High-Throughput Screening of Natural Product and Synthetic Molecule Libraries for Antibacterial Drug Discovery
Abstract
1. Introduction
2. Approaches and Strategies for Antibacterial High-Throughput Screening (HTS) Assays
2.1. Cellular and Molecular Target-Based HTS
2.2. Mechanism Informed Phenotypic HTS Screening (Reporter-Based HTS)
2.3. Virulence and Quorum-Sensing Targeting HTS
2.4. Genome Science, Molecular Target Identification, and HTS
2.5. Combination of HTS Strategies and Multi-Target Hits
2.6. Externally Interceded HTS
3. Natural Product Library (NPL) Screening for Antibacterial Drug Discovery
3.1. Historical Perspectives and Major Bottlenecks
3.2. Why Are Natural Products Still Preferred for Drug Discovery despite the Challenges in Screening Them?
3.3. Challenges of Collecting Natural Sources for Screening
3.4. Challenges in Growing Natural Sources under Laboratory Conditions
3.5. Challenges in Extracting Natural Products
3.6. Challenges in Preparing a Natural Product Library for HTS
3.7. Available Resources for NPL for Drug Screening Research and Campaigns
3.8. Examples of Successful HTS of NPL for Antibacterial Drug Discovery
3.9. Antibacterial Biofilm Inhibitory Compounds from HTS of NPL
3.10. Antibacterial Agents from HTS of Unconventional Natural Sources
3.11. Metagenomics and Metabologenomics Aided NPL HTS for Antibacterial Drug Discovery
3.12. Microfabricated Chip-Based HTS of NPL for Antibacterial Drug Discovery from Uncultivable Organisms
3.13. Coculture-Based HTS of NPL for Antibacterial Drug Discovery
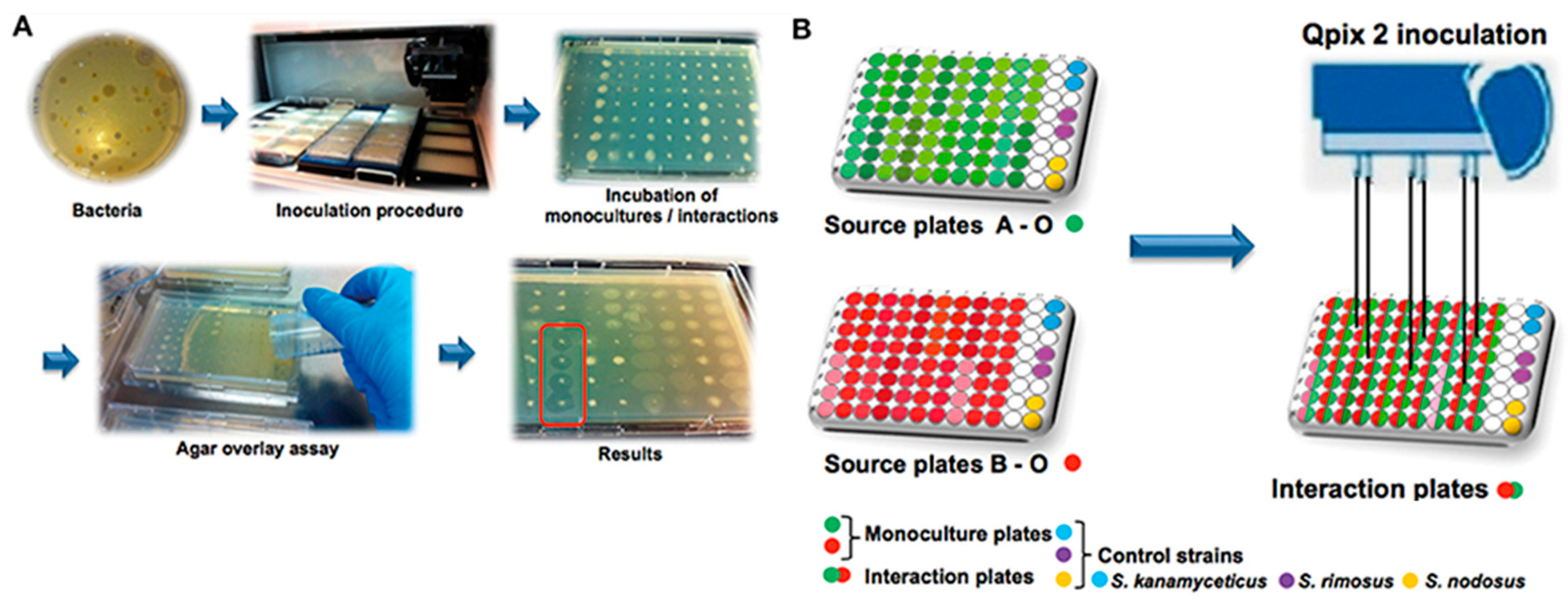
3.14. Integrated Platforms for HTS of NPL for Antibacterial Drug Discovery
4. Synthetic Molecule Library (SML) Screening for Antibacterial Drug Discovery
4.1. Historical Perspective and Available Resources
4.2. Cellular Target-Based HTS (CT-HTS) of the Synthetic Molecule Library (SML)
4.3. Molecular Target-Based HTS (MT-HTS) of the Synthetic Molecule Library (SML)
4.4. Other Miscellaneous HTS Assays Using the Synthetic Molecule Library (SML)
4.5. High-Throughput Synthetic Molecule Library Screening against Quorum-Sensing and Biofilm-Forming Bacteria
4.6. High-Throughput Synthetic Molecule Library Screening Using Biomimetic Conditions
4.7. High-Throughput Synthetic Molecule Library Screening, Drug Repurposing, and Synergy
4.8. The Library of Synthetic Peptides and Polymers and Antibacterial High-Throughput Screening
5. Technical Considerations for Designing High-Throughput Screening Assays for Antibacterial Drug Discovery
5.1. Library Selection
5.2. Logistics and Technology Platforms
5.3. Storage and Stability
5.4. Microorganisms and Culture Conditions
5.5. Orthogonal Assays for Hit Validation, Toxicity Screening, Dereplication, and Target Identification
5.6. Error Management, Quality Control, and HTS Triage
5.7. In Vivo Studies and Pharmacodynamic and Pharmacokinetic Characteristics
6. Technologies and Other Auxiliary Approaches for Antibacterial HTS Assays
6.1. Selective Screening
6.2. Genetic Engineering, Synthetic Biology, and Omics Technology
6.3. In Silico/Virtual Screening
6.4. Combinatorial Chemistry and the Focused Synthetic Approach
6.5. Microfluidic, Nanofluidic, and Imaging-Based Technologies
6.6. Phage Display and Antibody-Based Technologies
6.7. Metal Nanoparticles
6.8. Spectrometry, Cytometry, Spectroscopy, and Other Biophysical Approaches
7. Final Remarks and Future Perspectives
8. Conclusions
Funding
Conflicts of Interest
References
- Butler, M.S.; Blaskovich, M.A.; Cooper, M.A. Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. 2017, 70, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Frieden, T. Antibiotic Resistance Threats in the United States 2013; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Silver, L.L. Challenges of Antibacterial Discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting Antibiotic Resistance. Angew. Chem. Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Moore, A.M.; Patel, S.; Forsberg, K.J.; Wang, B.; Bentley, G.; Razia, Y.; Qin, X.; Tarr, P.I.; Dantas, G. Pediatric Fecal Microbiota Harbor Diverse and Novel Antibiotic Resistance Genes. PLOS ONE 2013, 8, e78822. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Waksman, S.A. Microbial Antagonisms and Antibiotic Substances; The Commonwealth Fund: New York, NY, USA, 1945. [Google Scholar]
- Mahady, G.B.; Huang, Y.; Doyle, B.J.; Locklear, T. Natural Products as Antibacterial Agents. Stud. Nat. Prod. Chem. 2008, 35, 423–444. [Google Scholar] [CrossRef]
- Nielsen, J.C.; Grijseels, S.; Prigent, S.; Ji, B.; Dainat, J.; Nielsen, K.F.; Frisvad, J.C.; Workman, M.; Nielsen, J. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2017, 2, 17044. [Google Scholar] [CrossRef]
- Coates, A.R.M.; Hu, Y. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 2007, 152, 1147–1154. [Google Scholar] [CrossRef]
- Lau, Q.Y.; Tan, Y.Y.F.; Goh, V.C.Y.; Lee, D.J.Q.; Ng, F.M.; Ong, E.H.Q.; Hill, J.; Chia, C.S.B. An FDA-Drug Library Screen for Compounds with Bioactivities against Meticillin-Resistant Staphylococcus aureus (MRSA). Antibiotics 2015, 4, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Younis, W.; Thangamani, S.; Seleem, M. Repurposing Non-Antimicrobial Drugs and Clinical Molecules to Treat Bacterial Infections. Curr. Pharm. Des. 2015, 21, 4106–4111. [Google Scholar] [CrossRef] [PubMed]
- Younis, W.; AbdelKhalek, A.; Mayhoub, A.S.; Seleem, M. In Vitro Screening of an FDA-Approved Library Against ESKAPE Pathogens. Curr. Pharm. Des. 2017, 23, 2147–2157. [Google Scholar] [CrossRef]
- Weissman, K.J.; Leadlay, P. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Genet. 2005, 3, 925–936. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2006, 6, 29–40. [Google Scholar] [CrossRef]
- Chan, P.F.; Holmes, D.J.; Payne, D.J. Finding the gems using genomic discovery: Antibacterial drug discovery strategies – the successes and the challenges. Drug Discov. Today Ther. Strat. 2004, 1, 519–527. [Google Scholar] [CrossRef]
- Thaker, M.N.; Waglechner, N.; Wright, G. Antibiotic resistance–mediated isolation of scaffold-specific natural product producers. Nat. Protoc. 2014, 9, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 2006, 33, 507–513. [Google Scholar] [CrossRef]
- Singh, S.B.; Young, K.; Miesel, L. Screening strategies for discovery of antibacterial natural products. Expert Rev. Anti-infective Ther. 2011, 9, 589–613. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Farha, M.A.; Brown, E.D. Unconventional screening approaches for antibiotic discovery. Ann. N. Y. Acad. Sci. 2015, 1354, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Lakemeyer, M.; Zhao, W.; Mandl, F.A.; Hammann, P.; Sieber, S.A. Thinking Outside the Box-Novel Antibacterials To Tackle the Resistance Crisis. Angew. Chem. Int. Ed. 2018, 57, 14440–14475. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, K.M.G.; Hodgkinson, J.T.; Sore, H.F.; Welch, M.; Salmond, G.P.C.; Spring, D.R. Combating Multidrug-Resistant Bacteria: Current Strategies for the Discovery of Novel Antibacterials. Angew. Chem. Int. Ed. 2013, 52, 10706–10733. [Google Scholar] [CrossRef]
- Schenone, M.; Dančík, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef]
- Finan, C.; Gaulton, A.; Kruger, F.A.; Lumbers, R.T.; Shah, T.; Engmann, J.; Casas, J.P. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9, eaag1166. [Google Scholar] [CrossRef]
- Yee, A.; Chang, X.; Pineda-Lucena, A.; Wu, B.; Semesi, A.; Le, B.; Ramelot, T.; Lee, G.M.; Bhattacharyya, S.; Gutierrez, P.; et al. An NMR approach to structural proteomics. Proc. Natl. Acad. Sci. USA 2002, 99, 1825–1830. [Google Scholar] [CrossRef] [PubMed]
- Reo, N.V. NMR-Based Metabolomics. Drug Chem. Toxicol. 2002, 25, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Gwynn, M.N.; Portnoy, A.; Rittenhouse, S.F.; Payne, D.J. Challenges of antibacterial discovery revisited. Ann. N. Y. Acad. Sci. 2010, 1213, 5–19. [Google Scholar] [CrossRef]
- Bharat, A.; Blanchard, J.E.; Brown, E.D. A High-Throughput Screen of the GTPase Activity of Escherichia coli EngA to Find an Inhibitor of Bacterial Ribosome Biogenesis. SLAS Discov. Adv. Sci. Drug Discov. 2013, 18, 830–836. [Google Scholar] [CrossRef]
- Brötz-Oesterhelt, H.; Sass, P. Postgenomic strategies in antibacterial drug discovery. Futur. Microbiol. 2010, 5, 1553–1579. [Google Scholar] [CrossRef]
- Walsh, C.T.; A Wencewicz, T. Prospects for new antibiotics: A molecule-centered perspective. J. Antibiot. 2013, 67, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Grohar, P.J.; Woldemichael, G.M.; Griffin, L.B.; Mendoza, A.; Chen, Q.-R.; Yeung, C.; Currier, D.; Davis, S.; Khanna, C.; Khan, J.; et al. Identification of an Inhibitor of the EWS-FLI1 Oncogenic Transcription Factor by High-Throughput Screening. Gynecol. Oncol. 2011, 103, 962–978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.G.; Lee, J.; Gallo, R.A.; Tao, W.; Tse, D.; Doddapaneni, R.; Pelaez, D. Therapeutic targeting of oncogenic transcription factors by natural products in eye cancer. Pharmacol. Res. 2017, 129, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Donadio, S.; Brandi, L.; Serina, S.; Sosio, M.; Stinchi, S. Discovering Novel Antibacterial Agents by High Throughput Screening. Front. Drug Des. Discov. 2005, 1, 3–16. [Google Scholar] [CrossRef]
- Navarro, G.; Cheng, A.T.; Peach, K.C.; Bray, W.M.; Bernan, V.S.; Yildiz, F.H.; Linington, R.G. Image-Based 384-Well High-Throughput Screening Method for the Discovery of Skyllamycins A to C as Biofilm Inhibitors and Inducers of Biofilm Detachment in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.C.; DiDone, L.; Jobson, J.; Sofia, M.K.; Krysan, D.; Dunman, P.M. Adenylate Kinase Release as a High-Throughput-Screening-Compatible Reporter of Bacterial Lysis for Identification of Antibacterial Agents. Antimicrob. Agents Chemother. 2013, 57, 26–36. [Google Scholar] [CrossRef]
- Forbes, L.; Ebsworth-Mojica, K.; DiDone, L.; Li, S.-G.; Freundlich, J.S.; Connell, N.; Dunman, P.M.; Krysan, D.J. A High Throughput Screening Assay for Anti-Mycobacterial Small Molecules Based on Adenylate Kinase Release as a Reporter of Cell Lysis. PLoS ONE 2015, 10, e0129234. [Google Scholar] [CrossRef]
- Boes, A.; Olatunji, S.; Mohammadi, T.; Breukink, E.; Terrak, M. Fluorescence anisotropy assays for high throughput screening of compounds binding to lipid II, PBP1b, FtsW and MurJ. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Rasko, D.A.; Moreira, C.G.; Li de, R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC signaling and virulence for antibiotic development. Science 2008, 321, 1078–1080. [Google Scholar] [CrossRef]
- Escaich, S. Novel agents to inhibit microbial virulence and pathogenicity. Expert Opin. Ther. Patents 2010, 20, 1401–1418. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Ioerger, T.R.; O’malley, T.; Liao, R.; Guinn, K.M.; Hickey, M.J.; Mohaideen, N.; Murphy, K.C.; Boshoff, H.I.M.; Mizrahi, V.; Rubin, E.J.; et al. Identification of New Drug Targets and Resistance Mechanisms in Mycobacterium tuberculosis. PLoS ONE 2013, 8, e75245. [Google Scholar] [CrossRef] [PubMed]
- Andries, K.; Verhasselt, P.; Guillemont, J.; Göhlmann, H.W.; Neefs, J.M.; Winkler, H.; Van Gestel, J.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Lea, W.A.; Simeonov, A. Fluorescence polarization assays in small molecule screening. Expert Opin. Drug Discov. 2010, 6, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Henrich, C.J.; Beutler, J.A. Matching the power of high throughput screening to the chemical diversity of natural products. Nat. Prod. Rep. 2013, 30, 1284–1298. [Google Scholar] [CrossRef]
- Liao, J.; Xu, G.; Mevers, E.E.; Clardy, J.; Watnick, P.I. A high-throughput, whole cell assay to identify compounds active against carbapenem-resistant Klebsiella pneumoniae. PLoS ONE 2018, 13, e0209389. [Google Scholar] [CrossRef]
- Li, X.; Zolli-Juran, M.; Cechetto, J.D.; Daigle, D.M.; Wright, G.D.; Brown, E.D. Multicopy suppressors for novel antibacterial compounds reveal targets and drug efflux susceptibility. Chem. Biol. 2004, 11, 1423–1430. [Google Scholar] [CrossRef]
- Stone, L.K.; Baym, M.; Lieberman, T.D.; Chait, R.; Clardy, J.; Kishony, R. Compounds that select against the tetracycline-resistance efflux pump. Nat. Chem. Biol. 2016, 12, 902–904. [Google Scholar] [CrossRef]
- Landeta, C.; Mejia-Santana, A. Union is strength: Target-based and whole-cell high throughput screens in antibacterial discovery. J. Bacteriol. 2021. [Google Scholar] [CrossRef]
- Matano, L.M.; Morris, H.G.; Wood, B.M.; Meredith, T.C.; Walker, S. Accelerating the discovery of antibacterial compounds using pathway-directed whole cell screening. Bioorganic Med. Chem. 2016, 24, 6307–6314. [Google Scholar] [CrossRef]
- Mak, P.A.; Rao, S.P.S.; Tan, M.P.; Lin, X.; Chyba, J.; Tay, J.; Ng, S.H.; Tan, B.H.; Cherian, J.; Duraiswamy, J.; et al. A High-Throughput Screen To Identify Inhibitors of ATP Homeostasis in Non-replicating Mycobacterium tuberculosis. ACS Chem. Biol. 2012, 7, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Isgut, M.; Rao, M.; Yang, C.; Subrahmanyam, V.; Rida, P.C.G.; Aneja, R. Application of Combination High-Throughput Phenotypic Screening and Target Identification Methods for the Discovery of Natural Product-Based Combination Drugs. Med. Res. Rev. 2017, 38, 504–524. [Google Scholar] [CrossRef] [PubMed]
- Grube, C.D.; Roy, H. A continuous assay for monitoring the synthetic and proofreading activities of multiple aminoacyl-tRNA synthetases for high-throughput drug discovery. RNA Biol. 2017, 15, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Kozuma, S.; Hirota-Takahata, Y.; Fukuda, D.; Kuraya, N.; Nakajima, M.; Ando, O. Screening and biological activities of pedopeptins, novel inhibitors of LPS produced by soil bacteria. J. Antibiot. 2013, 67, 237–242. [Google Scholar] [CrossRef]
- McGovern, S.L.; Caselli, E.; Grigorieff, N.; Shoichet, B.K. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J. Med. Chem. 2002, 45, 1712–1722. [Google Scholar] [CrossRef]
- Moon, K.; Xu, F.; Seyedsayamdost, M.R. Cebulantin, a Cryptic Lanthipeptide Antibiotic Uncovered Using Bioactivity-Coupled HiTES. Angew. Chem. Int. Ed. 2019, 58, 5973–5977. [Google Scholar] [CrossRef]
- Sharma, S.; Bhat, R.; Singh, R.; Sharma, S.; Wazir, P.; Singh, P.P.; Vishwakarma, R.A.; Khan, I.A. High-throughput screening of compounds library to identify novel inhibitors against latent Mycobacterium tuberculosis using streptomycin-dependent Mycobacterium tuberculosis 18b strain as a model. Tuberculosis 2020, 124, 101958. [Google Scholar] [CrossRef]
- Cho, S.H.; Warit, S.; Wan, B.; Hwang, C.H.; Pauli, G.F.; Franzblau, S.G. Low-Oxygen-Recovery Assay for High-Throughput Screening of Compounds against Nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007, 51, 1380–1385. [Google Scholar] [CrossRef]
- A Bauer, R.; Wurst, J.M.; Tan, D.S. Expanding the range of ‘druggable’ targets with natural product-based libraries: An academic perspective. Curr. Opin. Chem. Biol. 2010, 14, 308–314. [Google Scholar] [CrossRef]
- Moloney, M.G. Natural Products as a Source for Novel Antibiotics. Trends Pharmacol. Sci. 2016, 37, 689–701. [Google Scholar] [CrossRef]
- Santos, J.D.; Vitorino, I.; Reyes, F.; Vicente, F.; Lage, O.M. From Ocean to Medicine: Pharmaceutical Applications of Metabolites from Marine Bacteria. Antibiotics 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Saini, K.C.; Bast, F.; Varjani, S.; Mehariya, S.; Bhatia, S.K.; Sharma, N.; Funk, C. A Review on Microbial Products and Their Perspective Application as Antimicrobial Agents. Biomolecules 2021, 11, 1860. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.G.; Lister, T.; May-Dracka, T.L. New natural products as new leads for antibacterial drug discovery. Bioorganic Med. Chem. Lett. 2014, 24, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Koehn, F.E. New Strategies and Methods in the Discovery of Natural Product Anti-Infective Agents: The Mannopeptimycins. J. Med. Chem. 2008, 51, 2613–2617. [Google Scholar] [CrossRef]
- Wilson, B.A.P.; Thornburg, C.C.; Henrich, C.J.; Grkovic, T.; O’Keefe, B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [Google Scholar] [CrossRef]
- Li, J.W.-H.; Vederas, J.C. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef]
- Witting, K.; Sussmuth, R.D. Discovery of Antibacterials and Other Bioactive Compounds from Microorganisms—Evaluating Methodologies for Discovery and Generation of Non-Ribosomal Peptide Antibiotics. Curr. Drug Targets 2011, 12, 1547–1559. [Google Scholar] [CrossRef]
- Paterson, I.; Anderson, E.A. The Renaissance of Natural Products as Drug Candidates. Science 2005, 310, 451–453. [Google Scholar] [CrossRef]
- Kennedy, J.; Marchesi, J.R.; Dobson, A.D. Marine metagenomics: Strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Factories 2008, 7, 27. [Google Scholar] [CrossRef]
- Liu, X.; Ashforth, E.; Ren, B.; Song, F.; Dai, H.; Liu, M.; Wang, J.; Xie, Q.; Zhang, L. Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J. Antibiot. 2010, 63, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Li, Y.; Wang, J.; Song, F.-H.; Liu, M.; Dai, H.-Q.; Ren, B.; Gao, H.; Hu, X.; Liu, Z.-H.; et al. Amycolatopsis marina sp. nov., an actinomycete isolated from an ocean sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 477–481. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Li, Y.; Bian, J.; Tang, S.-K.; Ren, B.; Chen, M.; Li, W.-J.; Zhang, L.-X. Prauserella marina sp. nov., isolated from ocean sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 2010, 60, 985–989. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, L.; Zhang, J.; Ren, B.; Lu, W.; Hou, C.; Wang, J.; Ma, X.; Ma, R.; Liu, M.; Liu, Z.; et al. Characterization of anti-BCG benz[α]anthraquinones and new siderophores from a Xinjiang desert–isolated rare actinomycete Nocardia sp. XJ31. Appl. Microbiol. Biotechnol. 2020, 104, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Shen, Y. Harnessing the potential of chemical defenses from antimicrobial activities. Bioessays 2004, 26, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J. Antibiotics from Natural Sources. In Antibiotic Drug Resistance; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 311–341. [Google Scholar]
- Rienzo, M.; Jackson, S.J.; Chao, L.K.; Leaf, T.; Schmidt, T.J.; Navidi, A.H.; Nadler, D.C.; Ohler, M.; Leavell, M.D. High-throughput screening for high-efficiency small-molecule biosynthesis. Metab. Eng. 2020, 63, 102–125. [Google Scholar] [CrossRef]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofroňová, O.; Petříčková, K.; Bobek, J. Secondary Metabolites Produced during the Germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef]
- Butler, M.S.; Fontaine, F.; Cooper, M.A. Natural Product Libraries: Assembly, Maintenance, and Screening. Planta Medica 2013, 80, 1161–1170. [Google Scholar] [CrossRef]
- Richter, B.E.; Jones, B.A.; Ezzell, J.L.; Porter, N.L.; Avdalovic, N.; Pohl, C. Accelerated Solvent Extraction: A Technique for Sample Preparation. Anal. Chem. 1996, 68, 1033–1039. [Google Scholar] [CrossRef]
- Johnson, T.A.; Morgan, M.V.C.; Aratow, N.A.; Estee, S.A.; Sashidhara, K.V.; Loveridge, S.T.; Segraves, N.L.; Crews, P. Assessing Pressurized Liquid Extraction for the High-Throughput Extraction of Marine-Sponge-Derived Natural Products. J. Nat. Prod. 2009, 73, 359–364. [Google Scholar] [CrossRef]
- Chemat, F.; Rombaut, N.; Sicaire, A.-G.; Meullemiestre, A.; Fabiano-Tixier, A.-S.; Abert-Vian, M. Ultrasound assisted extraction of food and natural products. Mechanisms, techniques, combinations, protocols and applications. A review. Ultrason. Sonochem. 2017, 34, 540–560. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.-H.; Yusoff, R.; Ngoh, G.-C.; Kung, F.W.-L. Microwave-assisted extractions of active ingredients from plants. J. Chromatogr. A 2011, 1218, 6213–6225. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liang, J. Counter-current chromatography for high throughput analysis of natural products. Comb. Chem. High Throughput Screen. 2010, 13, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; del Pilar Sánchez-Camargo, A.; Cifuentes, A.; Ibáñez, E. Plants, seaweeds, microalgae and food by-products as natural sources of functional ingredients obtained using pressurized liquid extraction and supercritical fluid extraction. TrAC Trends Anal. Chem. 2015, 71, 26–38. [Google Scholar] [CrossRef]
- Potterat, O.; Hamburger, M. Concepts and technologies for tracking bioactive compounds in natural product extracts: Generation of libraries, and hyphenation of analytical processes with bioassays. Nat. Prod. Rep. 2013, 30, 546–564. [Google Scholar] [CrossRef]
- He, H.; Williamson, R.T.; Shen, B.; Graziani, E.I.; Yang, H.Y.; Sakya, S.M.; Petersen, P.J.; Carter, G.T. Mannopeptimycins, Novel Antibacterial Glycopeptides from Streptomyces hygroscopicus, LL-AC98. J. Am. Chem. Soc. 2002, 124, 9729–9736. [Google Scholar] [CrossRef]
- Thornburg, C.C.; Britt, J.R.; Evans, J.R.; Akee, R.K.; Whitt, J.A.; Trinh, S.K.; Harris, M.J.; Thompson, J.R.; Ewing, T.L.; Shipley, S.M.; et al. NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening. ACS Chem. Biol. 2018, 13, 2484–2497. [Google Scholar] [CrossRef]
- Camp, D.; Campitelli, M.; Carroll, A.R.; Davis, R.A.; Quinn, R.J. Front-Loading Natural-Product-Screening Libraries for logP: Background, Development, and Implementation. Chem. Biodivers. 2013, 10, 524–537. [Google Scholar] [CrossRef]
- Appleton, D.R.; Buss, A.D.; Butler, M.S. A Simple Method for High-Throughput Extract Prefractionation for Biological Screening. Chimia 2007, 61. [Google Scholar] [CrossRef]
- Funke, M.; Buchenauer, A.; Schnakenberg, U.; Mokwa, W.; Diederichs, S.; Mertens, A.; Müller, C.; Kensy, F.; Büchs, J. Microfluidic biolector-microfluidic bioprocess control in microtiter plates. Biotechnol. Bioeng. 2010, 107, 497–505. [Google Scholar] [CrossRef]
- Sandner, V.; Pybus, L.; McCreath, G.; Glassey, J. Scale-Down Model Development in ambr systems: An Industrial Perspective. Biotechnol. J. 2018, 14, e1700766. [Google Scholar] [CrossRef]
- Liu, X.; Bolla, K.; Ashforth, E.J.; Zhuo, Y.; Gao, H.; Huang, P.; Stanley, S.A.; Hung, D.T.; Zhang, L. Systematics-guided bioprospecting for bioactive microbial natural products. Antonie Van Leeuwenhoek 2011, 101, 55–66. [Google Scholar] [CrossRef] [PubMed]
- A Leeds, J.; Schmitt, E.K.; Krastel, P. Recent developments in antibacterial drug discovery: Microbe-derived natural products – from collection to the clinic. Expert Opin. Investig. Drugs 2006, 15, 211–226. [Google Scholar] [CrossRef]
- Bugni, T.S.; Richards, B.; Bhoite, L.; Cimbora, D.; Harper, M.K.; Ireland, C.M. Marine Natural Product Libraries for High-Throughput Screening and Rapid Drug Discovery. J. Nat. Prod. 2008, 71, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, G.R.; Vervoort, H.C.; Lee, C.M.; Cremin, P.A.; Williams, C.T.; Hart, S.M.; Goering, M.G.; O’Neil-Johnson, M.; Zeng, L. High-Throughput Method for the Production and Analysis of Large Natural Product Libraries for Drug Discovery. Anal. Chem. 2002, 74, 3963–3971. [Google Scholar] [CrossRef] [PubMed]
- Ardalani, H.; Anam, S.; Kromphardt, K.J.K.; Staerk, D.; Kongstad, K.T. Coupling Microplate-Based Antibacterial Assay with Liquid Chromatography for High-Resolution Growth Inhibition Profiling of Crude Extracts: Validation and Proof-of-Concept Study with Staphylococcus aureus. Molecules 2021, 26, 1550. [Google Scholar] [CrossRef]
- Adnani, N.; Michel, C.R.; Bugni, T.S. Universal Quantification of Structurally Diverse Natural Products Using an Evaporative Light Scattering Detector. J. Nat. Prod. 2012, 75, 802–806. [Google Scholar] [CrossRef]
- Johnson, T.A.; Sohn, J.; Inman, W.D.; Estee, S.A.; Loveridge, S.T.; Vervoort, H.C.; Tenney, K.; Liu, J.; Ang, K.K.-H.; Ratnam, J.; et al. Natural Product Libraries to Accelerate the High-Throughput Discovery of Therapeutic Leads. J. Nat. Prod. 2011, 74, 2545–2555. [Google Scholar] [CrossRef]
- Srivastava, N.; Sarethy, I.P.; Jeevanandam, J.; Danquah, M. Emerging strategies for microbial screening of novel chemotherapeutics. J. Mol. Struct. 2022, 1255, 132419. [Google Scholar] [CrossRef]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef]
- Singh, S.B.; Jayasuriya, H.; Ondeyka, J.G.; Herath, K.B.; Zhang, C.; Zink, D.L.; Tsou, N.N.; Ball, R.G.; Basilio, A.; Genilloud, O.; et al. Isolation, Structure, and Absolute Stereochemistry of Platensimycin, A Broad Spectrum Antibiotic Discovered Using an Antisense Differential Sensitivity Strategy. J. Am. Chem. Soc. 2006, 128, 11916–11920. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Steele, A.D.; Teijaro, C.N.; Yang, D.; Shen, B. Leveraging a large microbial strain collection for natural product discovery. J. Biol. Chem. 2019, 294, 16567–16576. [Google Scholar] [CrossRef]
- Wagenaar, M.M. Pre-fractionated Microbial Samples – The Second Generation Natural Products Library at Wyeth. Molecules 2008, 13, 1406–1426. [Google Scholar] [CrossRef]
- Paytubi, S.; de La Cruz, M.; Tormo, J.R.; Martín, J.; González, I.; González-Menendez, V.; Genilloud, O.; Reyes, F.; Vicente, F.; Madrid, C.; et al. A High-Throughput Screening Platform of Microbial Natural Products for the Discovery of Molecules with Antibiofilm Properties against Salmonella. Front. Microbiol. 2017, 8, 326. [Google Scholar] [CrossRef] [PubMed]
- Borghi, A.; Cavaletti, L.; Sponga, F.; Montanini, N.; Pollini, W.; Quarta, C.; Marinelli, F. The microbial product library for high-throughput screening at Lepetit Research center. In Studies in Organic Chemistry; Elsevier: Amsterdam, The Netherlands, 1998; pp. 235–238. [Google Scholar] [CrossRef]
- Camp, D.; A Davis, R.; A Evans-Illidge, E.; Quinn, R.J. Guiding principles for natural product drug discovery. Futur. Med. Chem. 2012, 4, 1067–1084. [Google Scholar] [CrossRef]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, N.; Williams, G.J. A high-throughput screen for directed evolution of aminocoumarin amide synthetases. Anal. Biochem. 2011, 419, 61–66. [Google Scholar] [CrossRef]
- Tan, D.S. Current Progress in Natural Product-like Libraries for Discovery Screening. Comb. Chem. High Throughput Screen. 2004, 7, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Nybond, S.; Karp, M.; Yrjönen, T.; Tammela, P. Bioluminescent whole-cell reporter gene assays as screening tools in the identification of antimicrobial natural product extracts. J. Microbiol. Methods 2015, 114, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Young, K.; Jayasuriya, H.; Ondeyka, J.G.; Herath, K.; Zhang, C.; Kodali, S.; Galgoci, A.; Painter, R.; Brown-Driver, V.; Yamamoto, R.; et al. Discovery of FabH/FabF Inhibitors from Natural Products. Antimicrob. Agents Chemother. 2006, 50, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Ymele-Leki, P.; Cao, S.; Sharp, J.; Lambert, K.G.; McAdam, A.J.; Husson, R.N.; Tamayo, G.; Clardy, J.; Watnick, P.I. A High-Throughput Screen Identifies a New Natural Product with Broad-Spectrum Antibacterial Activity. PLoS ONE 2012, 7, e31307. [Google Scholar] [CrossRef]
- Phillips, J.W.; Goetz, M.A.; Smith, S.K.; Zink, D.L.; Polishook, J.; Onishi, R.; Salowe, S.; Wiltsie, J.; Allocco, J.; Sigmund, J.; et al. Discovery of Kibdelomycin, a Potent New Class of Bacterial Type II Topoisomerase Inhibitor by Chemical-Genetic Profiling in Staphylococcus aureus. Chem. Biol. 2011, 18, 955–965. [Google Scholar] [CrossRef]
- Arai, M.A.; Kobatake, E.; Koyano, T.; Kowithayakorn, T.; Kato, S.; Ishibashi, M. A Method for the Rapid Discovery of Naturally Occurring Products by Proteins Immobilized on Magnetic Beads and Reverse Affinity Chromatography. Chem.–Asian J. 2009, 4, 1802–1808. [Google Scholar] [CrossRef]
- Kodali, S.; Galgoci, A.; Young, K.; Painter, R.; Silver, L.L.; Herath, K.B.; Singh, S.B.; Cully, D.; Barrett, J.F.; Schmatz, D.; et al. Determination of Selectivity and Efficacy of Fatty Acid Synthesis Inhibitors. J. Biol. Chem. 2005, 280, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Cao, S.; Vasquez, V.; Annamalai, T.; Tamayo-Castillo, G.; Clardy, J.; Tse-Dinh, Y.-C. Identification of Anziaic Acid, a Lichen Depside from Hypotrachyna sp., as a New Topoisomerase Poison Inhibitor. PLOS ONE 2013, 8, e60770. [Google Scholar] [CrossRef]
- Hu, Y.; Keniry, M.; Palmer, S.O.; Bullard, J.M. Discovery and Analysis of Natural-Product Compounds Inhibiting Protein Synthesis in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 4820–4829. [Google Scholar] [CrossRef]
- Zetterström, C.E.; Hasselgren, J.; Salin, O.; Davis, R.A.; Quinn, R.J.; Sundin, C.; Elofsson, M. The Resveratrol Tetramer (-)-Hopeaphenol Inhibits Type III Secretion in the Gram-Negative Pathogens Yersinia pseudotuberculosis and Pseudomonas aeruginosa. PLOS ONE 2013, 8, e81969. [Google Scholar] [CrossRef]
- Pal, R.; Dai, M.; Seleem, M.N. High-throughput screening identifies a novel natural product-inspired scaffold capable of inhibiting Clostridioides difficile in vitro. Sci. Rep. 2021, 11, 10913. [Google Scholar] [CrossRef]
- Guzman, J.D.; Gupta, A.; Evangelopoulos, D.; Basavannacharya, C.; Pabon, L.C.; Plazas, E.A.; Muñoz, D.R.; Delgado, W.A.; Cuca, L.E.; Ribon, W.; et al. Anti-tubercular screening of natural products from Colombian plants: 3-methoxynordomesticine, an inhibitor of MurE ligase of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2010, 65, 2101–2107. [Google Scholar] [CrossRef] [PubMed]
- Gilbert-Girard, S.; Reigada, I.; Savijoki, K.; Yli-Kauhaluoma, J.; Fallarero, A. Screening of natural compounds identifies ferutinin as an antibacterial and anti-biofilm compound. Biofouling 2021, 37, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; A Stremick, C.; Turner, R.J.; Allan, N.D.; Olson, M.; Ceri, H. Microtiter susceptibility testing of microbes growing on peg lids: A miniaturized biofilm model for high-throughput screening. Nat. Protoc. 2010, 5, 1236–1254. [Google Scholar] [CrossRef]
- Cooper, L.E.; McClerren, A.L.; Chary, A.; van der Donk, W.A. Structure-Activity Relationship Studies of the Two-Component Lantibiotic Haloduracin. Chem. Biol. 2008, 15, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.-W.; Balunas, M.J.; Chai, H.B.; Kinghorn, A.D. Drug discovery from natural sources. AAPS J. 2006, 8, E239–E253. [Google Scholar] [CrossRef] [PubMed]
- Simões, G.A.R. Novel Screening Platforms Construction for Detection of New Marine Microbial Bioactive Compounds. Ph.D. Thesis, Universidade de Lisboa, Lisbon, Portugal, 2018. [Google Scholar]
- Mickel, S.J.; Sedelmeier, G.H.; Niederer, D.; Daeffler, R.; Osmani, A.; Schreiner, K.; Seeger-Weibel, M.; Bérod, B.; Schaer, K.; Gamboni, R.; et al. Large-Scale Synthesis of the Anti-Cancer Marine Natural Product (+)-Discodermolide. Part 1: Synthetic Strategy and Preparation of a Common Precursor. Org. Process. Res. Dev. 2003, 8, 92–100. [Google Scholar] [CrossRef]
- Courtois, S.; Cappellano, C.M.; Ball, M.; Francou, F.-X.; Normand, P.; Helynck, G.; Martinez, A.; Kolvek, S.J.; Hopke, J.; Osburne, M.S.; et al. Recombinant Environmental Libraries Provide Access to Microbial Diversity for Drug Discovery from Natural Products. Appl. Environ. Microbiol. 2003, 69, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Scanlon, T.C.; Dostal, S.M.; Griswold, K.E. A high-throughput screen for antibiotic drug discovery. Biotechnol. Bioeng. 2013, 111, 232–243. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef] [PubMed]
- Eun, Y.J.; Utada, A.S.; Copeland, M.F.; Takeuchi, S.; Weibel, D.B. Encapsulating Bacteria in Agarose Microparticles Using Microfluidics for High-Throughput Cell Analysis and Isolation. ACS Chem. Biol. 2010, 6, 260–266. [Google Scholar] [CrossRef]
- Goering, A.W.; McClure, R.A.; Doroghazi, J.R.; Albright, J.C.; Haverland, N.A.; Zhang, Y.; Ju, K.-S.; Thomson, R.J.; Metcalf, W.W.; Kelleher, N.L. Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. ACS Central Sci. 2016, 2, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An interpreted atlas of biosynthetic gene clusters from 1000 fungal genomes. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Liu, X. Generate a bioactive natural product library by mining bacterial cytochrome P450 patterns. Synth. Syst. Biotechnol. 2016, 1, 95–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caesar, L.K.; Butun, F.A.; Robey, M.T.; Ayon, N.J.; Gupta, R.; Dainko, D.; Bok, J.W.; Nickles, G.; Stankey, R.J.; Johnson, D.; et al. Correlative metabologenomics of 110 fungi reveals metabolite–gene cluster pairs. Nat. Chem. Biol. 2023, 1–9. [Google Scholar] [CrossRef]
- Ateya, D.A.; Sachs, F.; Gottlieb, P.A.; Besch, S.; Hua, S.Z. Volume Cytometry: Microfluidic Sensor for High-Throughput Screening in Real Time. Anal. Chem. 2005, 77, 1290–1294. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef]
- Gavrish, E.; Sit, C.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Kloucek, P.; Smid, J.; Frankova, A.; Kokoska, L.; Valterova, I.; Pavela, R. Fast screening method for assessment of antimicrobial activity of essential oils in vapor phase. Food Res. Int. 2012, 47, 161–165. [Google Scholar] [CrossRef]
- Ingham, C.J.; Sprenkels, A.; Bomer, J.; Molenaar, D.; Berg, A.V.D.; Vlieg, J.E.T.V.H.; de Vos, W.M. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. USA 2007, 104, 18217–18222. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Demir, K.; Overhage, J.; Grunze, M.; Schwartz, T.; Levkin, P.A. Droplet-Microarray: Miniaturized Platform for High-Throughput Screening of Antimicrobial Compounds. Adv. Biosyst. 2020, 4. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.; Hassett, D.J.; Choi, S. A paper-based microbial fuel cell array for rapid and high-throughput screening of electricity-producing bacteria. Anal. 2015, 140, 4277–4283. [Google Scholar] [CrossRef]
- Yüce, I.; Morlock, G.E. Nanomole-scaled high-throughput chemistry plus direct bioautography on the same chromatography plate for drug discovery. Anal. Chim. Acta 2021, 1182, 338950. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.T.; Rotem, A.; Heyman, J.A.; Weitz, D.A. Droplet microfluidics for high-throughput biological assays. Lab A Chip 2012, 12, 2146–2155. [Google Scholar] [CrossRef]
- Brandi, L.; Fabbretti, A.; Milon, P.; Carotti, M.; Pon, C.L.; Gualerzi, C.O. Methods for Identifying Compounds that Specifically Target Translation. Methods Enzymol. 2007, 431, 229–267. [Google Scholar] [CrossRef]
- Brandi, L.; Fabbretti, A.; La Teana, A.; Abbondi, M.; Losi, D.; Donadio, S.; Gualerzi, C.O. Specific, efficient, and selective inhibition of prokaryotic translation initiation by a novel peptide antibiotic. Proc. Natl. Acad. Sci. USA 2005, 103, 39–44. [Google Scholar] [CrossRef]
- Brandi, L.; Fabbretti, A.; Di Stefano, M.; Lazzarini, A.; Abbondi, M.; Gualerzi, C.O. Characterization of GE82832, a peptide inhibitor of translocation interacting with bacterial 30S ribosomal subunits. RNA 2006, 12, 1262–1270. [Google Scholar] [CrossRef]
- Tyc, O.; van den Berg, M.; Gerards, S.; Van Veen, J.A.; Raaijmakers, J.M.; de Boer, W.; Garbeva, P. Impact of interspecific interactions on antimicrobial activity among soil bacteria. Front. Microbiol. 2014, 5, 567. [Google Scholar] [CrossRef]
- Murray, E.M.; Allen, C.F.; Handy, T.E.; Huffine, C.A.; Craig, W.R.; Seaton, S.C.; Wolfe, A.L. Development of a Robust and Quantitative High-Throughput Screening Method for Antibiotic Production in Bacterial Libraries. ACS Omega 2019, 4, 15414–15420. [Google Scholar] [CrossRef]
- Strege, M.A. High-performance liquid chromatographic–electrospray ionization mass spectrometric analyses for the integration of natural products with modern high-throughput screening. J. Chromatogr. B Biomed. Sci. Appl. 1999, 725, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Robinette, S.L.; Brüschweiler, R.; Schroeder, F.C.; Edison, A.S. NMR in Metabolomics and Natural Products Research: Two Sides of the Same Coin. Accounts Chem. Res. 2011, 45, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Kurita, K.L.; Glassey, E.; Linington, R.G. Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proc. Natl. Acad. Sci. USA 2015, 112, 11999–12004. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.R.; Oliver, A.G.; Linington, R.G. Development of Antibiotic Activity Profile Screening for the Classification and Discovery of Natural Product Antibiotics. Chem. Biol. 2012, 19, 1483–1495. [Google Scholar] [CrossRef]
- Wang, J.; Lyu, W.; Zhang, W.; Chen, Y.; Luo, F.; Wang, Y.; Ji, H.; Zhang, G. Discovery of natural products capable of inducing porcine host defense peptide gene expression using cell-based high throughput screening. J. Anim. Sci. Biotechnol. 2021, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Wang, J.; Lyu, W.; Wieneke, X.; Matts, R.; Ma, X.; Zhang, G. Development of a Cell-Based High-Throughput Screening Assay to Identify Porcine Host Defense Peptide-Inducing Compounds. J. Immunol. Res. 2018, 2018, 5492941. [Google Scholar] [CrossRef] [PubMed]
- Urban, A.; Eckermann, S.; Fast, B.; Metzger, S.; Gehling, M.; Ziegelbauer, K.; Rübsamen-Waigmann, H.; Freiberg, C. Novel Whole-Cell Antibiotic Biosensors for Compound Discovery. Appl. Environ. Microbiol. 2007, 73, 6436–6443. [Google Scholar] [CrossRef]
- Butler, M.S.; Buss, A.D. Natural products—The future scaffolds for novel antibiotics? Biochem. Pharmacol. 2006, 71, 919–929. [Google Scholar] [CrossRef]
- Von Nussbaum, F.; Brands, M.; Hinzen, B.; Weigand, S.; Häbich, D. Antibacterial Natural Products in Medicinal Chemistry—Exodus or Revival? Angew. Chem. Int. Ed. 2006, 45, 5072–5129. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef]
- Lam, K.S. New aspects of natural products in drug discovery. Trends Microbiol. 2007, 15, 279–289. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.P.; Brady, L.S.; Insel, T.R.; Collins, F.S. NIH Molecular Libraries Initiative. Science 2004, 306, 1138–1139. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A. A community-based approach to new antibiotic discovery. Nat. Rev. Drug Discov. 2015, 14, 587–588. [Google Scholar] [CrossRef]
- Blaskovich, M.A.T.; Zuegg, J.; Elliott, A.G.; Cooper, M.A. Helping Chemists Discover New Antibiotics. ACS Infect. Dis. 2015, 1, 285–287. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, R.; Sonawane, N.D.; Arumainayagam, D.; Verkman, A.S. Small molecules with antimicrobial activity against E. coli and P. aeruginosa identified by high-throughput screening. Br. J. Pharmacol. 2006, 149, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Malapaka, V.R.; Barrese, I.A.A.; Tripp, B.C. High-Throughput Screening for Antimicrobial Compounds Using a 96-Well Format Bacterial Motility Absorbance Assay. SLAS Discov. Adv. Sci. Drug Discov. 2007, 12, 849–854. [Google Scholar] [CrossRef]
- Wang, N.R. The Identification and Characterization Of Novel Antibacterial Compounds via Target-Based and Whole Cell Screening Approaches; University of Illinois at Urbana-Champaign: Champaign, IL, USA, 2011. [Google Scholar]
- Maddry, J.A.; Ananthan, S.; Goldman, R.C.; Hobrath, J.V.; Kwong, C.D.; Maddox, C.; Rasmussen, L.; Reynolds, R.C.; Secrist, J.A.; Sosa, M.I.; et al. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis 2009, 89, 354–363. [Google Scholar] [CrossRef]
- Ananthan, S.; Faaleolea, E.R.; Goldman, R.C.; Hobrath, J.V.; Kwong, C.D.; Laughon, B.E.; Maddry, J.A.; Mehta, A.; Rasmussen, L.; Reynolds, R.C.; et al. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis 2009, 89, 334–353. [Google Scholar] [CrossRef]
- Kotapalli, S.S.; Nallam, S.S.A.; Nadella, L.; Banerjee, T.; Rode, H.B.; Mainkar, P.S.; Ummanni, R. Identification of New Molecular Entities (NMEs) as Potential Leads against Tuberculosis from Open Source Compound Repository. PLoS ONE 2015, 10, e0144018. [Google Scholar] [CrossRef]
- Morgan, H.P.; Walsh, M.J.; Blackburn, E.A.; Wear, M.A.; Boxer, M.B.; Shen, M.; Veith, H.; McNae, I.W.; Nowicki, M.W.; Michels, P.A.M.; et al. A new family of covalent inhibitors block nucleotide binding to the active site of pyruvate kinase. Biochem. J. 2012, 448, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.-J.R.; Sung, M.-T.; Liao, H.-Y.; Chang, Y.-F.; Chen, C.-W.; Huang, C.-Y.; Chou, L.-Y.; Wu, Y.-D.; Chen, Y.-H.; Cheng, Y.-S.E.; et al. Domain requirement of moenomycin binding to bifunctional transglycosylases and development of high-throughput discovery of antibiotics. Proc. Natl. Acad. Sci. USA 2008, 105, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, L.; Goldberg, A.B.; Chen, C.; Turk, B.E. Identification of Exosite-Targeting Inhibitors of Anthrax Lethal Factor by High-Throughput Screening. Chem. Biol. 2012, 19, 875–882. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Duncan, M.C.; Wong, W.R.; Dupzyk, A.J.; Bray, W.M.; Linington, R.G.; Auerbuch, V. An NF-κB-Based High-Throughput Screen Identifies Piericidins as Inhibitors of the Yersinia pseudotuberculosis Type III Secretion System. Antimicrob. Agents Chemother. 2014, 58, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Schepetkin, I.A.; Khlebnikov, A.I.; Kirpotina, L.N.; Quinn, M.T. Novel Small-Molecule Inhibitors of Anthrax Lethal Factor Identified by High-Throughput Screening. J. Med. Chem. 2006, 49, 5232–5244. [Google Scholar] [CrossRef]
- Brown, A.S.; Owen, J.G.; Jung, J.; Baker, E.N.; Ackerley, D.F. Inhibition of Indigoidine Synthesis as a High-Throughput Colourimetric Screen for Antibiotics Targeting the Essential Mycobacterium tuberculosis Phosphopantetheinyl Transferase PptT. Pharmaceutics 2021, 13, 1066. [Google Scholar] [CrossRef]
- Brott, A.S.; Jones, C.S.; Clarke, A.J. Development of a High Throughput Screen for the Identification of Inhibitors of Peptidoglycan O-Acetyltransferases, New Potential Antibacterial Targets. Antibiotics 2019, 8, 65. [Google Scholar] [CrossRef]
- Lefas, G.; Chaconas, G. High-Throughput Screening Identifies Three Inhibitor Classes of the Telomere Resolvase from the Lyme Disease Spirochete. Antimicrob. Agents Chemother. 2009, 53, 4441–4449. [Google Scholar] [CrossRef]
- Spicer, T.; Fernandez-Vega, V.; Chase, P.; Scampavia, L.; To, J.; Dalton, J.P.; Da Silva, F.L.; Skinner-Adams, T.S.; Gardiner, D.L.; Trenholme, K.R.; et al. Identification of Potent and Selective Inhibitors of the Plasmodium falciparum M18 Aspartyl Aminopeptidase (PfM18AAP) of Human Malaria via High-Throughput Screening. SLAS Discov. Adv. Sci. Drug Discov. 2014, 19, 1107–1115. [Google Scholar] [CrossRef][Green Version]
- Nayar, A.S.; Dougherty, T.J.; Ferguson, K.E.; Granger, B.A.; McWilliams, L.; Stacey, C.; Leach, L.J.; Narita, S.-I.; Tokuda, H.; Miller, A.A.; et al. Novel Antibacterial Targets and Compounds Revealed by a High-Throughput Cell Wall Reporter Assay. J. Bacteriol. 2015, 197, 1726–1734. [Google Scholar] [CrossRef]
- Olson, M.E.; Abate-Pella, D.; Perkins, A.L.; Li, M.; Carpenter, M.A.; Rathore, A.; Harris, R.S.; Harki, D.A. Oxidative Reactivities of 2-Furylquinolines: Ubiquitous Scaffolds in Common High-Throughput Screening Libraries. J. Med. Chem. 2015, 58, 7419–7430. [Google Scholar] [CrossRef] [PubMed]
- Bageshwar, U.K.; VerPlank, L.; Baker, D.; Dong, W.; Hamsanathan, S.; Whitaker, N.; Sacchettini, J.C.; Musser, S.M. High Throughput Screen for Escherichia coli Twin Arginine Translocation (Tat) Inhibitors. PLoS ONE 2016, 11, e0149659. [Google Scholar] [CrossRef]
- Sergeev, G.; Roy, S.; Jarek, M.; Zapolskii, V.; E Kaufmann, D.; Nandy, R.K.; Tegge, W. High-throughput screening and whole genome sequencing identifies an antimicrobially active inhibitor of Vibrio cholerae. BMC Microbiol. 2014, 14, 49. [Google Scholar] [CrossRef]
- Rani, C.; Mehra, R.; Sharma, R.; Chib, R.; Wazir, P.; Nargotra, A.; Khan, I.A. High-throughput screen identifies small molecule inhibitors targeting acetyltransferase activity of Mycobacterium tuberculosis GlmU. Tuberculosis 2015, 95, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.P.; Blanchard, J.E.; Murphy, C.; Roderick, S.L.; Brown, E.D. High-Throughput Screening Identifies Novel Inhibitors of the Acetyltransferase Activity of Escherichia coli GlmU. Antimicrob. Agents Chemother. 2009, 53, 2306–2311. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.A.; Grant, S.S.; Kawate, T.; Iwase, N.; Shimizu, M.; Wivagg, C.; Silvis, M.; Kazyanskaya, E.; Aquadro, J.; Golas, A.; et al. Identification of Novel Inhibitors of M. tuberculosis Growth Using Whole Cell Based High-Throughput Screening. ACS Chem. Biol. 2012, 7, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Christophe, T.; Jackson, M.; Jeon, H.K.; Fenistein, D.; Contreras-Dominguez, M.; Kim, J.; Genovesio, A.; Carralot, J.-P.; Ewann, F.; Kim, E.H.; et al. High Content Screening Identifies Decaprenyl-Phosphoribose 2′ Epimerase as a Target for Intracellular Antimycobacterial Inhibitors. PLOS Pathog. 2009, 5, e1000645. [Google Scholar] [CrossRef]
- Ivanenkov, Y.A.; Yamidanov, R.S.; Osterman, I.; Sergiev, P.V.; Aladinskiy, V.A.; Aladinskaya, A.V.; Terentiev, V.A.; Veselov, M.S.; Ayginin, A.A.; Skvortsov, D.A.; et al. 2-Pyrazol-1-yl-thiazole derivatives as novel highly potent antibacterials. J. Antibiot. 2019, 72, 827–833. [Google Scholar] [CrossRef]
- Reithuber, E.; Wixe, T.; Ludwig, K.C.; Müller, A.; Uvell, H.; Grein, F.; Lindgren, A.E.G.; Muschiol, S.; Nannapaneni, P.; Eriksson, A.; et al. THCz: Small molecules with antimicrobial activity that block cell wall lipid intermediates. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Bolger, G.; Roy, S.; Zapolskii, V.; Kaufmann, D.E.; Schnürch, M.; Mihovilovic, M.D.; Nandy, R.K.; Tegge, W. Targeting aphA: A new high-throughput screening assay identifies compounds that reduce prime virulence factors of Vibrio cholerae. J. Med Microbiol. 2016, 65, 678–687. [Google Scholar] [CrossRef]
- Lacriola, C.J.; Falk, S.P.; Weisblum, B. Screen for Agents That Induce Autolysis in Bacillus subtilis. Antimicrob. Agents Chemother. 2013, 57, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Langsdorf, E.F.; Malikzay, A.; Lamarr, W.A.; Daubaras, D.; Kravec, C.; Zhang, R.; Hart, R.; Monsma, F.; Black, T.; Ozbal, C.C.; et al. Screening for Antibacterial Inhibitors of the UDP-3-O-(R-3-Hydroxymyristoyl)-N-Acetylglucosamine Deacetylase (LpxC) Using a High-Throughput Mass Spectrometry Assay. SLAS Discov. Adv. Sci. Drug Discov. 2009, 15, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Rajamuthiah, R.; Fuchs, B.B.; Jayamani, E.; Kim, Y.; Larkins-Ford, J.; Conery, A.; Ausubel, F.M.; Mylonakis, E. Whole Animal Automated Platform for Drug Discovery against Multi-Drug Resistant Staphylococcus aureus. PLOS ONE 2014, 9, e89189. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef]
- Gatta, V.; Ilina, P.; Porter, A.; McElroy, S.; Tammela, P. Targeting Quorum Sensing: High-Throughput Screening to Identify Novel LsrK Inhibitors. Int. J. Mol. Sci. 2019, 20, 3112. [Google Scholar] [CrossRef] [PubMed]
- Gray, B.; Hall, P.; Gresham, H. Targeting agr- and agr-Like Quorum Sensing Systems for Development of Common Therapeutics to Treat Multiple Gram-Positive Bacterial Infections. Sensors 2013, 13, 5130–5166. [Google Scholar] [CrossRef]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective Chemical Inhibition of agr Quorum Sensing in Staphylococcus aureus Promotes Host Defense with Minimal Impact on Resistance. PLOS Pathog. 2014, 10, e1004174. [Google Scholar] [CrossRef]
- Ishii, S.; Fukui, K.; Yokoshima, S.; Kumagai, K.; Beniyama, Y.; Kodama, T.; Fukuyama, T.; Okabe, T.; Nagano, T.; Kojima, H.; et al. High-throughput Screening of Small Molecule Inhibitors of the Streptococcus Quorum-sensing Signal Pathway. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Torres, N.S.; Abercrombie, J.J.; Srinivasan, A.; Lopez-Ribot, J.L.; Ramasubramanian, A.K.; Leung, K.P. Screening a Commercial Library of Pharmacologically Active Small Molecules against Staphylococcus aureus Biofilms. Antimicrob. Agents Chemother. 2016, 60, 5663–5672. [Google Scholar] [CrossRef]
- Gilbert-Girard, S.; Savijoki, K.; Yli-Kauhaluoma, J.; Fallarero, A. Optimization of a High-Throughput 384-Well Plate-Based Screening Platform with Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 15442 Biofilms. Int. J. Mol. Sci. 2020, 21, 3034. [Google Scholar] [CrossRef]
- Gilbert-Girard, S.; Savijoki, K.; Yli-Kauhaluoma, J.; Fallarero, A. Screening of FDA-Approved Drugs Using a 384-Well Plate-Based Biofilm Platform: The Case of Fingolimod. Microorganisms 2020, 8, 1834. [Google Scholar] [CrossRef]
- Weber, B.S.; De Jong, A.M.; Guo, A.B.; Dharavath, S.; French, S.; Fiebig-Comyn, A.A.; Coombes, B.K.; Magolan, J.; Brown, E.D. Genetic and Chemical Screening in Human Blood Serum Reveals Unique Antibacterial Targets and Compounds against Klebsiella pneumoniae. Cell Rep. 2020, 32. [Google Scholar] [CrossRef]
- Colquhoun, J.M.; Wozniak, R.A.F.; Dunman, P.M. Clinically Relevant Growth Conditions Alter Acinetobacter baumannii Antibiotic Susceptibility and Promote Identification of Novel Antibacterial Agents. PLoS ONE 2015, 10, e0143033. [Google Scholar] [CrossRef]
- Sarigul, N.; Korkmaz, F.; Kurultak, I. A New Artificial Urine Protocol to Better Imitate Human Urine. Sci. Rep. 2019, 9, 20159. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Nemeth, E. Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 2015, 15, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Umland, T.C.; Schultz, L.W.; MacDonald, U.; Beanan, J.M.; Olson, R.; Russo, T.A. In Vivo -Validated Essential Genes Identified in Acinetobacter baumannii by Using Human Ascites Overlap Poorly with Essential Genes Detected on Laboratory Media. mBio 2012, 3. [Google Scholar] [CrossRef]
- Zlitni, S.; Ferruccio, L.F.; Brown, E.D. Metabolic suppression identifies new antibacterial inhibitors under nutrient limitation. Nat. Chem. Biol. 2013, 9, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Tsai, C.N.; Johnson, J.W.; French, S.; Elhenawy, W.; Porwollik, S.; Andrews-Polymenis, H.; McClelland, M.; Magolan, J.; Coombes, B.K.; et al. A macrophage-based screen identifies antibacterial compounds selective for intracellular Salmonella Typhimurium. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kleymann, G.; Werling, H.-O. A Generally Applicable, High-Throughput Screening–Compatible Assay to Identify, Evaluate, and Optimize Antimicrobial Agents for Drug Therapy. SLAS Discov. Adv. Sci. Drug Discov. 2004, 9, 578–587. [Google Scholar] [CrossRef]
- Fahnoe, K.C.; Flanagan, M.E.; Gibson, G.; Shanmugasundaram, V.; Che, Y.; Tomaras, A.P. Non-Traditional Antibacterial Screening Approaches for the Identification of Novel Inhibitors of the Glyoxylate Shunt in Gram-Negative Pathogens. PLOS ONE 2012, 7, e51732. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Sun, W.; Xu, M.; Shen, M.; Khraiwesh, M.; Sciotti, R.; Zheng, W. Repurposing Screen Identifies Unconventional Drugs With Activity Against Multidrug Resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 2019, 8, 438. [Google Scholar] [CrossRef]
- Miller, J.R.; Dunham, S.; Mochalkin, I.; Banotai, C.; Bowman, M.; Buist, S.; Dunkle, B.; Hanna, D.; Harwood, H.J.; Huband, M.D.; et al. A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc. Natl. Acad. Sci. USA 2009, 106, 1737–1742. [Google Scholar] [CrossRef] [PubMed]
- Ayon, N.J.; Gutheil, W.G. Dimensionally Enhanced Antibacterial Library Screening. ACS Chem. Biol. 2019, 14, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Jubaer, N. Qualification of Human Liver Microsomes for Antibacterial Activity Screening of Drug Metabolites. Appl. Microbiol. 2023, 3, 104–118. [Google Scholar] [CrossRef]
- Ayon, N.J. Metabolomics and Chemical Library Screening for Antibacterial Drug Discovery. Ph.D. Thesis, Pharmaceutical Sciences (UMKC), Kansas City, MO, USA, 2020. [Google Scholar]
- McLeod, J.R.; Harvey, P.A.; Detweiler, C.S. An Oral Fluorouracil Prodrug, Capecitabine, Mitigates a Gram-Positive Systemic Infection in Mice. Microbiol. Spectr. 2021, 9, e0027521. [Google Scholar] [CrossRef]
- Zulauf, K.E.; Kirby, J.E. Discovery of small-molecule inhibitors of multidrug-resistance plasmid maintenance using a high-throughput screening approach. Proc. Natl. Acad. Sci. USA 2020, 117, 29839–29850. [Google Scholar] [CrossRef]
- Mattingly, A.E.; Cox, K.E.; Smith, R.; Melander, R.J.; Ernst, R.K.; Melander, C. Screening an Established Natural Product Library Identifies Secondary Metabolites That Potentiate Conventional Antibiotics. ACS Infect. Dis. 2020, 6, 2629–2640. [Google Scholar] [CrossRef]
- Farha, M.A.; Brown, E.D. Chemical Probes of Escherichia coli Uncovered through Chemical-Chemical Interaction Profiling with Compounds of Known Biological Activity. Chem. Biol. 2010, 17, 852–862. [Google Scholar] [CrossRef]
- Ayon, N.J.; Gutheil, W.G. Correction to “Dimensionally Enhanced Antibacterial Library Screening”. ACS Chem. Biol. 2021, 16, 1610–1611. [Google Scholar] [CrossRef]
- Sharma, A.D.; Gutheil, W.G. Synergistic Combinations of FDA-Approved Drugs with Ceftobiprole against Methicillin-Resistant Staphylococcus aureus. Microbiol. Spectr. 2022. [Google Scholar] [CrossRef]
- Farha, M.A.; Leung, A.; Sewell, E.W.; D’elia, M.A.; Allison, S.E.; Ejim, L.; Pereira, P.M.; Pinho, M.G.; Wright, G.D.; Brown, E.D. Inhibition of WTA Synthesis Blocks the Cooperative Action of PBPs and Sensitizes MRSA to β-Lactams. ACS Chem. Biol. 2012, 8, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, P.R.; Pesesky, M.W.; Bouley, R.; Ballard, A.; Biddy, B.A.; Suckow, M.; Wolter, W.R.; Schroeder, V.A.; Burnham, C.-A.D.; Mobashery, S.; et al. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 2015, 11, 855–861. [Google Scholar] [CrossRef]
- Saini, M.; Gaurav, A.; Kothari, A.; Omar, B.J.; Gupta, V.; Bhattacharjee, A.; Pathania, R. Small Molecule IITR00693 (2-Aminoperimidine) Synergizes Polymyxin B Activity against Staphylococcus aureus and Pseudomonas aeruginosa. ACS Infect. Dis. 2023. [Google Scholar] [CrossRef]
- Yam, J.K.H.; Tan, L.Z.W.; Hong, Z.; Salido, M.M.S.; Woo, B.Y.; Yong, A.M.H.; Tan, C.A.Z.; Li, S.F.Y.; Yang, L.; Givskov, M.; et al. Auranofin inhibits virulence pathways in Pseudomonas aeruginosa. Bioorganic Med. Chem. 2023, 79. [Google Scholar] [CrossRef]
- Fleeman, R.M.; Debevec, G.; Antonen, K.; Adams, J.L.; Santos, R.G.; Welmaker, G.S.; Houghten, R.A.; Giulianotti, M.A.; Shaw, L.N. Identification of a Novel Polyamine Scaffold with Potent Efflux Pump Inhibition Activity Toward Multi-Drug Resistant Bacterial Pathogens. Front. Microbiol. 2018, 9, 1301. [Google Scholar] [CrossRef]
- Mishra, A.; Dobritsa, S.V.; Crouch, M.-L.; Rabenstein, J.; Lee, J.X.Y.; Dhakshinamoorthy, S. Establishment and validation of a 384-well antibacterial assay amenable for high-throughput screening and combination testing. J. Microbiol. Methods 2015, 118, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Peyclit, L.; Baron, S.A.; Hadjadj, L.; Rolain, J.-M. In Vitro Screening of a 1280 FDA-Approved Drugs Library against Multidrug-Resistant and Extensively Drug-Resistant Bacteria. Antibiotics 2022, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Ramón-García, S.; Ng, C.; Anderson, H.; Chao, J.D.; Zheng, X.; Pfeifer, T.; Av-Gay, Y.; Roberge, M.; Thompson, C.J. Synergistic Drug Combinations for Tuberculosis Therapy Identified by a Novel High-Throughput Screen. Antimicrob. Agents Chemother. 2011, 55, 3861–3869. [Google Scholar] [CrossRef]
- Chiaraviglio, L.; Kirby, J.E. High-Throughput Intracellular Antimicrobial Susceptibility Testing of Legionella pneumophila. Antimicrob. Agents Chemother. 2015, 59, 7517–7529. [Google Scholar] [CrossRef]
- Smith, K.P.; Kirby, J.E. Validation of a High-Throughput Screening Assay for Identification of Adjunctive and Directly Acting Antimicrobials Targeting Carbapenem-Resistant Enterobacteriaceae. ASSAY Drug Dev. Technol. 2016, 14, 194–206. [Google Scholar] [CrossRef]
- Wambaugh, M.A.; Shakya, V.P.S.; Lewis, A.J.; Mulvey, M.A.; Brown, J.C.S. High-throughput identification and rational design of synergistic small-molecule pairs for combating and bypassing antibiotic resistance. PLoS Biol. 2017, 15, e2001644. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.L.; Rossi, L.; De Pascale, G.; Wright, G.D. A Forward Chemical Screen Identifies Antibiotic Adjuvants in Escherichia coli. ACS Chem. Biol. 2012, 7, 1547–1555. [Google Scholar] [CrossRef]
- King, A.M.; Reid-Yu, S.A.; Wang, W.; King, D.T.; De Pascale, G.; Strynadka, N.C.; Walsh, T.R.; Coombes, B.K.; Wright, G.D. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; She, P.; Zhou, L.; Zeng, X.; Xu, L.; Liu, Y.; Chen, L.; Wu, Y. High-Throughput Identification of Antibacterials Against Pseudomonas aeruginosa. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Chen, C.H.; Ulmschneider, J.P.; Ulmschneider, M.B. Mechanisms of a Small Membrane-Active Antimicrobial Peptide from Hyla punctata. Aust. J. Chem. 2020, 73, 236. [Google Scholar] [CrossRef]
- Dings, R.P.M.; Mayo, K.H. A Journey in Structure-Based Drug Discovery: From Designed Peptides to Protein Surface Topomimetics as Antibiotic and Antiangiogenic Agents. Accounts Chem. Res. 2007, 40, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Judzewitsch, P.R.; Corrigan, N.; Trujillo, F.; Xu, J.; Moad, G.; Hawker, C.J.; Wong, E.H.H.; Boyer, C. High-Throughput Process for the Discovery of Antimicrobial Polymers and Their Upscaled Production via Flow Polymerization. Macromolecules 2020, 53, 631–639. [Google Scholar] [CrossRef]
- Raventos, D.; Taboureau, O.; Mygind, P.H.; Nielsen, J.D.; Sonksen, C.P.; Kristensen, H.-H. Improving on Natures Defenses: Optimization & High Throughput Screening of Antimicrobial Peptides. Comb. Chem. High Throughput Screen. 2005, 8, 219–233. [Google Scholar] [CrossRef]
- Chen, C.H.; Starr, C.G.; Guha, S.; Wimley, W.C.; Ulmschneider, M.B.; Ulmschneider, J.P. Tuning of a Membrane-Perforating Antimicrobial Peptide to Selectively Target Membranes of Different Lipid Composition. J. Membr. Biol. 2021, 254, 75–96. [Google Scholar] [CrossRef]
- Xie, Q.; Matsunaga, S.; Wen, Z.; Niimi, S.; Kumano, M.; Sakakibara, Y.; Machida, S. In vitro system for high-throughput screening of random peptide libraries for antimicrobial peptides that recognize bacterial membranes. J. Pept. Sci. 2006, 12, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Watkins, D.; Jin, Y.; Gong, C.; King, A.; Washington, A.Z.; Green, K.D.; Garneau-Tsodikova, S.; Oyelere, A.K.; Arya, D.P. Rapid Synthesis, RNA Binding, and Antibacterial Screening of a Peptidic-Aminosugar (PA) Library. ACS Chem. Biol. 2015, 10, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, K.; Volkmer-Engert, R.; Walter, T.; Hancock, R. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005, 23, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Dalecki, A.G.; Zorn, K.M.; Clark, A.M.; Ekins, S.; Narmore, W.T.; Tower, N.; Rasmussen, L.; Bostwick, R.; Kutsch, O.; Wolschendorf, F. High-throughput screening and Bayesian machine learning for copper-dependent inhibitors of Staphylococcus aureus. Metallomics 2019, 11, 696–706. [Google Scholar] [CrossRef]
- Dalecki, A.G.; Malalasekera, A.P.; Schaaf, K.; Kutsch, O.; Bossmann, S.H.; Wolschendorf, F. Combinatorial phenotypic screen uncovers unrecognized family of extended thiourea inhibitors with copper-dependent anti-staphylococcal activity. Metallomics 2016, 8, 412–421. [Google Scholar] [CrossRef]
- Tucker, A.T.; Leonard, S.P.; DuBois, C.D.; Knauf, G.A.; Cunningham, A.L.; Wilke, C.O.; Trent, M.S.; Davies, B.W. Discovery of Next-Generation Antimicrobials through Bacterial Self-Screening of Surface-Displayed Peptide Libraries. Cell 2018, 172, 618–628.e13. [Google Scholar] [CrossRef]
- Foster, A.D.; Ingram, J.D.; Leitch, E.K.; Lennard, K.R.; Osher, E.L.; Tavassoli, A. Methods for the Creation of Cyclic Peptide Libraries for Use in Lead Discovery. SLAS Discov. Adv. Sci. Drug Discov. 2015, 20, 563–576. [Google Scholar] [CrossRef]
- A Desimmie, B.; Humbert, M.; Lescrinier, E.; Hendrix, J.; Vets, S.; Gijsbers, R.; Ruprecht, R.M.; Dietrich, U.; Debyser, Z.; Christ, F. Phage Display-directed Discovery of LEDGF/p75 Binding Cyclic Peptide Inhibitors of HIV Replication. Mol. Ther. 2012, 20, 2064–2075. [Google Scholar] [CrossRef]
- Horswill, A.R.; Savinov, S.N.; Benkovic, S.J. A systematic method for identifying small-molecule modulators of protein–protein interactions. Proc. Natl. Acad. Sci. USA 2004, 101, 15591–15596. [Google Scholar] [CrossRef]
- Ma, Z.; Hartman, M.C.T. In Vitro Selection of Unnatural Cyclic Peptide Libraries via mRNA Display. Methods Mol Biol. 2011, 805, 367–390. [Google Scholar] [CrossRef]
- Lee, O.W.; Austin, S.; Gamma, M.; Cheff, D.M.; Lee, T.D.; Wilson, K.M.; Johnson, J.; Travers, J.; Braisted, J.C.; Guha, R.; et al. Cytotoxic Profiling of Annotated and Diverse Chemical Libraries Using Quantitative High-Throughput Screening. SLAS Discov. Adv. Sci. Drug Discov. 2019, 25, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, T. A Practical Guide to Assay Development and High-Throughput Screening in Drug Discovery; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar] [CrossRef]
- Reymond, J.-L.; van Deursen, R.; Blum, L.C.; Ruddigkeit, L. Chemical space as a source for new drugs. Med. Chem. Comm. 2010, 1, 30–38. [Google Scholar] [CrossRef]
- Paricharak, S.; Méndez-Lucio, O.; Ravindranath, A.C.; Bender, A.; Ijzerman, A.P.; Van Westen, G.J.P. Data-driven approaches used for compound library design, hit triage and bioactivity modeling in high-throughput screening. Briefings Bioinform. 2016, 19, 277–285. [Google Scholar] [CrossRef]
- Renner, S.; Popov, M.; Schuffenhauer, A.; Roth, H.-J.; Breitenstein, W.; Marzinzik, A.; Lewis, I.; Krastel, P.; Nigsch, F.; Jenkins, J.; et al. Recent trends and observations in the design of high-quality screening collections. Futur. Med. Chem. 2011, 3, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- O’shea, R.; Moser, H.E. Physicochemical Properties of Antibacterial Compounds: Implications for Drug Discovery. J. Med. Chem. 2008, 51, 2871–2878. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Netherton, M.; Byrd, T.F.; Rohde, K.H. Reporter-Based Assays for High-Throughput Drug Screening against Mycobacterium abscessus. Front. Microbiol. 2017, 8, 2204. [Google Scholar] [CrossRef]
- Peláez, F. The historical delivery of antibiotics from microbial natural products—Can history repeat? Biochem. Pharmacol. 2006, 71, 981–990. [Google Scholar] [CrossRef]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Central Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Inglese, J.; Johnson, R.L.; Simeonov, A.; Xia, M.; Zheng, W.; Austin, C.P.; Auld, D.S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007, 3, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Macarrón, R.; Hertzberg, R.P. Design and Implementation of High-Throughput Screening Assays. Methods Mol Biol. 2009, 565, 1–32. [Google Scholar] [CrossRef]
- Moir, D.T.; Di, M.; Opperman, T.; Schweizer, H.P.; Bowlin, T.L. A High-Throughput, Homogeneous, Bioluminescent Assay for Pseudomonas aeruginosa Gyrase Inhibitors and Other DNA-Damaging Agents. SLAS Discov. Adv. Sci. Drug Discov. 2007, 12, 855–864. [Google Scholar] [CrossRef]
- Radnai, L.; Stremel, R.F.; Vaissiere, T.; Lin, L.; Cameron, M.; Martin, W.H.; Rumbaugh, G.; Kamenecka, T.M.; Griffin, P.R.; Miller, C. A simple and robust cell-based assay for the discovery of novel cytokinesis inhibitors. J. Biol. Methods 2020, 7, e136. [Google Scholar] [CrossRef]
- Szymański, P.; Markowicz, M.; Mikiciuk-Olasik, E. Adaptation of High-Throughput Screening in Drug Discovery—Toxicological Screening Tests. Int. J. Mol. Sci. 2011, 13, 427–452. [Google Scholar] [CrossRef] [PubMed]
- Mayr, L.M.; Fuerst, P. The Future of High-Throughput Screening. SLAS Discov. Adv. Sci. Drug Discov. 2008, 13, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Michael, S.; Auld, D.; Klumpp, C.; Jadhav, A.; Zheng, W.; Thorne, N.; Austin, C.P.; Inglese, J.; Simeonov, A. A Robotic Platform for Quantitative High-Throughput Screening. ASSAY Drug Dev. Technol. 2008, 6, 637–657. [Google Scholar] [CrossRef]
- Feng, B.Y.; Shoichet, B.K. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 2006, 1, 550–553. [Google Scholar] [CrossRef]
- Johnston, P.A.; Soares, K.M.; Shinde, S.N.; Foster, C.A.; Shun, T.Y.; Takyi, H.K.; Wipf, P.; Lazo, J.S. Development of a 384-Well Colorimetric Assay to Quantify Hydrogen Peroxide Generated by the Redox Cycling of Compounds in the Presence of Reducing Agents. ASSAY Drug Dev. Technol. 2008, 6, 505–518. [Google Scholar] [CrossRef]
- Rishton, G.M. Natural Products as a Robust Source of New Drugs and Drug Leads: Past Successes and Present Day Issues. Am. J. Cardiol. 2008, 101, S43–S49. [Google Scholar] [CrossRef]
- Dahlin, J.L.; Nissink, J.W.M.; Strasser, J.M.; Francis, S.; Higgins, L.; Zhou, H.; Zhang, Z.; Walters, M.A. PAINS in the Assay: Chemical Mechanisms of Assay Interference and Promiscuous Enzymatic Inhibition Observed during a Sulfhydryl-Scavenging HTS. J. Med. Chem. 2015, 58, 2091–2113. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.D.; Wilson, B.A.P.; Henrich, C.J.; Staudt, L.M.; Krumpe, L.R.H.; Smith, E.A.; King, J.; Wendt, K.L.; Stchigel, A.M.; Miller, A.N.; et al. Secondary Metabolites from the Fungus Dictyosporium sp. and Their MALT1 Inhibitory Activities. J. Nat. Prod. 2019, 82, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Fiala, J.; Schöbel, H.; Vrabl, P.; Dietrich, D.; Hammerle, F.; Artmann, D.J.; Stärz, R.; Peintner, U.; Siewert, B. A New High-Throughput-Screening-Assay for Photoantimicrobials Based on EUCAST Revealed Unknown Photoantimicrobials in Cortinariaceae. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Ollinger, J.; Kumar, A.; Roberts, D.M.; Bailey, M.A.; Casey, A.; Parish, T. A high-throughput whole cell screen to identify inhibitors of Mycobacterium tuberculosis. PLoS ONE 2019, 14, e0205479. [Google Scholar] [CrossRef]
- Simeonov, A.; Jadhav, A.; Thomas, C.J.; Wang, Y.; Huang, R.; Southall, N.; Shinn, P.; Smith, J.; Austin, C.P.; Auld, D.S.; et al. Fluorescence Spectroscopic Profiling of Compound Libraries. J. Med. Chem. 2008, 51, 2363–2371. [Google Scholar] [CrossRef]
- Auld, D.S.; Southall, N.T.; Jadhav, A.; Johnson, R.L.; Diller, D.J.; Simeonov, A.; Austin, C.P.; Inglese, J. Characterization of Chemical Libraries for Luciferase Inhibitory Activity. J. Med. Chem. 2008, 51, 2372–2386. [Google Scholar] [CrossRef]
- Simeonov, A.; Yasgar, A.; Jadhav, A.; Lokesh, G.; Klumpp, C.; Michael, S.; Austin, C.P.; Natarajan, A.; Inglese, J. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT–phosphoprotein interaction. Anal. Biochem. 2008, 375, 60–70. [Google Scholar] [CrossRef][Green Version]
- Fen, W.; Sicen, W.; Xilan, G. A review for cell-based screening methods in drug discovery. Biophys. Rep. 2021, 7, 504–516. [Google Scholar] [CrossRef]
- Kessel, S.; Cribbes, S.; Déry, O.; Kuksin, D.; Sincoff, E.; Qiu, J.; Chan, L.L.-Y. High-Throughput 3D Tumor Spheroid Screening Method for Cancer Drug Discovery Using Celigo Image Cytometry. SLAS Technol. Transl. Life Sci. Innov. 2016, 22, 454–465. [Google Scholar] [CrossRef]
- Foster, N.C.; Hall, M.N.M.; El Haj, A.J. Two-Dimensional and Three-Dimensional Cartilage Model Platforms for Drug Evaluation and High-Throughput Screening Assays. Tissue Eng. Part B Rev. 2022, 28, 421–436. [Google Scholar] [CrossRef]
- Graham, A.D.; Pandey, R.; Tsancheva, V.S.; Candeo, A.; Botchway, S.W.; Allan, A.J.; Teboul, L.; Madi, K.; Babra, T.S.; Zolkiewski, L.A.K.; et al. The development of a high throughput drug-responsive model of white adipose tissue comprising adipogenic 3T3-L1 cells in a 3D matrix. Biofabrication 2019, 12, 015018. [Google Scholar] [CrossRef] [PubMed]
- Kozikowski, B.A.; Burt, T.M.; Tirey, D.A.; Williams, L.E.; Kuzmak, B.R.; Stanton, D.T.; Morand, K.L.; Nelson, S.L. The Effect of Room-Temperature Storage on the Stability of Compounds in DMSO. SLAS Discov. Adv. Sci. Drug Discov. 2003, 8, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.; Miller, C.H.; Bellows, D.S.; O’toole, R. Evaluation of the Mycobacterium smegmatis and BCG models for the discovery of Mycobacterium tuberculosis inhibitors. Tuberculosis 2010, 90, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Bassett, I.M.; Lun, S.; Bishai, W.R.; Guo, H.; Kirman, J.R.; Altaf, M.; O’toole, R.F. Detection of inhibitors of phenotypically drug-tolerant Mycobacterium tuberculosis using an in vitro bactericidal screen. J. Microbiol. 2013, 51, 651–658. [Google Scholar] [CrossRef]
- Zhang, Y.; Wade, M.M.; Scorpio, A.; Zhang, H.; Sun, Z. Mode of action of pyrazinamide: Disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother. 2003, 52, 790–795. [Google Scholar] [CrossRef]
- Arhin, F.; Bélanger, O.; Ciblat, S.; Dehbi, M.; Delorme, D.; Dietrich, E.; Dixit, D.; Lafontaine, Y.; Lehoux, D.; Liu, J.; et al. A new class of small molecule RNA polymerase inhibitors with activity against Rifampicin-resistant Staphylococcus aureus. Bioorganic Med. Chem. 2006, 14, 5812–5832. [Google Scholar] [CrossRef]
- Choi, K.-J.; Yu, Y.G.; Hahn, H.G.; Choi, J.-D.; Yoon, M.-Y. Characterization of acetohydroxyacid synthase from Mycobacterium tuberculosis and the identification of its new inhibitor from the screening of a chemical library. FEBS Lett. 2005, 579, 4903–4910. [Google Scholar] [CrossRef]
- Han, C.; Wang, L.; Yu, K.; Chen, L.; Hu, L.; Chen, K.; Jiang, H.; Shen, X. Biochemical characterization and inhibitor discovery of shikimate dehydrogenase from Helicobacter pylori. FEBS J. 2006, 273, 4682–4692. [Google Scholar] [CrossRef]
- Miller, C.H.; Nisa, S.; Dempsey, S.; Jack, C.; O’Toole, R. Modifying Culture Conditions in Chemical Library Screening Identifies Alternative Inhibitors of Mycobacteria. Antimicrob. Agents Chemother. 2009, 53, 5279–5283. [Google Scholar] [CrossRef][Green Version]
- Ersoy, S.C.; Heithoff, D.M.; Barnes, L.; Tripp, G.K.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. Ebiomedicine 2017, 20, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Gordhan, B.G.; Peters, J.; Kana, B.D. Chapter Three—Application of model systems to study adaptive responses of Mycobacterium tuberculosis during infection and disease. Adv. Appl. Microbiol. 2019, 108, 115–161. [Google Scholar] [PubMed]
- Proctor, R.A.; Von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Heitmann, V.; Hussain, M.; Viemann, D.; Roth, J.; von Eiff, C.; Peters, G.; Becker, K.; Löffler, B. Staphylococcus aureus Small-Colony Variants Are Adapted Phenotypes for Intracellular Persistence. J. Infect. Dis. 2010, 202, 1031–1040. [Google Scholar] [CrossRef]
- Baumert, N.; von Eiff, C.; Schaaff, F.; Peters, G.; Proctor, R.A.; Sahl, H.-G. Physiology and Antibiotic Susceptibility of Staphylococcus aureus Small Colony Variants. Microb. Drug Resist. 2002, 8, 253–260. [Google Scholar] [CrossRef]
- Trombetta, R.P.; Dunman, P.M.; Schwarz, E.M.; Kates, S.L.; Awad, H. A High-Throughput Screening Approach To Repurpose FDA-Approved Drugs for Bactericidal Applications against Staphylococcus aureus Small-Colony Variants. Msphere 2018, 3, e00422-18. [Google Scholar] [CrossRef]
- Blanchard, C.; Brooks, L.; Ebsworth-Mojica, K.; Didione, L.; Wucher, B.; Dewhurst, S.; Krysan, D.; Dunman, P.M.; Wozniak, R.A.F. Zinc Pyrithione Improves the Antibacterial Activity of Silver Sulfadiazine Ointment. Msphere 2016, 1. [Google Scholar] [CrossRef]
- Kumar, S.S.; Ghosh, A.R. Assessment of bacterial viability: A comprehensive review on recent advances and challenges. Microbiology 2019, 165, 593–610. [Google Scholar] [CrossRef]
- Dahlin, J.L.; A Walters, M. The essential roles of chemistry in high-throughput screening triage. Futur. Med. Chem. 2014, 6, 1265–1290. [Google Scholar] [CrossRef] [PubMed]
- Weston, G.S.; Blázquez, J.; Baquero, F.; Shoichet, B.K. Structure-Based Enhancement of Boronic Acid-Based Inhibitors of AmpC β-Lactamase. J. Med. Chem. 1998, 41, 4577–4586. [Google Scholar] [CrossRef]
- Bisson, J.; McAlpine, J.B.; Friesen, J.B.; Chen, S.-N.; Graham, J.; Pauli, G.F. Can Invalid Bioactives Undermine Natural Product-Based Drug Discovery? J. Med. Chem. 2015, 59, 1671–1690. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B. Feeling Nature’s PAINS: Natural Products, Natural Product Drugs, and Pan Assay Interference Compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Goktug, A.N.; Chai, S.C.; Chen, T. Data Analysis Approaches in High Throughput Screening. In Drug Discovery; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Caraus, I.; Alsuwailem, A.A.; Nadon, R.; Makarenkov, V. Detecting and overcoming systematic bias in high-throughput screening technologies: A comprehensive review of practical issues and methodological solutions. Briefings Bioinform. 2015, 16, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Folmer, R.H. Integrating biophysics with HTS-driven drug discovery projects. Drug Discov. Today 2016, 21, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Hackbarth, C.; Ni, Z.J.; Wu, C.; Wang, W.; Jain, R.; He, Y.; Bracken, K.; Weidmann, B.; Patel, D.V.; et al. Peptide Deformylase Inhibitors as Antibacterial Agents: Identification of VRC3375, a Proline-3-Alkylsuccinyl Hydroxamate Derivative, by Using an Integrated Combinatorial and Medicinal Chemistry Approach. Antimicrob. Agents Chemother. 2004, 48, 250–261. [Google Scholar] [CrossRef]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Farha, M.A.; French, S.; Stokes, J.M.; Brown, E.D. Bicarbonate Alters Bacterial Susceptibility to Antibiotics by Targeting the Proton Motive Force. ACS Infect. Dis. 2017, 4, 382–390. [Google Scholar] [CrossRef]
- Heesterbeek, D.A.C.; Martin, N.I.; Velthuizen, A.; Duijst, M.; Ruyken, M.; Wubbolts, R.; Rooijakkers, S.H.M.; Bardoel, B.W. Complement-dependent outer membrane perturbation sensitizes Gram-negative bacteria to Gram-positive specific antibiotics. Sci. Rep. 2019, 9, 3074. [Google Scholar] [CrossRef]
- Moy, T.I.; Ball, A.R.; Anklesaria, Z.; Casadei, G.; Lewis, K.; Ausubel, F.M. Identification of novel antimicrobials using a live-animal infection model. Proc. Natl. Acad. Sci. USA 2006, 103, 10414–10419. [Google Scholar] [CrossRef]
- Hanna, N.; Kicka, S.; Chiriano, G.; Harrison, C.; Sakouhi, H.O.; Trofimov, V.; Kranjc, A.; Nitschke, J.; Pagni, M.; Cosson, P.; et al. Identification of Anti-Mycobacterium and Anti-Legionella Compounds with Potential Distinctive Structural Scaffolds From an HD-PBL Using Phenotypic Screens in Amoebae Host Models. Front. Microbiol. 2020, 11, 266. [Google Scholar] [CrossRef]
- Moy, T.I.; Conery, A.L.; Larkins-Ford, J.; Wu, G.; Mazitschek, R.; Casadei, G.; Lewis, K.; Carpenter, A.E.; Ausubel, F.M. High-Throughput Screen for Novel Antimicrobials using a Whole Animal Infection Model. ACS Chem. Biol. 2009, 4, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.F.; Kicka, S.; Trofimov, V.; Berschl, K.; Ouertatani-Sakouhi, H.; Ackermann, N.; Hedberg, C.; Cosson, P.; Soldati, T.; Hilbi, H. Exploring Anti-Bacterial Compounds against Intracellular Legionella. PLoS ONE 2013, 8, e74813. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.F.; Chiriano, G.; Finsel, I.; Manske, C.; Hoffmann, C.; Steiner, B.; Kranjc, A.; Patthey-Vuadens, O.; Kicka, S.; Trofimov, V.; et al. Amoebae-Based Screening Reveals a Novel Family of Compounds Restricting Intracellular Legionella pneumophila. ACS Infect. Dis. 2015, 1, 327–338. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Cadieux, B.; Colavecchio, A.; Jeukens, J.; Freschi, L.; Emond-Rheault, J.-G.; Kukavica-Ibrulj, I.; Levesque, R.C.; Bekal, S.; Chandler, J.C.; Coleman, S.M.; et al. Prophage induction reduces Shiga toxin producing Escherichia coli (STEC) and Salmonella enterica on tomatoes and spinach: A model study. Food Control. 2018, 89, 250–259. [Google Scholar] [CrossRef]
- Tompkins, E. Development and Application of a High-Throughput Luminescent Prophage Induction Assay for the Identification of Temperate Bacteriophage-Inducing Food-Grade Compounds; McGill University Libraries: Montreal, QC, Canada, 2019. [Google Scholar]
- Firestine, S.M.; Paritala, H.; Mcdonnell, J.E.; Thoden, J.B.; Holden, H.M. Identification of inhibitors of N5-carboxyaminoimidazole ribonucleotide synthetase by high-throughput screening. Bioorganic Med. Chem. 2009, 17, 3317–3323. [Google Scholar] [CrossRef]
- Woods, A.L.; Parker, D.; Glick, M.M.; Peng, Y.; Lenoir, F.; Mulligan, E.; Yu, V.; Piizzi, G.; Lister, T.; Lilly, M.-D.; et al. High-Throughput Screen for Inhibitors of Klebsiella pneumoniae Virulence Using a Tetrahymena pyriformis Co-Culture Surrogate Host Model. ACS Omega 2022, 7, 5401–5414. [Google Scholar] [CrossRef]
- Thaker, M.N.; Wang, W.; Spanogiannopoulos, P.; Waglechner, N.; King, A.M.; Medina, R.; Wright, G. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 2013, 31, 922–927. [Google Scholar] [CrossRef]
- Choudhary, P.; Rao, A. SELECT-GLYCOCIN: A recombinant microbial system for expression and high-throughput screening of glycocins. Glycoconj. J. 2020, 38, 233–250. [Google Scholar] [CrossRef]
- Caesar, L.K.; Montaser, R.; Keller, N.P.; Kelleher, N.L. Metabolomics and genomics in natural products research: Complementary tools for targeting new chemical entities. Nat. Prod. Rep. 2021, 38, 2041–2065. [Google Scholar] [CrossRef]
- Wencewicz, T.A. New antibiotics from Nature’s chemical inventory. Bioorganic Med. Chem. 2016, 24, 6227–6252. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Lai, J.R.; Roche, E.D.; Walsh, C.T.; Liu, D.R. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc. Natl. Acad. Sci. 2007, 104, 11951–11956. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, K.D.; Bok, J.W.; Ye, R.; Miley, G.P.; Verdan, M.H.; Velk, T.; Chen, C.; Yang, K.; Robey, M.T.; Gao, P.; et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol. 2017, 13, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Keasling, J.D. Synthetic Biology for Synthetic Chemistry. ACS Chem. Biol. 2008, 3, 64–76. [Google Scholar] [CrossRef]
- Schorn, M.A.; Verhoeven, S.; Ridder, L.; Huber, F.; Acharya, D.D.; Aksenov, A.A.; Aleti, G.; Moghaddam, J.A.; Aron, A.T.; Aziz, S.; et al. A community resource for paired genomic and metabolomic data mining. Nat. Chem. Biol. 2021, 17, 363–368. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.-S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10, 963–968. [Google Scholar] [CrossRef]
- Evans, B.S.; Zhao, C.; Gao, J.; Evans, C.M.; Ju, K.-S.; Doroghazi, J.R.; van der Donk, W.; Kelleher, N.L.; Metcalf, W.W. Discovery of the Antibiotic Phosacetamycin via a New Mass Spectrometry-Based Method for Phosphonic Acid Detection. ACS Chem. Biol. 2013, 8, 908–913. [Google Scholar] [CrossRef]
- Albright, J.C.; Goering, A.W.; Doroghazi, J.R.; Metcalf, W.W.; Kelleher, N.L. Strain-specific proteogenomics accelerates the discovery of natural products via their biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2013, 41, 451–459. [Google Scholar] [CrossRef]
- Crofts, T.S.; Gasparrini, A.J.; Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 2017, 15, 422–434. [Google Scholar] [CrossRef]
- Cochrane, W.G.; Fitzgerald, P.R.; Paegel, B.M. Antibacterial Discovery via Phenotypic DNA-Encoded Library Screening. ACS Chem. Biol. 2021. [Google Scholar] [CrossRef]
- Nandi, S.; Ahmed, S.; Saxena, A.K. Combinatorial design and virtual screening of potent anti-tubercular fluoroquinolone and isothiazoloquinolone compounds utilizing QSAR and pharmacophore modelling. SAR QSAR Environ. Res. 2018, 29, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.; Lendebol, E.; Shen, A.; Philipp, M.; Clement, C.C. In Silico-Mediated Virtual Screening and Molecular Docking Platforms for Discovery of Non β-Lactam Inhibitors of Y-49 β-Lactamase from Mycobacterium Tuberculosis. MOJ Proteom. Bioinform. 2018, 7, 1–14. [Google Scholar] [CrossRef][Green Version]
- Pradhan, D.; Priyadarshini, V.; Munikumar, M.; Swargam, S.; Umamaheswari, A.; Bitla, A. Para-(benzoyl)-phenylalanine as a potential inhibitor against LpxC of Leptospira spp.: Homology modeling, docking, and molecular dynamics study. J. Biomol. Struct. Dyn. 2013, 32, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Babaoglu, K.; Simeonov, A.; Irwin, J.; Nelson, M.E.; Feng, B.; Thomas, C.J.; Cancian, L.; Costi, M.P.; Maltby, D.A.; Jadhav, A.; et al. Comprehensive Mechanistic Analysis of Hits from High-Throughput and Docking Screens against β-Lactamase. J. Med. Chem. 2008, 51, 2502–2511. [Google Scholar] [CrossRef]
- Sahner, J.H.; Groh, M.; Negri, M.; Haupenthal, J.; Hartmann, R.W. Novel small molecule inhibitors targeting the “switch region” of bacterial RNAP: Structure-based optimization of a virtual screening hit. Eur. J. Med. Chem. 2013, 65, 223–231. [Google Scholar] [CrossRef]
- Chan, A.H.; Wereszczynski, J.; Amer, B.R.; Yi, S.W.; Jung, M.E.; McCammon, J.A.; Clubb, R.T. Discovery of S taphylococcus aureus Sortase A Inhibitors Using Virtual Screening and the Relaxed Complex Scheme. Chem. Biol. Drug Des. 2013, 82, 418–428. [Google Scholar] [CrossRef]
- Chan, F.-Y.; Sun, N.; Neves, M.A.C.; Lam, P.C.-H.; Chung, W.-H.; Wong, L.-K.; Chow, H.-Y.; Ma, D.-L.; Chan, P.-H.; Leung, Y.-C.; et al. Identification of a New Class of FtsZ Inhibitors by Structure-Based Design and in Vitro Screening. J. Chem. Inf. Model. 2013, 53, 2131–2140. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, S.; Balius, T.E.; Singh, I.; Levit, A.; Moroz, Y.S.; O’meara, M.J.; Che, T.; Algaa, E.; Tolmachova, K.; et al. Ultra-large library docking for discovering new chemotypes. Nature 2019, 566, 224–229. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; Macnair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef]
- Hu, X.; Compton, J.R.; AbdulHameed, M.D.M.; Marchand, C.L.; Robertson, K.L.; Leary, D.H.; Jadhav, A.; Hershfield, J.R.; Wallqvist, A.; Friedlander, A.M.; et al. 3-Substituted Indole Inhibitors Against Francisella tularensis FabI Identified by Structure-Based Virtual Screening. J. Med. Chem. 2013, 56, 5275–5287. [Google Scholar] [CrossRef]
- Kinjo, T.; Koseki, Y.; Kobayashi, M.; Yamada, A.; Morita, K.; Yamaguchi, K.; Tsurusawa, R.; Gulten, G.; Komatsu, H.; Sakamoto, H.; et al. Identification of Compounds with Potential Antibacterial Activity against Mycobacterium through Structure-Based Drug Screening. J. Chem. Inf. Model. 2013, 53, 1200–1212. [Google Scholar] [CrossRef]
- Brynildsen, M.P.; A Winkler, J.; Spina, C.S.; MacDonald, I.C.; Collins, J.J. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat. Biotechnol. 2013, 31, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Ma, H.; Wu, M.; Welsch, T.A.; Vora, S.R.; Ren, D.; Nangia, S. Development of the computational antibiotic screening platform (CLASP) to aid in the discovery of new antibiotics. Soft Matter 2021, 17, 2725–2736. [Google Scholar] [CrossRef] [PubMed]
- Look, G.C.; Vacin, C.; Dias, T.M.; Ho, S.; Tran, T.H.; Lee, L.L.; Wiesner, C.; Fang, F.; Marra, A.; Westmacott, D.; et al. The discovery of biaryl acids and amides exhibiting antibacterial activity against Gram-positive bacteria. Bioorganic Med. Chem. Lett. 2004, 14, 1423–1426. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.J.; Critchlow, R.E. Beyond Mere Diversity: Tailoring Combinatorial Libraries for Drug Discovery. J. Comb. Chem. 1998, 1, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Fleeman, R.; LaVoi, T.M.; Santos, R.G.; Morales, A.; Nefzi, A.; Welmaker, G.S.; Medina-Franco, J.L.; Giulianotti, M.A.; Houghten, R.A.; Shaw, L.N. Combinatorial Libraries As a Tool for the Discovery of Novel, Broad-Spectrum Antibacterial Agents Targeting the ESKAPE Pathogens. J. Med. Chem. 2015, 58, 3340–3355. [Google Scholar] [CrossRef]
- Itoh, H.; Tokumoto, K.; Kaji, T.; Paudel, A.; Panthee, S.; Hamamoto, H.; Sekimizu, K.; Inoue, M. Development of a high-throughput strategy for discovery of potent analogues of antibiotic lysocin E. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Ansari, F.L.; Nazir, S.; Noureen, H.; Mirza, B. Combinatorial Synthesis and Antibacterial Evaluation of an Indexed Chalcone Library. Chem. Biodivers. 2005, 2, 1656–1664. [Google Scholar] [CrossRef]
- McClay, K.; Wan, B.; Wang, Y.; Cho, S.; Yu, J.; Santarsiero, B.; Mehboob, S.; Johnson, M.; Franzblau, S.; Steffan, R. A novel combinatorial biocatalytic approach for producing antibacterial compounds effective against Mycobacterium tuberculosis (TB). Appl. Microbiol. Biotechnol. 2013, 97, 7151–7163. [Google Scholar] [CrossRef]
- Takada, Y.; Itoh, H.; Paudel, A.; Panthee, S.; Hamamoto, H.; Sekimizu, K.; Inoue, M. Discovery of gramicidin A analogues with altered activities by multidimensional screening of a one-bead-one-compound library. Nat. Commun. 2020, 11, 4935. [Google Scholar] [CrossRef]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef]
- Over, B.; Wetzel, S.; Grütter, C.; Nakai, Y.; Renner, S.; Rauh, D.; Waldmann, H. Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 2012, 5, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Painter, R.E.; Enesa, K.; Holmes, D.; Whyte, G.; Garlisi, C.G.; Monsma, F.J.; Rehak, M.; Craig, F.F.; Smith, C.A. High-throughput screening of antibiotic-resistant bacteria in picodroplets. Lab. Chip. 2016, 16, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Watterson, W.J.; Tanyeri, M.; Watson, A.R.; Cham, C.M.; Shan, Y.; Chang, E.B.; Eren, A.M.; Tay, S. Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. Elife 2020, 9, e56998. [Google Scholar] [CrossRef] [PubMed]
- Mocciaro, A.; Roth, T.L.; Bennett, H.M.; Soumillon, M.; Shah, A.; Hiatt, J.; Chapman, K.; Marson, A.; Lavieu, G. Light-activated cell identification and sorting (LACIS) for selection of edited clones on a nanofluidic device. Commun. Biol. 2018, 1, 41. [Google Scholar] [CrossRef]
- Haney, S.A.; LaPan, P.; Pan, J.; Zhang, J. High-content screening moves to the front of the line. Drug Discov. Today 2006, 11, 889–894. [Google Scholar] [CrossRef]
- Korn, K.; Krausz, E. Cell-based high-content screening of small-molecule libraries. Curr. Opin. Chem. Biol. 2007, 11, 503–510. [Google Scholar] [CrossRef]
- Bray, M.-A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; E Carpenter, A. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016, 11, 1757–1774. [Google Scholar] [CrossRef]
- Njikan, S.; Manning, A.J.; Ovechkina, Y.; Awasthi, D.; Parish, T. High content, high-throughput screening for small molecule inducers of NF-κB translocation. PLOS ONE 2018, 13, e0199966. [Google Scholar] [CrossRef]
- Turan, N.B. Chapter 8—New approaches to antibacterial drug discovery. In Drug Discovery Targeting Drug-Resistant Bacteria; Kesharwani, P., Chopra, S., Dasgupta, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 223–248. [Google Scholar]
- Kummerfeldt, C. Raxibacumab: Potential role in the treatment of inhalational anthrax. Infect. Drug Resist. 2014, 7, 101–109. [Google Scholar] [CrossRef]
- Jin, X.; Li, M.; Wang, J.; Marambio-Jones, C.; Peng, F.; Huang, X.; Damoiseaux, R.; Hoek, E.M.V. High-Throughput Screening of Silver Nanoparticle Stability and Bacterial Inactivation in Aquatic Media: Influence of Specific Ions. Environ. Sci. Technol. 2010, 44, 7321–7328. [Google Scholar] [CrossRef] [PubMed]
- Correa, M.G.; Martínez, F.B.; Vidal, C.P.; Streitt, C.; Escrig, J.; de Dicastillo, C.L. Antimicrobial metal-based nanoparticles: A review on their synthesis, types and antimicrobial action. Beilstein J. Nanotechnol. 2020, 11, 1450–1469. [Google Scholar] [CrossRef] [PubMed]
- Karaman, D.; Manner, S.; Fallarero, A.; Rosenholm, J.M. Current Approaches for Exploration of Nanoparticles as Antibacterial Agents. In Antibacterial Agents; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Li, H.; Chen, Q.; Zhao, J.; Urmila, K. Enhancing the antimicrobial activity of natural extraction using the synthetic ultrasmall metal nanoparticles. Sci. Rep. 2015, 5, srep11033. [Google Scholar] [CrossRef] [PubMed]
- Bankier, C.; Cheong, Y.-K.; Mahalingam, S.; Edirisinghe, M.; Ren, G.; Cloutman-Green, E.; Ciric, L. A comparison of methods to assess the antimicrobial activity of nanoparticle combinations on bacterial cells. PLOS ONE 2018, 13, e0192093. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.L.; Garcia, C.B.; Gallon, S.N.; Webster, T.J. Novel Silver-Platinum Nanoparticles for Anticancer and Antimicrobial Applications. Int. J. Nanomed. 2020, ume 15, 169–179. [Google Scholar] [CrossRef]
- Watson, C.; Ge, J.; Cohen, J.; Pyrgiotakis, G.; Engelward, B.P.; Demokritou, P. High-Throughput Screening Platform for Engineered Nanoparticle-Mediated Genotoxicity Using CometChip Technology. ACS Nano 2014, 8, 2118–2133. [Google Scholar] [CrossRef]
- da Cunha, B.R.; Fonseca, L.P.; Calado, C.R.C. Simultaneous elucidation of antibiotic mechanism of action and potency with high-throughput Fourier-transform infrared (FTIR) spectroscopy and machine learning. Appl. Microbiol. Biotechnol. 2021, 105, 1269–1286. [Google Scholar] [CrossRef]
- Apostolos, A.J.; Ferraro, N.J.; Dalesandro, B.E.; Pires, M.M. SaccuFlow: A High-Throughput Analysis Platform to Investigate Bacterial Cell Wall Interactions. ACS Infect. Dis. 2021, 7, 2483–2491. [Google Scholar] [CrossRef]
- Barker, J.J. Antibacterial drug discovery and structure-based design. Drug Discov. Today 2006, 11, 391–404. [Google Scholar] [CrossRef]
- McLaren, D.G.; Shah, V.; Wisniewski, T.; Ghislain, L.; Liu, C.; Zhang, H.; Saldanha, S.A. High-Throughput Mass Spectrometry for Hit Identification: Current Landscape and Future Perspectives. SLAS Discov. Adv. Sci. Drug Discov. 2021, 26, 168–191. [Google Scholar] [CrossRef]
- Quercia, A.K.; Lamarr, W.A.; Myung, J.; Özbal, C.C.; Landro, J.A.; Lumb, K.J. High-Throughput Screening by Mass Spectrometry: Comparison with the Scintillation Proximity Assay with a Focused-File Screen of AKT1/PKBα. SLAS Discov. Adv. Sci. Drug Discov. 2007, 12, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, I.; Stearns, R.; Pringle, S.; Wingfield, J.; Datwani, S.; Hall, E.; Ghislain, L.; Majlof, L.; Bachman, M. Novel Acoustic Loading of a Mass Spectrometer: Toward Next-Generation High-Throughput MS Screening. SLAS Technol. Transl. Life Sci. Innov. 2016, 21, 19–26. [Google Scholar] [CrossRef] [PubMed]
- VanderPorten, E.C.; Scholle, M.D.; Sherrill, J.; Tran, J.C.; Liu, Y. Identification of Small-Molecule Noncovalent Binders Utilizing SAMDI Technology. SLAS Discov. Adv. Sci. Drug Discov. 2017, 22, 1211–1217. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Annis, A.; Chuang, C.; Nazef, N. ALIS: An Affinity Selection–Mass Spectrometry System for the Discovery and Characterization of Protein–Ligand Interactions. Methods Princ. Med. Chem. 2007, 121–156. [Google Scholar] [CrossRef]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wang, J.; Wu, R.P.; Gomez, F.; Loo, J.A.; et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef]
- Bandow, J.E.; Brötz, H.; Leichert, L.I.O.; Labischinski, H.; Hecker, M. Proteomic Approach to Understanding Antibiotic Action. Antimicrob. Agents Chemother. 2003, 47, 948–955. [Google Scholar] [CrossRef]
- Zampieri, M.; Szappanos, B.; Buchieri, M.V.; Trauner, A.; Piazza, I.; Picotti, P.; Gagneux, S.; Borrell, S.; Gicquel, B.; Lelievre, J.; et al. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci. Transl. Med. 2018, 10, eaal3973. [Google Scholar] [CrossRef]
- Putty, S.; Rai, A.; Jamindar, D.; Pagano, P.; Quinn, C.L.; Mima, T.; Schweizer, H.P.; Gutheil, W.G. Characterization of d-boroAla as a Novel Broad-Spectrum Antibacterial Agent Targeting d-Ala-d-Ala Ligase. Chem. Biol. Drug Des. 2011, 78, 757–763. [Google Scholar] [CrossRef]
- Vemula, H.; Bobba, S.; Putty, S.; Barbara, J.E.; Gutheil, W.G. Ion-pairing liquid chromatography–tandem mass spectrometry-based quantification of uridine diphosphate-linked intermediates in the Staphylococcus aureus cell wall biosynthesis pathway. Anal. Biochem. 2014, 465, 12–19. [Google Scholar] [CrossRef]
- Vemula, H.; Ayon, N.J.; Gutheil, W.G. Cytoplasmic peptidoglycan intermediate levels in Staphylococcus aureus. Biochimie 2015, 121, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Vemula, H.; Ayon, N.J.; Burton, A.; Gutheil, W.G. Antibiotic Effects on Methicillin-Resistant Staphylococcus aureus Cytoplasmic Peptidoglycan Intermediate Levels and Evidence for Potential Metabolite Level Regulatory Loops. Antimicrob. Agents Chemother. 2017, 61, e02253-16. [Google Scholar] [CrossRef] [PubMed]
- Ayon, N.J.; Sharma, A.D.; Gutheil, W.G. LC-MS/MS-Based Separation and Quantification of Marfey’s Reagent Derivatized Proteinogenic Amino Acid dl-Stereoisomers. J. Am. Soc. Mass Spectrom. 2018, 30, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Viable screening targets related to the bacterial cell wall. Ann. N. Y. Acad. Sci. 2012, 1277, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.; Braddick, D.; Dowson, C.; Roper, D. Bacterial cell wall assembly: Still an attractive antibacterial target. Trends Biotechnol. 2011, 29, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Graham, D.; Kastenmüller, G.; Alharbi, N.H.; AlSanosi, S.M.; McBride, M.; Mangino, M.; Titcombe, P.; Shin, S.-Y.; Psatha, M.; et al. Metabolomic Identification of a Novel Pathway of Blood Pressure Regulation Involving Hexadecanedioate. Hypertension 2015, 66, 422–429. [Google Scholar] [CrossRef]
- Yu, W.; Yang, W.; Zhao, M.-Y.; Meng, X.-L. Functional Metabolomics Analysis Elucidating the Metabolic Biomarker and Key Pathway Change Associated with the Chronic Glomerulonephritis and Revealing Action Mechanism of Rhein. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]
- Elnaas, A.R.; Grice, D.; Han, J.; Feng, Y.; Di Capua, A.; Mak, T.; Laureanti, J.A.; Buchko, G.W.; Myler, P.J.; Cook, G.; et al. Discovery of a Natural Product That Binds to the Mycobacterium tuberculosis Protein Rv1466 Using Native Mass Spectrometry. Molecules 2020, 25, 2384. [Google Scholar] [CrossRef]
- Vemula, H.; Kitase, Y.; Ayon, N.J.; Bonewald, L.; Gutheil, W.G. Gaussian and linear deconvolution of LC-MS/MS chromatograms of the eight aminobutyric acid isomers. Anal. Biochem. 2017, 516, 75–85. [Google Scholar] [CrossRef]
- Ayon, N.J. Features, roles and chiral analyses of proteinogenic amino acids. AIMS Mol. Sci. 2020, 7, 229–268. [Google Scholar] [CrossRef]
- Cummins, L.L.; Chen, S.; Blyn, L.B.; Sannes-Lowery, K.A.; Drader, J.J.; Griffey, R.H.; Hofstadler, S.A. Multitarget Affinity/Specificity Screening of Natural Products: Finding and Characterizing High-Affinity Ligands from Complex Mixtures by Using High-Performance Mass Spectrometry. J. Nat. Prod. 2003, 66, 1186–1190. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.T.H.; Bennett, J.L.; Liu, S.; Hancock, S.E.; Winter, D.L.; Glover, D.J.; Donald, W.A. Multiplexed Screening of Thousands of Natural Products for Protein–Ligand Binding in Native Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 21379–21387. [Google Scholar] [CrossRef] [PubMed]
- Si, T.; Li, B.; Comi, T.J.; Wu, Y.; Hu, P.; Wu, Y.; Min, Y.; Mitchell, D.A.; Zhao, H.; Sweedler, J.V. Profiling of Microbial Colonies for High-Throughput Engineering of Multistep Enzymatic Reactions via Optically Guided Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Am. Chem. Soc. 2017, 139, 12466–12473. [Google Scholar] [CrossRef]
- Gurard-Levin, Z.A.; Scholle, M.D.; Eisenberg, A.H.; Mrksich, M. High-Throughput Screening of Small Molecule Libraries using SAMDI Mass Spectrometry. ACS Comb. Sci. 2011, 13, 347–350. [Google Scholar] [CrossRef]
- Yao, H.; Zhao, H.; Zhao, X.; Pan, X.; Feng, J.; Xu, F.; Zhang, S.; Zhang, X. Label-free Mass Cytometry for Unveiling Cellular Metabolic Heterogeneity. Anal. Chem. 2019, 91, 9777–9783. [Google Scholar] [CrossRef] [PubMed]
- Lamoree, B.; Hubbard, R.E. Using Fragment-Based Approaches to Discover New Antibiotics. SLAS Discov. Adv. Sci. Drug Discov. 2018, 23, 495–510. [Google Scholar] [CrossRef]
- Genick, C.C.; Wright, S.K. Biophysics: For HTS hit validation, chemical lead optimization, and beyond. Expert Opin. Drug Discov. 2017, 12, 897–907. [Google Scholar] [CrossRef]
- Hall, M.D.; Yasgar, A.; Peryea, T.; Braisted, J.C.; Jadhav, A.; Simeonov, A.; Coussens, N.P. Fluorescence polarization assays in high-throughput screening and drug discovery: A review. Methods Appl. Fluoresc. 2016, 4, 022001. [Google Scholar] [CrossRef] [PubMed]
- Freedberg, D.I.; Selenko, P. Live Cell NMR. Annu. Rev. Biophys. 2014, 43, 171–192. [Google Scholar] [CrossRef]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundbäck, T.; Nordlund, P.; Molina, D.M. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef]
- Ciulli, A. Biophysical Screening for the Discovery of Small-Molecule Ligands. Methods Mol Biol. 2013, 1008, 357–388. [Google Scholar] [CrossRef]
- Renaud, J.-P.; Chung, C.-W.; Danielson, U.H.; Egner, U.; Hennig, M.; Hubbard, R.E.; Nar, H. Biophysics in drug discovery: Impact, challenges and opportunities. Nat. Rev. Drug Discov. 2016, 15, 679–698. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Li, B.; Xu, Y.; Mao, F.; Li, X.; Lan, L.; Zhu, J.; Li, J. Targeting virulence factors as an antimicrobial approach: Pigment inhibitors. Med. Res. Rev. 2019, 40, 293–338. [Google Scholar] [CrossRef] [PubMed]
- da Cunha, B.R.; Zoio, P.; Fonseca, L.P.; Calado, C.R.C. Technologies for High-Throughput Identification of Antibiotic Mechanism of Action. Antibiotics 2021, 10, 565. [Google Scholar] [CrossRef] [PubMed]
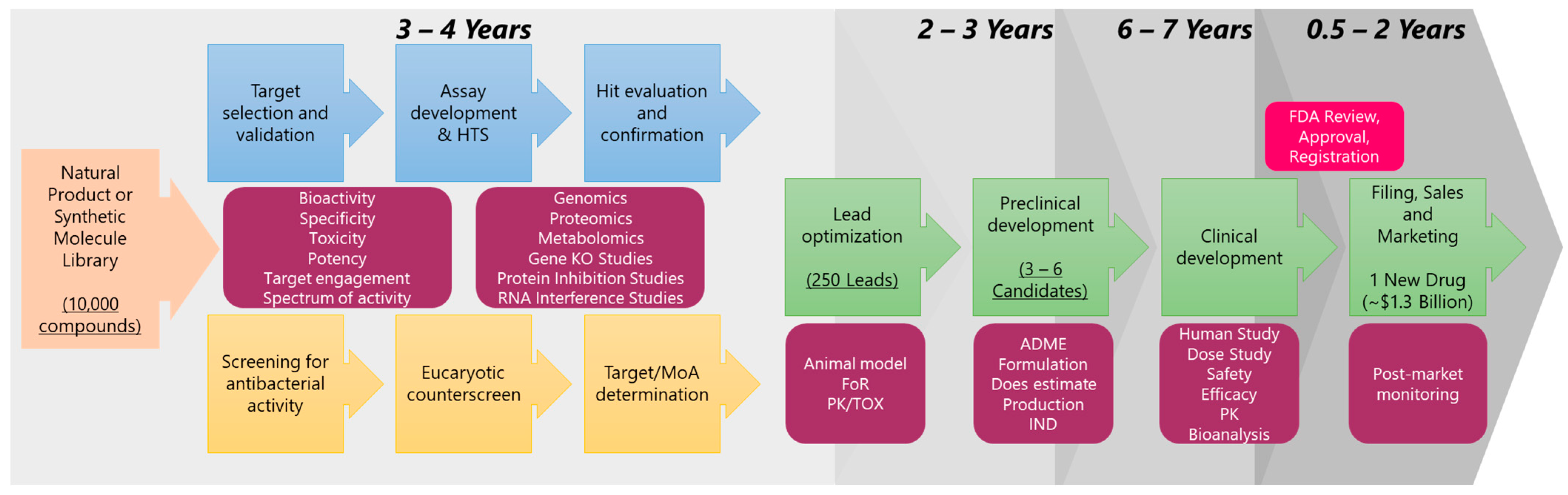
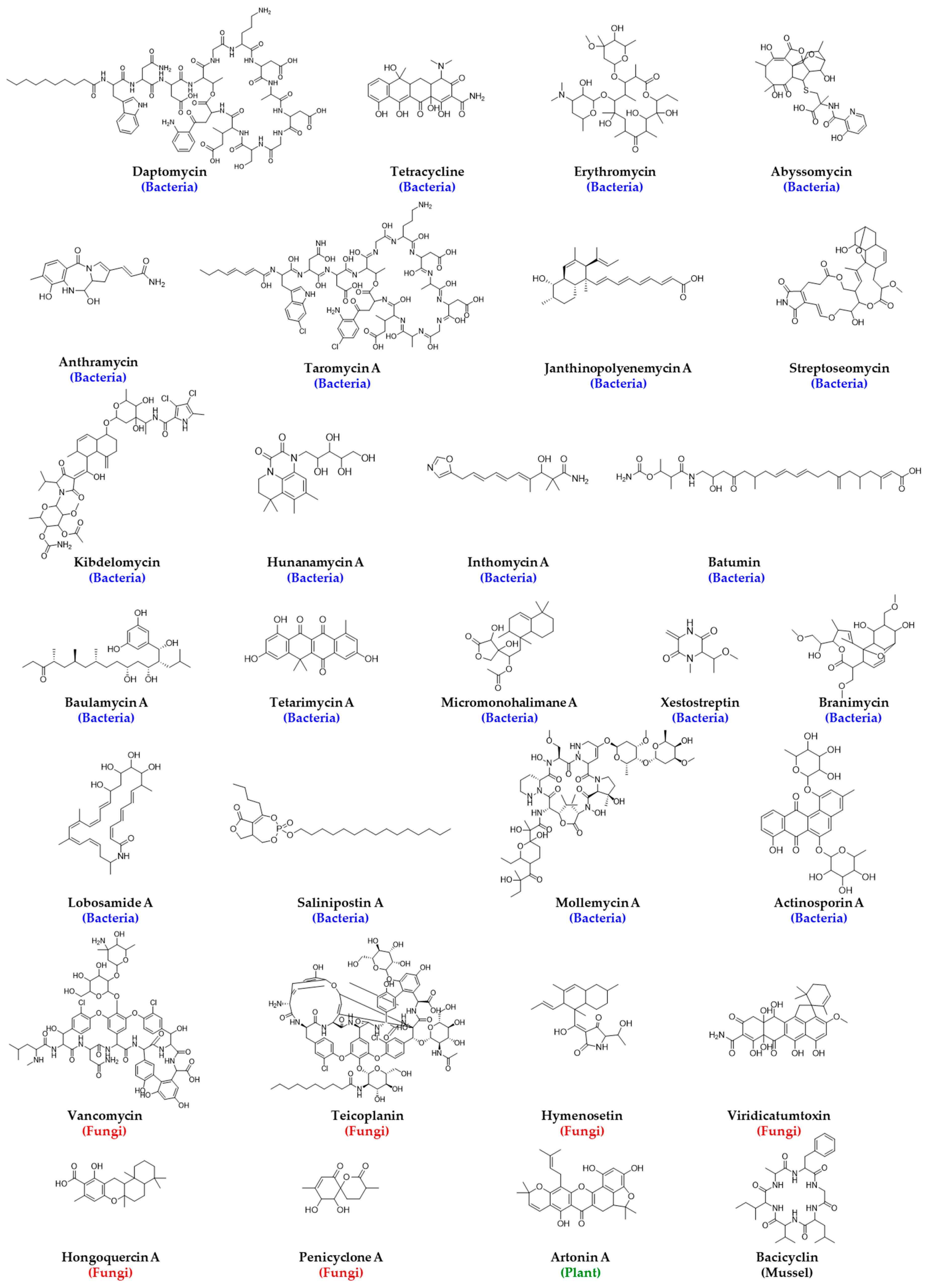
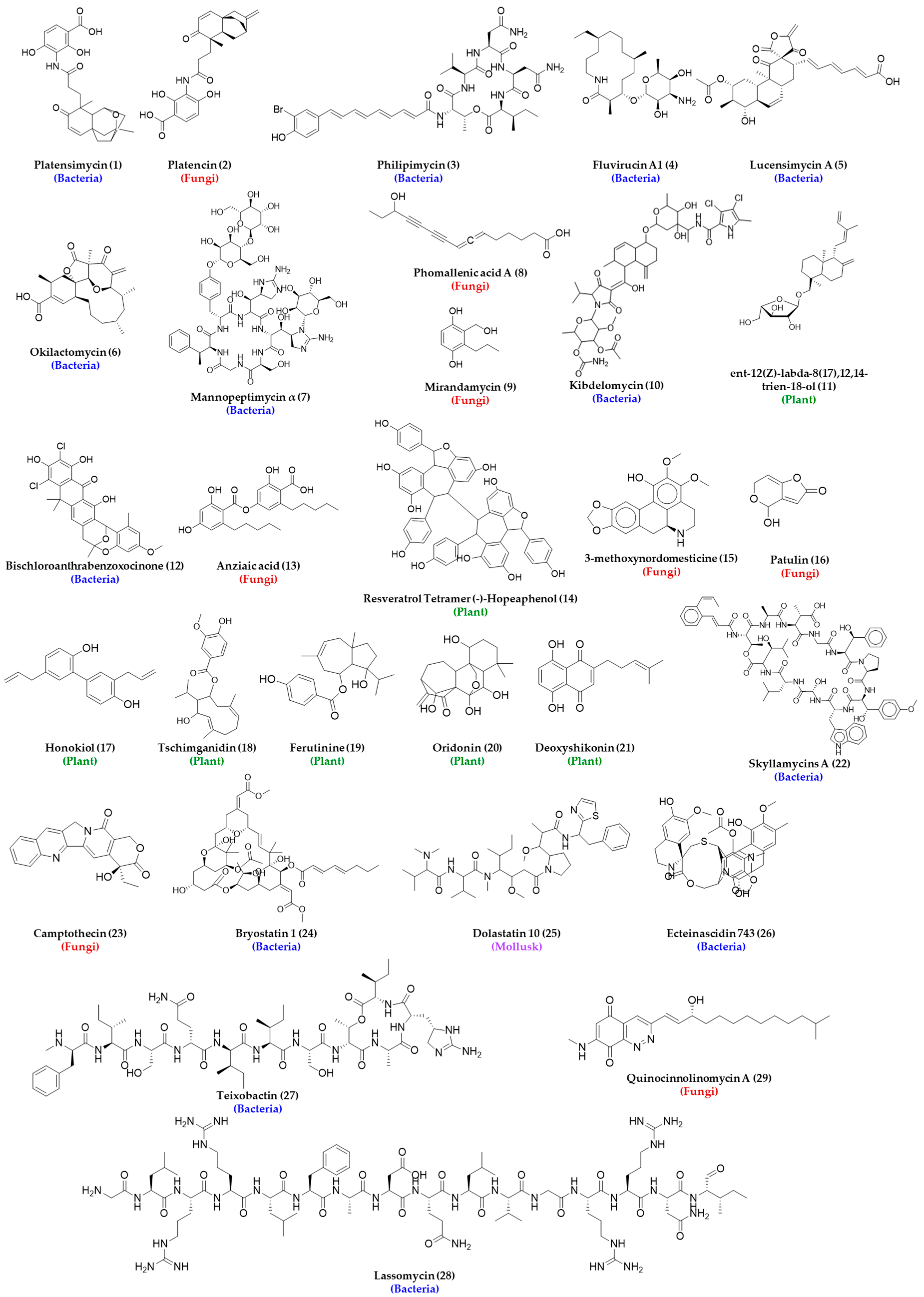
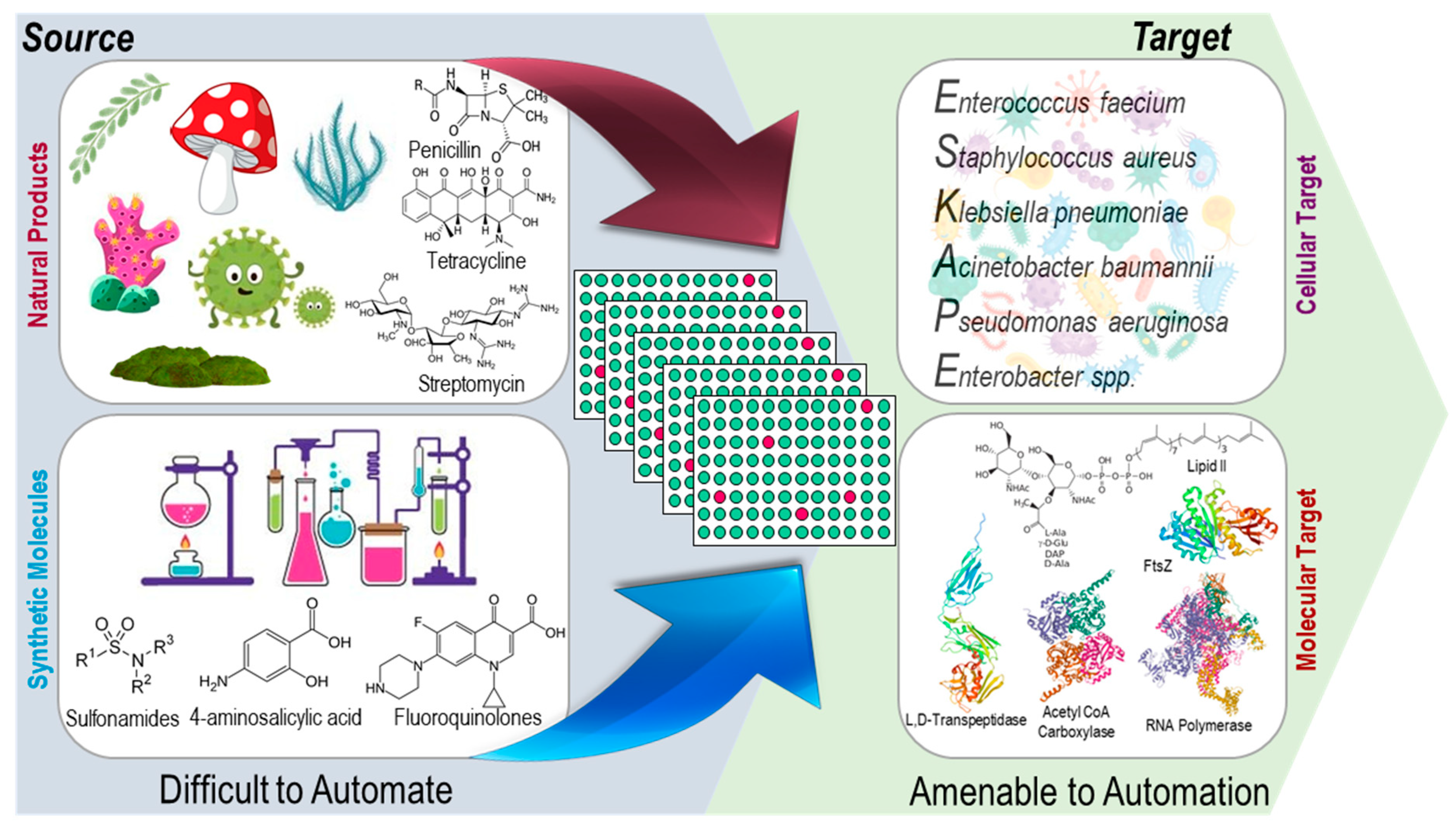

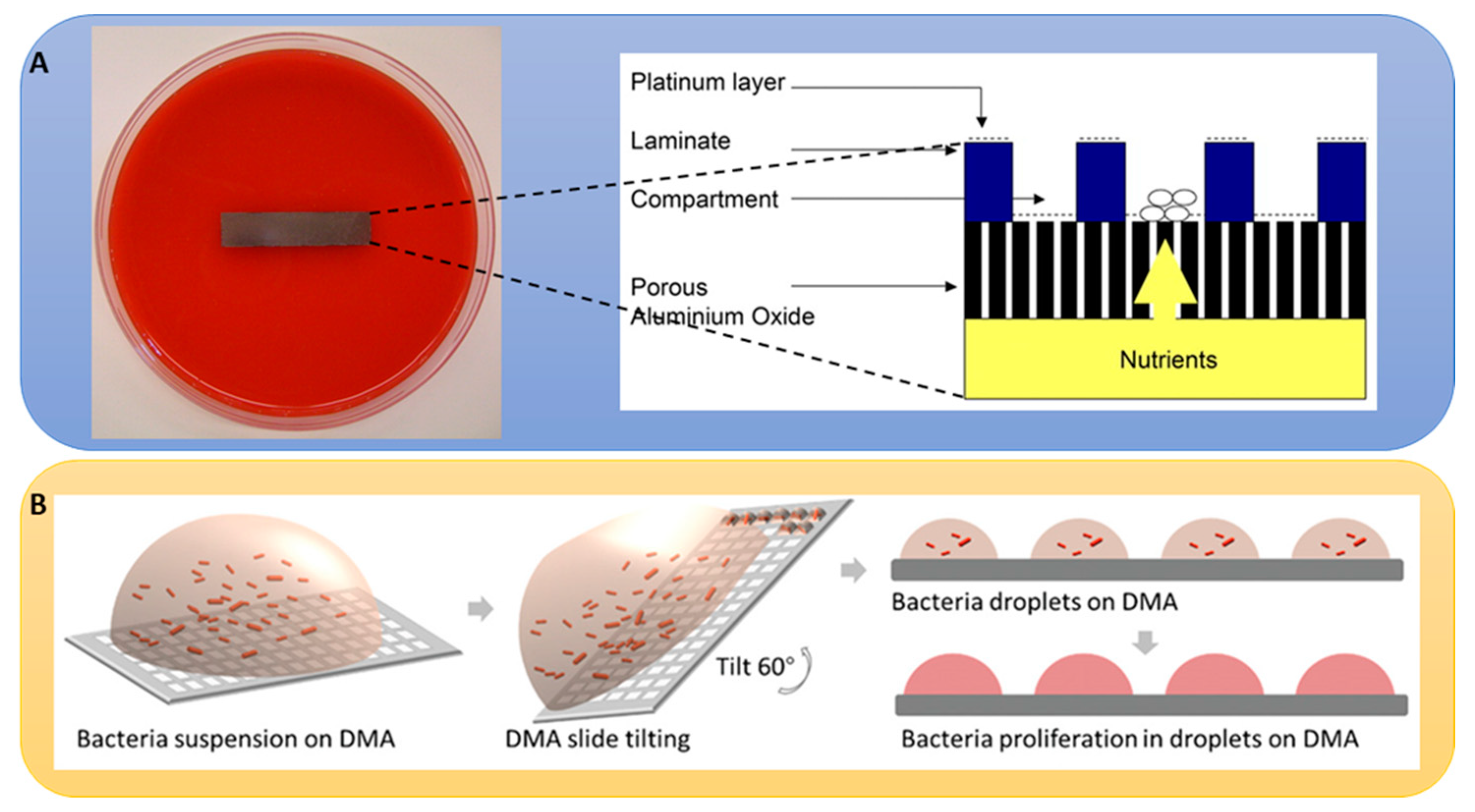
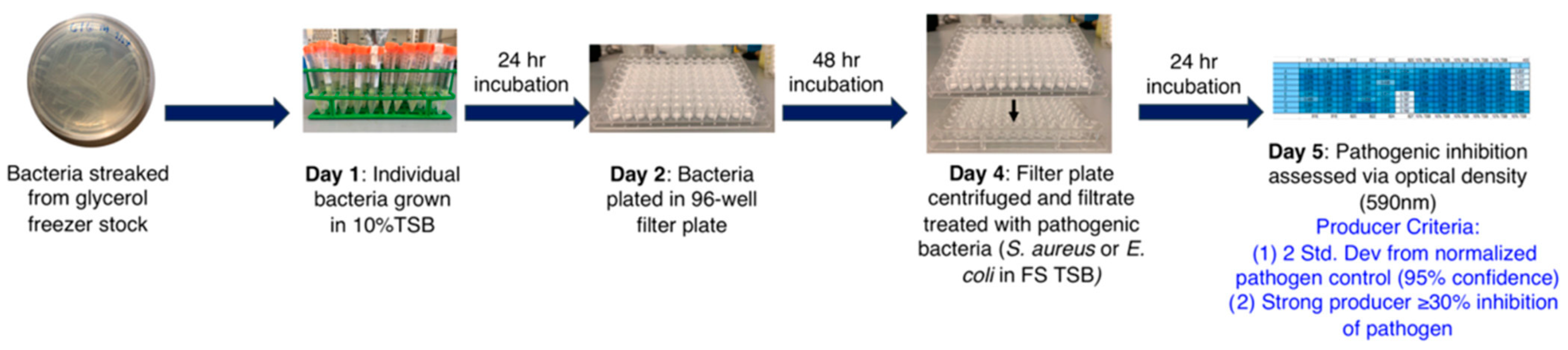


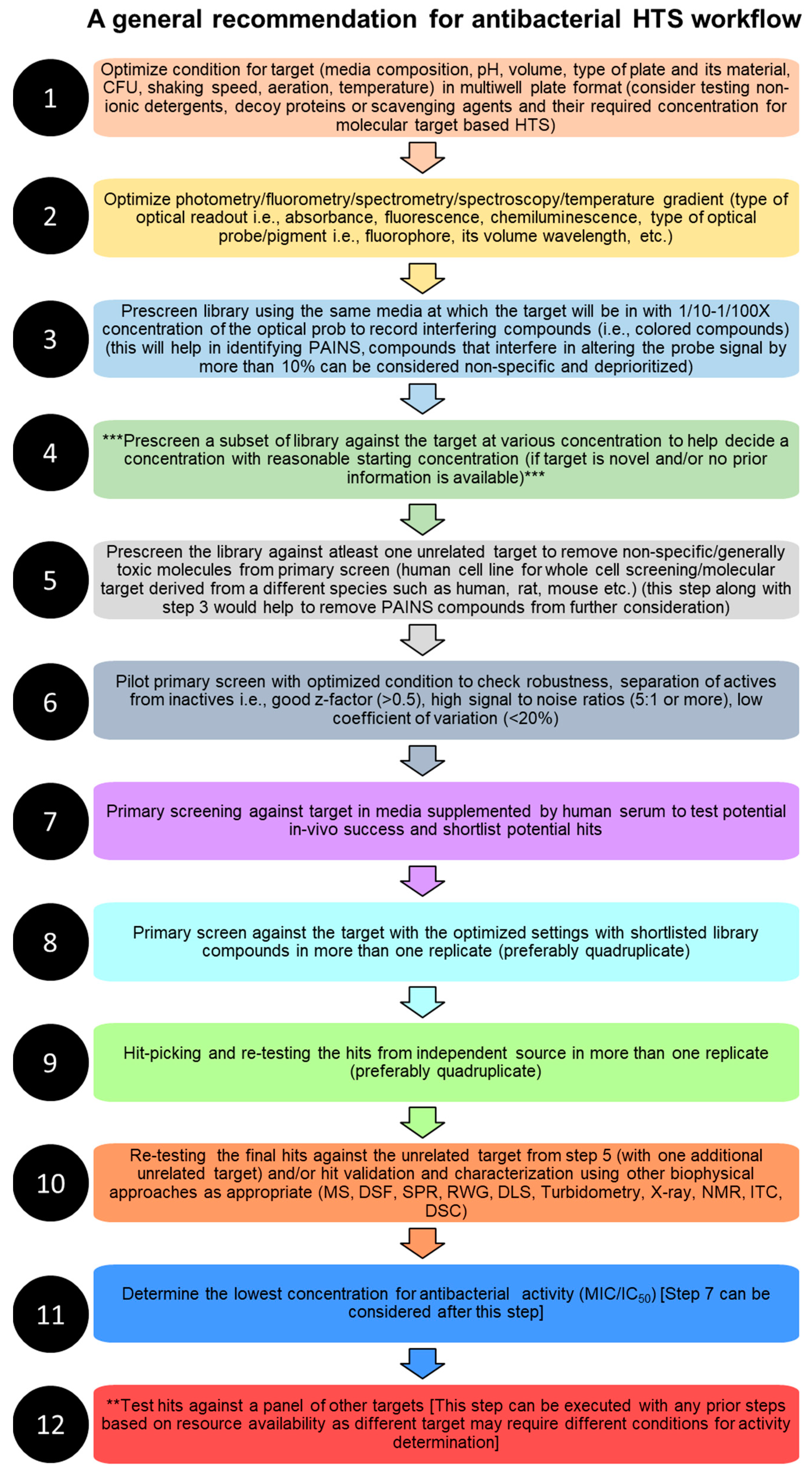
|
 | ||||
|---|---|---|---|---|
| Manufacturing and Pilot Scale Bioreactor | Lab-Scale Bioreactor | Microplate Culture Model | Microfluidic Culture Model | |
| Volume | 100–200,000 L | 0.25–30 L | 0.1–1.0 mL | <100 nL |
| Throughput (genotypes/year) | <10 | ~103 | ~106 | ~109 |
| Cost per fermentation | USD 10 K–USD 200 K | ~USD 1 K | <USD 0.5 | <USD 0.0001 |
| Advantages | Scalable, cost-effective | High-confident strain Manufacturing with high control | High parallelization to rank genotypes for further testing | Highest throughput with the smallest cost |
| Limitations | Hardware constraints hinder process control | Non-homogenous chemical environment | High false positives, irreproducibility, or unreliable readouts | Highly sophisticated instruments are required, very low sample size requires high sensitivity |
| Predictive power for commercial scale | – | Reliable | Variable | Unexplored |
| Successful Hit | Target Organism | Protein Target | MIC a/IC50 b | Reference |
|---|---|---|---|---|
| Bischloroanthrabenzoxocinone (BABX) (12) | E. coli (MB4902) S. aureus (MB2985) | Fatty acid synthesis Type II (FASII) | 0.2–0.4 μg/mLa | [120] |
| Anziaic acid (13) | B. subtilis (ATCC 6633) E. coli (BAS3023) | Topoisomerase I | 6 μg/mL a 12 μg/mL a | [121] |
| 04E04 03F11 04E04 02F09, 02H08 | E. faecalis H. influenzae M. catarrhalis P. aeruginosa hypersensitive (ATCC 35151) | Phenylalanyl-tRNA Synthetases (PheRS) | 8 μg/mL a 4 μg/mL a 1 μg/mL a 4 μg/mL a | [122] |
| Resveratrol Tetramer (-)-Hopeaphenol (14) | Y. pseudotuberculosis | Type III secretion | 6.6 μM b | [123] |
| NAT13-338148 NAT18-355531 NAT18-355768 | Clostridioides difficile (ATCC BAA 1870) | ND | 0.5–2 μg/mL a | [124] |
| 3-methoxynordomesticine (15) | Mycobacterium tuberculosis H37Rv Mycobacterium bovis BCG | MurE (IC50 < 100 µM) | ≤5 μg/mL a | [125] |
| HTS Strategy | Library Size and Name | Z/Z′-Score | Target | Hits (ID/name) | Activity * | Cytotoxicity Tested | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| MT | CT | C | |||||||
| ✔ | ✔ | 20,000 (ChemBridge) | 0.82 | N-acetylglucosamine-1-phosphate uridyltransferase (GlmU)(acetyltransferase domain) | 6624116 a 5655606 a 5810599 a 6012954 a | 65.15 ± 3.31 µM b 18.58 ± 0.81 µM b 9.01 ± 0.04 µM b 38.84 ± 0.29 µM b | Human liver hepatocellular carcinoma cells (HepG2 cells) | [190] | |
| M. tuberculosis H37Rv | 6624116 a 5655606 a 5810599 a 6012954 a | 16 µg/mL c 2 µg/mL c >32 µg/mL c >32 µg/mL c | |||||||
| ✔ | 50,000 (Maybridge) | 0.71 | N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) (acetyltransferase domain) | MAC0021939 MAC0008028 MAC0029665 | 189 ± 17.7 nM d 1.11 ± 0.08 ×103 nM d 28.2 ± 1.83 nM d | Not tested | [191] | ||
| ✔ | 20,502 (Broad Institute) | 0.7–0.8 | M. tuberculosis H37Rv, M. bovis BCG strain Pasteur, and M. smegmatis MC2155 | Benzimidazole derivative e Nitro-triazole derivative f | 37.5 µM c 0.488 µM g | Not tested | [192] | ||
| Membrane protein large 3 (MmpL3) (target for ‘e’)Decaprenylphosphoryl-β-d-ribose 2′-epimerase (DprE1) (target for ‘f’) | |||||||||
| ✔ | 57,000 (Timtec, Cerep, and ChemBridge) | <0.5 | Mycobacterium tuberculosis H37Rv, H37Ra and BCG Pasteur | DNB1 h DNB2 i | 0.2 µM c 0.2 µM c | Mouse bone marrow-derived macrophages | [193] | ||
| Decaprenylphosphoryl-β-D-ribose 2′-epimerase (DprE1/DprE2) | |||||||||
| ✔ | 125,000 | Not reported | ΔtolC Escherichia coli | Eight compounds containing 2-pyrazol-1-yl-thiazole scaffold | 0.037–8 µg/mL c | HEK293 cells A549 cells MCF7 cells | [194] | ||
| ✔ | 17,500 (Chemical Biology Consortium Sweden Primary Screening Set Collection) | Not reported | Streptococcus pneumonia T4 | THCz-1 (1-amino substituted Tetrahydrocarbazole) | 1.3 µg/mLc | Lung epithelial A549 cells | [195] | ||
| Undecaprenyl pyrophosphate-containing lipid intermediates | |||||||||
| ✔ | 28,300
| 0.5–0.9 | Vibrio cholera N16961 and NM06-058 | vz0825 vz0500 1541-0004 | 1.6 µM c 3.1 µM c 6.3 µM c | Mouse fibroblast cell line L929 | [189] | ||
| K+-channel sensor histidine kinase (KdpD) | |||||||||
| ✔ | 20,338
| 0.5–0.9 | 0139 V. cholerae MO10 (aphA transcript) | vz0761 vz0852 53760866 | 10 µM c 6 µM c 38 µM c | Mouse fibroblast cell line L929 j | [196] | ||
| Target Bacteria | Compounds Showing Synergy | Reference |
|---|---|---|
| Acinetobacter baumannii strain AB5075 | 5-fluorouracil and azithromycin Colistin sulfate with fluspirilene and Bay 11-7082 | [218] |
| MRSA (ATCC 43300) | Cefoxitin with floxuridine, gemcitabine, novobiocin, rifaximin, 4-quinazolinediamine, celastrol, and teniposide | [220,222,227] |
| Ceftobiprole with cloxacillin, cefotaxime, oxacilline, flucloxacillin, dicloxacillin, nafcillin, imipenem, meropenem, cefoxitin, piperacillin, and tazobactam | [228] | |
| Ceftaroline with cloxacillin | ||
| MRSA USA300 | Cefuroxime and ticlopidine | [229] |
| MRSA N315 | Meropenem, piperacillin, and tazobactam | [230] |
| MRSA ATCC 29213 Pseudomonas aeruginosa | IITR00693 (2-Aminoperimidine) and polymyxin B | [231] |
| Pseudomonas aeruginosa lasB-gfp (ASV) | Auranofin and colistin | [232] |
| P. aeruginosa (Clinical isolates) | Tetracycline and polyamine scaffolds containing molecules | [233] |
| P. aeruginosa ATCC 27853 | Amikacin and ceftriaxone | [234] |
| P. aeruginosa P6540 | Rifampicin and imipenem Colistin and imipenem | [235] |
| M. smegmatis mc2155 M. tuberculosis H37Rv (ATCC 27294) | Spectinomycin and bromperidol | [236] |
| M. tuberculosis H37Rv (ATCC 27294) | Rifampicin with Compound 5655606 | [190] |
| Legionella pneumophila serogroup 1 (Lp02::flaA::lux) | Rifampin, azithromycin, and minocycline in any combination | [237] |
| Carbapenem-resistant enterobacteriaceae | Triclosan and meropenem | [238] |
| Trimethoprim-resistant clinical E. coli | Azidothymidine with trimethoprim and sulfamethizole | [239] |
| Pandoraea nosoerga P8103 | Rifampicin and minocycline | [235] |
| Burkholderia multivorans P6539 | Fluoroquinolones and β-lactams | |
| E. coli BW25113 P. aeruginosa PA01 | Novobiocin with pivmecillinam and echinomycin Novobiocin with niridazole | [240] |
| Klebsiella pneumoniae N11-2218 | Meropenem with aspergillomarasmine A | [241] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayon, N.J. High-Throughput Screening of Natural Product and Synthetic Molecule Libraries for Antibacterial Drug Discovery. Metabolites 2023, 13, 625. https://doi.org/10.3390/metabo13050625
Ayon NJ. High-Throughput Screening of Natural Product and Synthetic Molecule Libraries for Antibacterial Drug Discovery. Metabolites. 2023; 13(5):625. https://doi.org/10.3390/metabo13050625
Chicago/Turabian StyleAyon, Navid J. 2023. "High-Throughput Screening of Natural Product and Synthetic Molecule Libraries for Antibacterial Drug Discovery" Metabolites 13, no. 5: 625. https://doi.org/10.3390/metabo13050625
APA StyleAyon, N. J. (2023). High-Throughput Screening of Natural Product and Synthetic Molecule Libraries for Antibacterial Drug Discovery. Metabolites, 13(5), 625. https://doi.org/10.3390/metabo13050625





