades in FHN Pathophysiology. Metabolites 2023, 13, 662. https://doi.org/10.3390/metabo13050662
Ramser A, Hawken R, Greene E, Okimoto R, Flack B, Christopher CJ, Campagna SR, Dridi S. Bone Metabolite Profile Differs between Normal and Femur Head Necrosis (FHN/BCO)-Affected Broilers: Implications for Dysregulated Metabolic Cascades in FHN Pathophysiology. Metabolites. 2023; 13(5):662. https://doi.org/10.3390/metabo13050662
Chicago/Turabian StyleRamser, Alison, Rachel Hawken, Elizabeth Greene, Ron Okimoto, Brenda Flack, Courtney J. Christopher, Shawn R. Campagna, and Sami Dridi. 2023. "Bone Metabolite Profile Differs between Normal and Femur Head Necrosis (FHN/BCO)-Affected Broilers: Implications for Dysregulated Metabolic Cascades in FHN Pathophysiology" Metabolites 13, no. 5: 662. https://doi.org/10.3390/metabo13050662
APA StyleRamser, A., Hawken, R., Greene, E., Okimoto, R., Flack, B., Christopher, C. J., Campagna, S. R., & Dridi, S. (2023). Bone Metabolite Profile Differs between Normal and Femur Head Necrosis (FHN/BCO)-Affected Broilers: Implications for Dysregulated Metabolic Cascades in FHN Pathophysiology. Metabolites, 13(5), 662. https://doi.org/10.3390/metabo13050662







