Cytotoxicity and Identification of Antibacterial Compounds from Baillonella toxisperma Bark Using a LC-MS/MS and Molecular Networking Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Extraction and Fractionation
2.3. Data-Dependent LC-HR-ESI-MS2 Analysis
2.4. Bacterial Germs Tested
2.5. Antibacterial Assays
2.6. Cell Culture
2.7. Cell Viability CCK-8 Assays
3. Results and Discussion
3.1. Metabolite Profiling of B. toxisperma Bark from LC-MS/MS Analysis
3.2. MS/MS–Molecular Networking-Based Dereplication
3.3. Antibacterial Activity
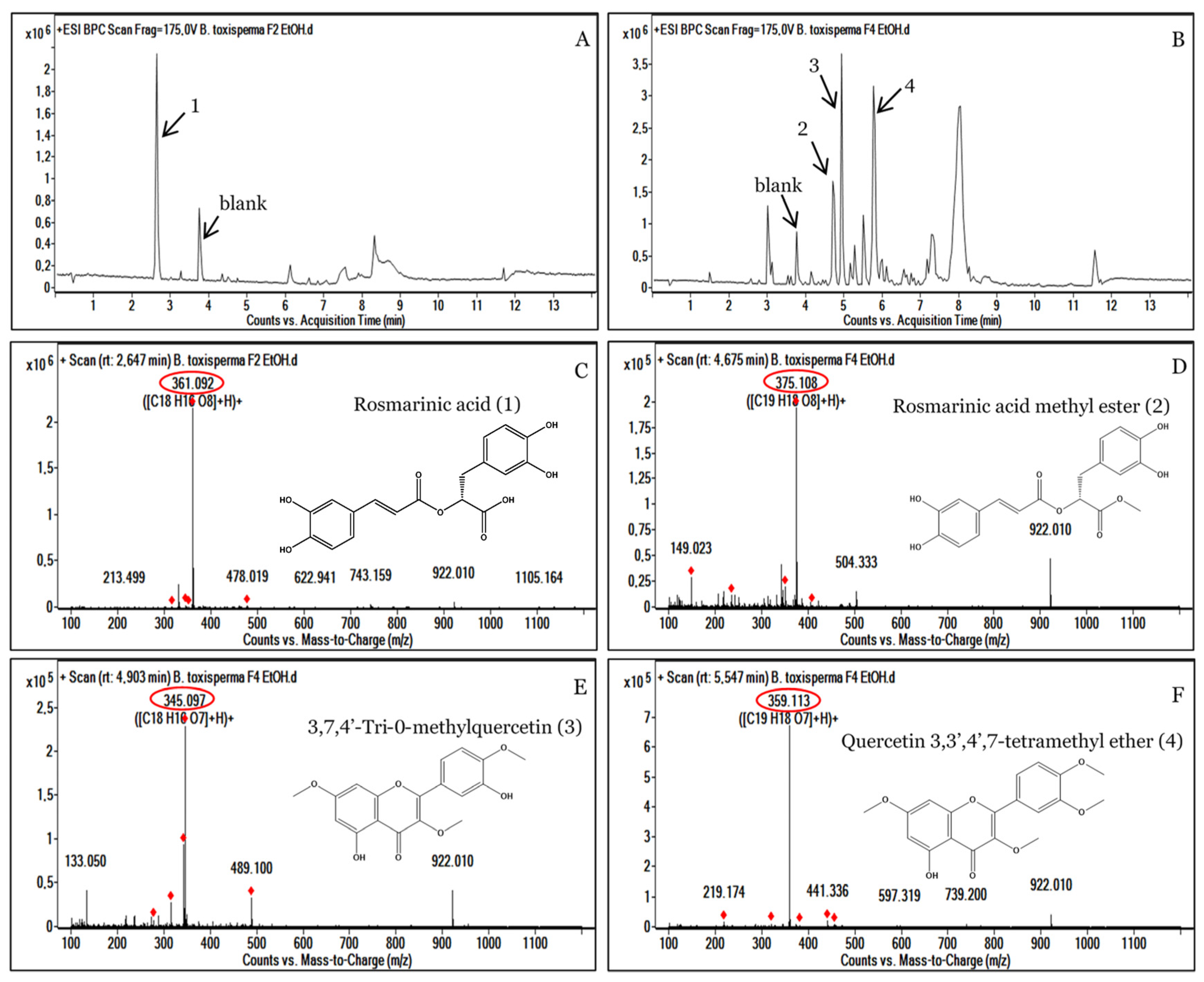
3.4. Analysis of the Most Active Fractions, F2 and F4
3.5. Cytotoxicity of the ethanolic crude extract of B. toxisperma
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Segers, A. Détermination des Visiteurs Floraux de Trois Essences Ligneuses à Haute Valeur Commerciale: Baillonella toxisperma Pierre, Afzelia Bipindensis Harms et Erythrophleum Suaveolens (Guill. & Perr.) Brenan. 72. Available online: https://www.semanticscholar.org/paper/D%C3%A9termination-des-visiteurs-floraux-de-trois-%C3%A0-%3A-et-Segers/66554ccb59f9560950888148134b2600bef7b542 (accessed on 19 February 2023).
- Ngueguim, J.R.; Dondjang, J.P.; Onana, J.; Ijang, P.T.; Zapfack, L.; Noumi, V.N.; Kengne, O.C.; Solefack, C.M. Moabi (Baillonella toxisperma Pierre): Arbre à Usage Multiple de Forêt Dense Humide Du Cameroun. Int. J. Biol. Chem. Sci. 2012, 5, 2395–2406. [Google Scholar] [CrossRef]
- Simo, R.; Ateba, S.B.; Zingue, S.; Pieme, A.; Njamen, D. Baillonella toxisperma Improves Sexual Performance and Protects against Stress-Induced Reproductive Dysfunction in Male Wistar Rats. J. Phytopharm. 2019, 8, 117–123. [Google Scholar] [CrossRef]
- Saha, J.-B.T.; Abia, D.; Dumarçay, S.; Ndikontar, M.K.; Gérardin, P.; Ngamveng Noah, J.; Perrin, D. Antioxidant Activities, Total Phenolic Contents and Chemical Compositions of Extracts from Four Cameroonian Woods: Padouk (Pterocarpus Soyauxii Taubb), Tali (Erythrophleum Suaveolens), Moabi (Baillonella toxisperma), and Movingui (Distemonanthus Benthamianus). Ind. Crops Prod. 2013, 41, 71–77. [Google Scholar] [CrossRef]
- Ndiade-Bourobou, D.; Vaillant, A.; Favreau, B.; Gayrin, E.; Bouvet, J.M. Isolation and Characterization of 15 Nuclear Microsatellite Markers for Baillonella toxisperma Pierre (Sapotaceae), a Low-Density Tree Species of Central Africa. Mol. Ecol. Resour. 2009, 9, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Pennington, T.D. Sapotaceae. In Flowering Plants Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales; Kubitzki, K., Ed.; la série de Livres The Families and Genera of Vascular Plants; Springer: Berlin/Heidelberg, Germany, 2004; pp. 390–421. ISBN 978-3-662-07257-8. [Google Scholar]
- Khayi, S.; Gaboun, F.; Pirro, S.; Tatusova, T.; El Mousadik, A.; Ghazal, H.; Mentag, R. Complete Chloroplast Genome of Argania Spinosa: Structural Organization and Phylogenetic Relationships in Sapotaceae. Plants 2020, 9, 1354. [Google Scholar] [CrossRef] [PubMed]
- Swenson, U.; Richardson, J.E.; Bartish, I.V. Multi-Gene Phylogeny of the Pantropical Subfamily Chrysophylloideae (Sapotaceae): Evidence of Generic Polyphyly and Extensive Morphological Homoplasy. Cladistics 2008, 24, 1006–1031. [Google Scholar] [CrossRef]
- Essama, S.H.R.; Nyegue, M.A.; Foe, C.N.; Tamo, S.P.B.; Etoa, F.X. In Vitro Evaluation of the Antifungal Activity of Extracts of Baillonella toxisperma (Pierre), a Sapotaceae, on the Growth of Some Human Pathogenic Yeasts. AJPP 2015, 9, 299–306. [Google Scholar] [CrossRef]
- Fodouop, M.; Tamo, S.P.B.; Pegnyemb, D.E.; Etoa, F.X.; de Paula, R.A.; Barbosa, A.A.T.; Machado, S.R.S.; Saraiva, M.A.F.; de Moraes, C.A.; Mantovani, H.C.; et al. In Vitro Antibacterial Activity of Baillonella toxisperma (Pierre) Extracts against Staphylococcus Aureus, Salmonella Typhi, Proteus Mirabilis and Bacillus Cereus F3748. AJMR 2015, 9, 2088–2094. [Google Scholar] [CrossRef]
- Guy Roussel, T.N.; Martin, F.; Janvier Aimé, Y.F.; Ferdinand Lanvin, E.E.; Ruth Edwige, D.K.; Boris, A.K.; Laure, N.J.; Enyong, O.J. Antihyperglycemic and Antihyperlipidemic Activities of Hydroethanolic Extract of the Fruit of Baillonella toxisperma in Streptozotocin-Induced Diabetic Rats. Metab. Open 2022, 15, 100199. [Google Scholar] [CrossRef]
- Benoit, M.Z.; Florence, N.T.; Raceline, G.K.; Yannick, F.B.; Alide, W.N.M.; Theophile, D. Antidiabetic Effects of Aqueous Extract of Baillonella toxisperma Pierre (Sapotacae) in Streptozotocin-Induced Diabetic Rats. J. Med. Plants Stud. 2021, 9, 28–37. [Google Scholar] [CrossRef]
- Ohigashi, H.; Kaji, M.; Sakaki, M.; Koshimizu, K. 3-Hydroxyuridine, an Allelopathic Factor of an African Tree, Baillonella toxisperma. Phytochemistry 1989, 28, 1365–1368. [Google Scholar] [CrossRef]
- Kim, J.G.; Le, T.P.L.; Han, J.S.; Cho, Y.B.; Lee, D.; Lee, M.K.; Hwang, B.Y. Molecular Networking-Guided Isolation of Cycloartane-Type Triterpenoids from Curculigo Orchioides and Their Inhibitory Effect on Nitric Oxide Production. ACS Omega. 2022, 7, 26853–26862. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, C.; Miyata, R.; Wakayama, S.; Kumazawa, S. New Acylated Flavonoid Isolated from Thai Bee Pollen Using Molecular Networking Analysis and Determination of Its Catechol-O-Methyltransferase Inhibitory Activity. Phytochem. Lett. 2023, 53, 239–244. [Google Scholar] [CrossRef]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Essono Mintsa, M.; Otogo N’nang, E.; Choque, É.; Siah, A.; Jacquin, J.; Muchembled, J.; Molinié, R.; Roulard, R.; Cailleu, D.; Beniddir, M.A.; et al. Combined LC-MS/MS and Molecular Networking Approach Reveals Antioxidant and Antimicrobial Compounds from Erismadelphus Exsul Bark. Plants 2022, 11, 1505. [Google Scholar] [CrossRef]
- Lang, G.; Mayhudin, N.A.; Mitova, M.I.; Sun, L.; van der Sar, S.; Blunt, J.W.; Cole, A.L.J.; Ellis, G.; Laatsch, H.; Munro, M.H.G. Evolving Trends in the Dereplication of Natural Product Extracts: New Methodology for Rapid, Small-Scale Investigation of Natural Product Extracts. J. Nat. Prod. 2008, 71, 1595–1599. [Google Scholar] [CrossRef]
- Lučić, D.; Pavlović, I.; Brkljačić, L.; Bogdanović, S.; Farkaš, V.; Cedilak, A.; Nanić, L.; Rubelj, I.; Salopek-Sondi, B. Antioxidant and Antiproliferative Activities of Kale (Brassica Oleracea L. Var. Acephala DC.) and Wild Cabbage (Brassica Incana Ten.) Polyphenolic Extracts. Molecules 2023, 28, 1840. [Google Scholar] [CrossRef]
- Jilani, H.; Cilla, A.; Barberá, R.; Hamdi, M. Antiproliferative Activity of Green, Black Tea and Olive Leaves Polyphenols Subjected to Biosorption and in Vitro Gastrointestinal Digestion in Caco-2 Cells. Food Res. Int. 2020, 136, 109317. [Google Scholar] [CrossRef]
- Huang, D.; Jiang, Y.; Chen, W.; Yao, F.; Sun, L. Polyphenols with Anti-Proliferative Activities from Penthorum Chinense Pursh. Molecules 2014, 19, 11045–11055. [Google Scholar] [CrossRef]
- Yanez, J.; Vicente, V.; Alcaraz, M.; Castillo, J.; Benavente-Garcia, O.; Canteras, M.; Teruel, J.A.L. Cytotoxicity and Antiproliferative Activities of Several Phenolic Compounds Against Three Melanocytes Cell Lines: Relationship Between Structure and Activity. Nutr. Cancer 2004, 49, 191–199. [Google Scholar] [CrossRef]
- Otogo N’Nang, E.; Bernadat, G.; Mouray, E.; Kumulungui, B.; Grellier, P.; Poupon, E.; Champy, P.; Beniddir, M.A. Theionbrunonines A and B: Dimeric Vobasine Alkaloids Tethered by a Thioether Bridge from Mostuea brunonis. Org. Lett. 2018, 20, 6596–6600. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Obiang, C.S.; Misso, R.L.N.M.; Atome, G.R.N.; Obame, R.B.M.; Ondo, J.P.; Engonga, L.C.O.; Emvo, E.N. Antimicrobial, Antioxidant, Anti-Inflammatory and Cytotoxic Study of Extracts of Guibourtia Tessmanii (Harms) J. Léonard from Gabon. Clin. Phytosci. 2021, 7, 45. [Google Scholar] [CrossRef]
- Moldovan, C.V.; Savu, M.; Dussert, E.; Aboubacar, H.; Sarbu, L.G.; Matiut, S.; Cudennec, B.; Krier, F.; Ravallec, R.; Birsa, L.M.; et al. Synthetic Flavonoid BrCl-Flav—An Alternative Solution to Combat ESKAPE Pathogens. Antibiotics 2022, 11, 1389. [Google Scholar] [CrossRef]
- Buckingham, J. Dictionary of Natural Products, 1st ed.; Chapman & Hall: London, UK; New York, NY, USA, 1994; ISBN 978-0-412-46620-5. [Google Scholar]
- Baky, M.H.; Kamal, A.M.; Elgindi, M.R.; Haggag, E.G. A Review on Phenolic Compounds from Family Sapotaceae. 8. Available online: https://www.phytojournal.com/archives/2016/vol5issue2/PartD/5-2-15-216.pdf (accessed on 19 February 2023).
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Ma, X.; Zheng, C.; Hu, C.; Rahman, K.; Qin, L. The Genus Desmodium (Fabaceae)-Traditional Uses in Chinese Medicine, Phytochemistry and Pharmacology. J. Ethnopharmacol. 2011, 138, 314–332. [Google Scholar] [CrossRef]
- Metsämuuronen, S.; Sirén, H. Bioactive Phenolic Compounds, Metabolism and Properties: A Review on Valuable Chemical Compounds in Scots Pine and Norway Spruce. Phytochem. Rev. 2019, 18, 623–664. [Google Scholar] [CrossRef]
- Ma, J.; Yang, H.; Basile, M.J.; Kennelly, E.J. Analysis of Polyphenolic Antioxidants from the Fruits of Three Pouteria Species by Selected Ion Monitoring Liquid Chromatography−Mass Spectrometry. J. Agric. Food Chem. 2004, 52, 5873–5878. [Google Scholar] [CrossRef] [PubMed]
- Sarfraz, A.; Javeed, M.; Shah, M.A.; Hussain, G.; Shafiq, N.; Sarfraz, I.; Riaz, A.; Sadiqa, A.; Zara, R.; Zafar, S.; et al. Biochanin A: A Novel Bioactive Multifunctional Compound from Nature. Sci. Total Environ. 2020, 722, 137907. [Google Scholar] [CrossRef] [PubMed]
- Paulo, L.; Ferreira, S.; Gallardo, E.; Queiroz, J.A.; Domingues, F. Antimicrobial Activity and Effects of Resveratrol on Human Pathogenic Bacteria. World J. Microbiol. Biotechnol. 2010, 26, 1533–1538. [Google Scholar] [CrossRef]
- Mattio, L.M.; Dallavalle, S.; Musso, L.; Filardi, R.; Franzetti, L.; Pellegrino, L.; D’Incecco, P.; Mora, D.; Pinto, A.; Arioli, S. Antimicrobial Activity of Resveratrol-Derived Monomers and Dimers against Foodborne Pathogens. Sci. Rep. 2019, 9, 19525. [Google Scholar] [CrossRef] [PubMed]
- Demir, T. Effects of Green Tea Powder, Pomegranate Peel Powder, Epicatechin and Punicalagin Additives on Antimicrobial, Antioxidant Potential and Quality Properties of Raw Meatballs. Molecules 2021, 26, 4052. [Google Scholar] [CrossRef]
- Daglia, M. Polyphenols as Antimicrobial Agents. Curr. Opin. Biotechnol. 2012, 23, 174–181. [Google Scholar] [CrossRef]
- Pei, J.; Yu, H.; Qiu, W.; Mei, J.; Xie, J. Antimicrobial Effect of Epigallocatechin Gallate Against Shewanella Putrefaciens ATCC 8071: A Study Based on Cell Membrane and Biofilm. Curr. Microbiol. 2022, 79, 297. [Google Scholar] [CrossRef]
- Wang, J.; Pan, X.; Han, Y.; Guo, D.; Guo, Q.; Li, R. Rosmarinic Acid from Eelgrass Shows Nematicidal and Antibacterial Activities against Pine Wood Nematode and Its Carrying Bacteria. Mar. Drugs 2012, 10, 2729–2740. [Google Scholar] [CrossRef]
- Iqbal, H.; Wright, C.L.; Jones, S.; da Silva, G.R.; McKillen, J.; Gilmore, B.F.; Kavanagh, O.; Green, B.D. Extracts of Sida Cordifolia Contain Polysaccharides Possessing Immunomodulatory Activity and Rosmarinic Acid Compounds with Antibacterial Activity. BMC Complement. Med. 2022, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Yeo, H.J.; Kwon, M.J.; Han, S.Y.; Jeong, J.C.; Kim, C.Y.; Park, S.U.; Park, C.H. Effects of Carbohydrates on Rosmarinic Acid Production and In Vitro Antimicrobial Activities in Hairy Root Cultures of Agastache Rugosa. Plants 2023, 12, 797. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, X.; Zhang, M.; Bai, B.; Yang, Y.; Fan, S. The Antibacterial Mechanism of Perilla Rosmarinic Acid. Biotechnol. Appl. Biochem. 2022, 69, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Jaisinghani, R.N. Antibacterial Properties of Quercetin. Microbiol. Res. 2017, 8, 6877. [Google Scholar] [CrossRef]
- Chittasupho, C.; Manthaisong, A.; Okonogi, S.; Tadtong, S.; Samee, W. Effects of Quercetin and Curcumin Combination on Antibacterial, Antioxidant, In Vitro Wound Healing and Migration of Human Dermal Fibroblast Cells. Int. J. Mol. Sci. 2022, 23, 142. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.A.; Bhattacharya, D. Antimicrobial Activity of Quercetin: An Approach to Its Mechanistic Principle. Molecules 2022, 27, 2494. [Google Scholar] [CrossRef]
- Podolak, I.; Galanty, A.; Sobolewska, D. Saponins as Cytotoxic Agents: A Review. Phytochem. Rev. 2010, 9, 425–474. [Google Scholar] [CrossRef] [PubMed]
- Kumla, J.; Suwannarach, N.; Tanruean, K.; Lumyong, S. Comparative Evaluation of Chemical Composition, Phenolic Compounds, and Antioxidant and Antimicrobial Activities of Tropical Black Bolete Mushroom Using Different Preservation Methods. Foods 2021, 10, 781. [Google Scholar] [CrossRef] [PubMed]
- Csepregi, R.; Temesfői, V.; Das, S.; Alberti, Á.; Tóth, C.A.; Herczeg, R.; Papp, N.; Kőszegi, T. Cytotoxic, Antimicrobial, Antioxidant Properties and Effects on Cell Migration of Phenolic Compounds of Selected Transylvanian Medicinal Plants. Antioxidants 2020, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Corona, A.V.; Valencia-Espinosa, I.; González-Sánchez, F.A.; Sánchez-López, A.L.; Garcia-Amezquita, L.E.; Garcia-Varela, R. Antioxidant, Anti-Inflammatory and Cytotoxic Activity of Phenolic Compound Family Extracted from Raspberries (Rubus Idaeus): A General Review. Antioxidants 2022, 11, 1192. [Google Scholar] [CrossRef]
- Xie, P.; Cecchi, L.; Bellumori, M.; Balli, D.; Giovannelli, L.; Huang, L.; Mulinacci, N. Phenolic Compounds and Triterpenes in Different Olive Tissues and Olive Oil By-Products, and Cytotoxicity on Human Colorectal Cancer Cells: The Case of Frantoio, Moraiolo and Leccino Cultivars (Olea Europaea L.). Foods 2021, 10, 2823. [Google Scholar] [CrossRef]
- Hsu, Y.-N.; Shyu, H.-W.; Hu, T.-W.; Yeh, J.-P.; Lin, Y.-W.; Lee, L.-Y.; Yeh, Y.-T.; Dai, H.-Y.; Perng, D.-S.; Su, S.-H.; et al. Anti-Proliferative Activity of Biochanin A in Human Osteosarcoma Cells via Mitochondrial-Involved Apoptosis. Food Chem. Toxicol. 2018, 112, 194–204. [Google Scholar] [CrossRef]
- Kole, L.; Giri, B.; Manna, S.K.; Pal, B.; Ghosh, S. Biochanin-A, an Isoflavon, Showed Anti-Proliferative and Anti-Inflammatory Activities through the Inhibition of INOS Expression, P38-MAPK and ATF-2 Phosphorylation and Blocking NFκB Nuclear Translocation. Eur. J. Pharmacol. 2011, 653, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Storniolo, C.E.; Moreno, J.J. Resveratrol Analogs with Antioxidant Activity Inhibit Intestinal Epithelial Cancer Caco-2 Cell Growth by Modulating Arachidonic Acid Cascade. J. Agric. Food Chem. 2019, 67, 819–828. [Google Scholar] [CrossRef]
- Aboushanab, S.A.; Shevyrin, V.A.; Melekhin, V.V.; Andreeva, E.I.; Makeev, O.G.; Kovaleva, E.G. Cytotoxic Activity and Phytochemical Screening of Eco-Friendly Extracted Flavonoids from Pueraria Montana Var. Lobata (Willd.) Sanjappa & Pradeep and Trifolium Pratense L. Flowers Using HPLC-DAD-MS/HRMS. AppliedChem 2023, 3, 119–140. [Google Scholar] [CrossRef]
- Gurgul, A.; Nauman, M.C.; Wu, Z.; Shetye, G.; Ma, R.; Youn, I.; Souliya, O.; Bisson, J.; Johnson, J.J.; Che, C.-T. Chemical Constituents of the Stem of Marsypopetalum Modestum and Their Bioactivities. Nat. Prod. Res. 2022, 2022, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yesil-Celiktas, O.; Sevimli, C.; Bedir, E.; Vardar-Sukan, F. Inhibitory Effects of Rosemary Extracts, Carnosic Acid and Rosmarinic Acid on the Growth of Various Human Cancer Cell Lines. Plant Foods Hum. Nutr. 2010, 65, 158–163. [Google Scholar] [CrossRef] [PubMed]
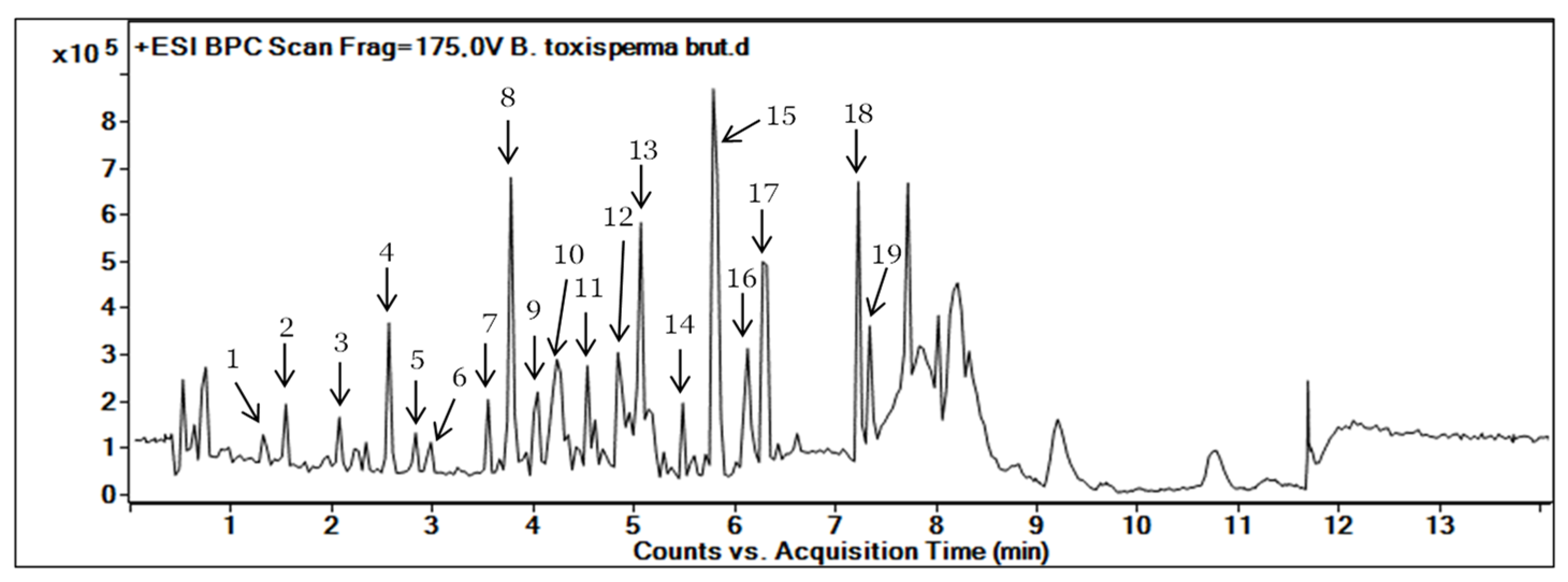
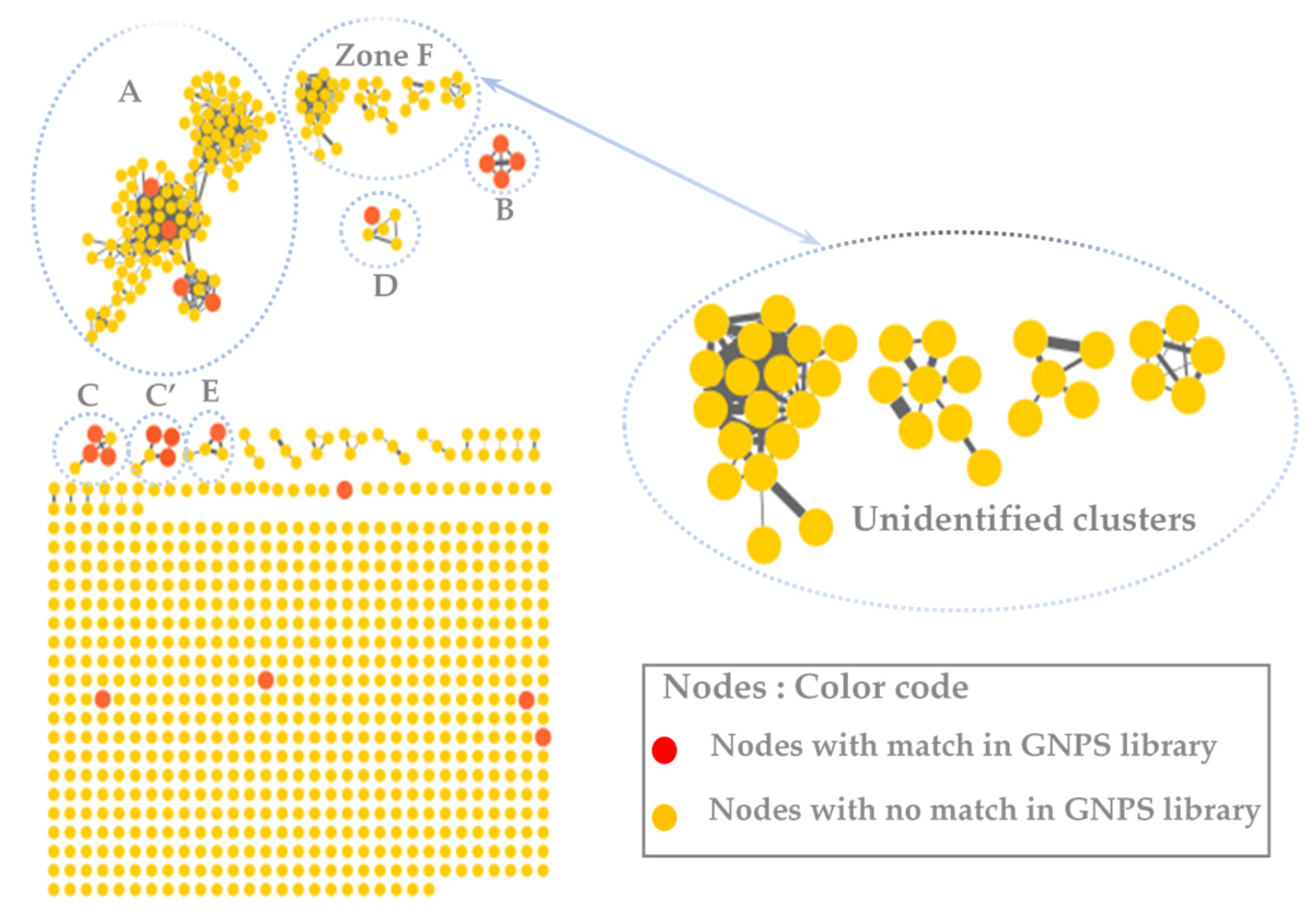
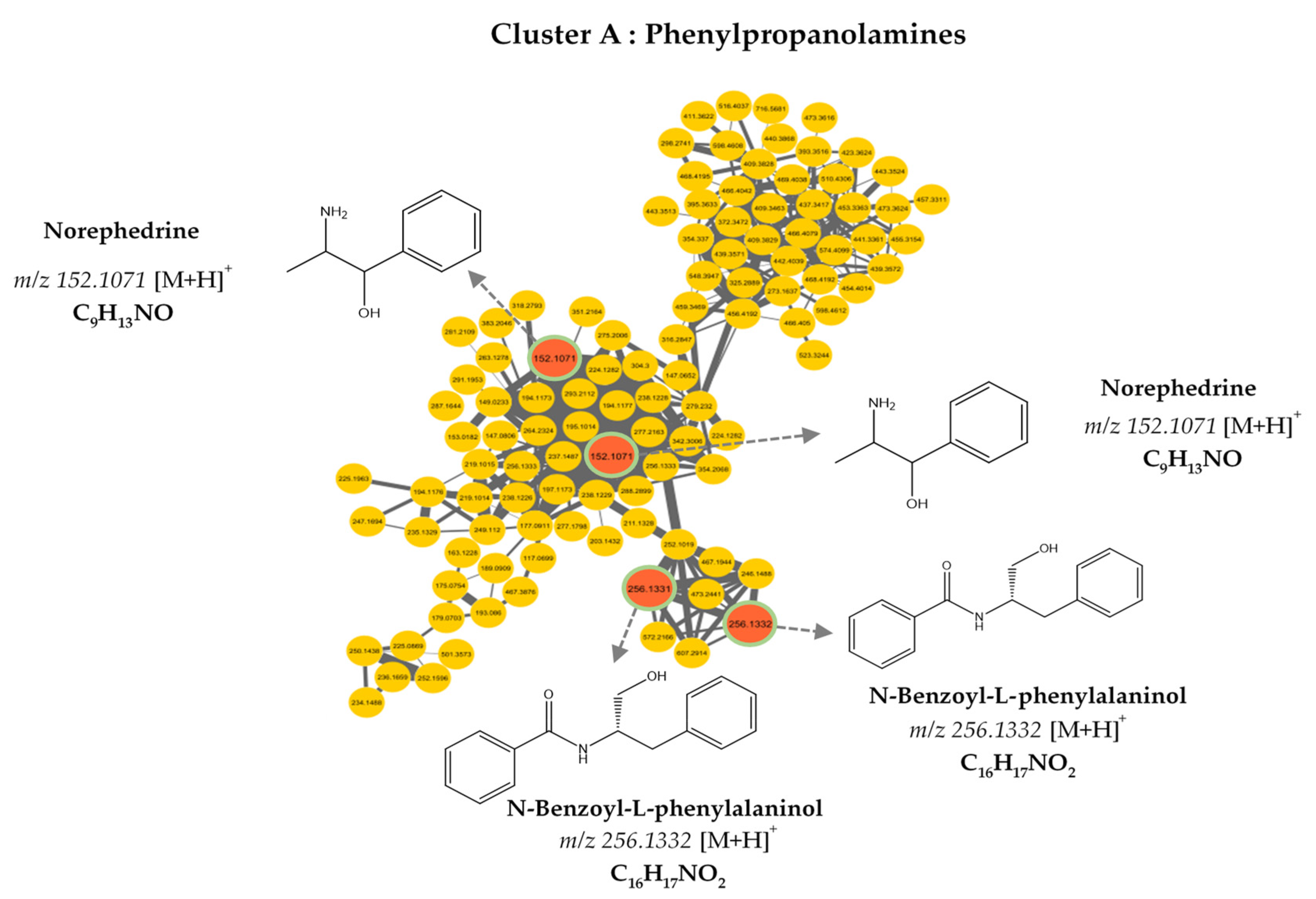
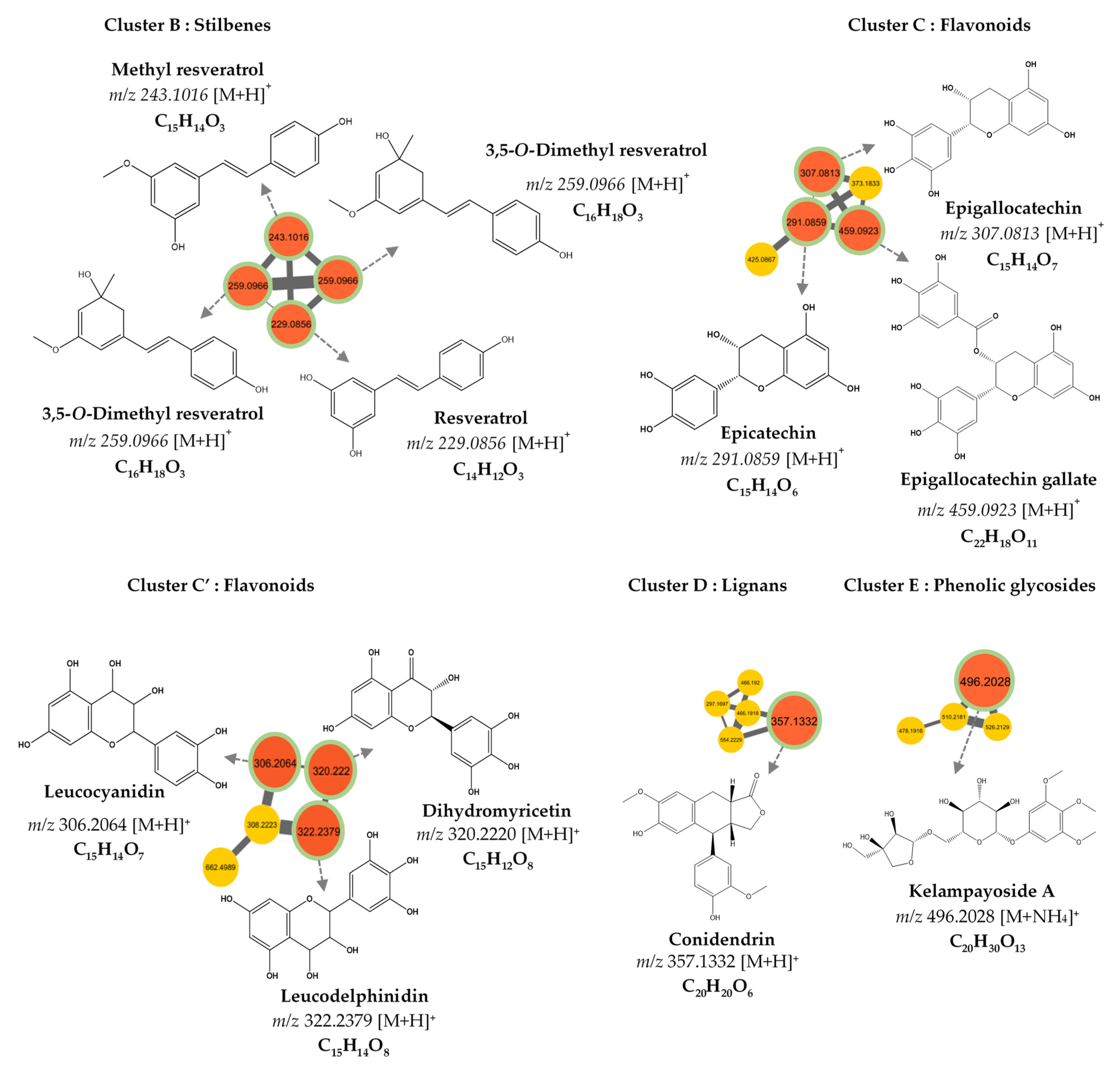

| Compound Number | RT (min) | MS (m/z) [M+H]+ | Molecular Formula | Fragments | Proposed Identification | Sources | Confidence Level |
|---|---|---|---|---|---|---|---|
| 1 | 1.450 | 307.081 | C15H14O7 | 307.081, 165.070 | Epigallocatechin | PubChem, DNP | 3 |
| 2 | 1.701 | 185.084 | C9H12O4 | 185.081, 123.007 | Unknown | - | 4 |
| 3 | 1.710 | 496.203 | C20H30O13 | 496.203, 295.097, 327.123 | Kelampayoside A | PubChem, ChemSpider | 3 |
| 4 | 1.938 | 459.092 | C22H18O11 | 459.092, 165.069 | Epigallocatechin gallate | PubChem, DNP | 3 |
| 5 | 2.830 | 289.071 | C14H6N7O | - | Unknown | - | 4 |
| 6 | 2.851 | 255.065 | C15H10O4 | - | Unknown | - | 4 |
| 7 | 3.550 | 229.086 | C14H12O3 | 229.086, 165.070 | Resveratrol | DNP | 3 |
| 8 | 3.777 | 357.133 | C20H20O6 | 357.133, 295.097, 327.123 | β-Conidendrin | PubChem | 3 |
| 9 | 3.427 | 259.097 | C16H18O3 | 259.097, 152.062, 165.070 | 3,5-O-Dimethyl resveratrol | PubChem, DNP | 3 |
| 10 | 4.042 | 243.102 | C15H14O3 | 243.102, 165.069 | Methyl resveratrol | PubChem | 3 |
| 11 | 4.233 | 420.186 | C21H24O9 | - | Isorhapontin | PubChem | 3 |
| 12 | 4.536 | 316.285 | C18H37NO3 | - | Unknown | - | 4 |
| 13 | 5.066 | 453.337 | C30H44O3 | 453.337, 305.159, 213.165 | Unknown | - | 4 |
| 14 | 5.483 | 194.117 | C11H15NO2 | 194.117, 163.210 | Unknown | - | 4 |
| 15 | 5.786 | 238.123 | C16H15NO | - | Unknown | - | 4 |
| 16 | 6.127 | 452.322 | C22H45NO8 | 452.322, 294.700, 155.070 | Unknown | - | 4 |
| 17 | 6.278 | 301.141 | C16H18N3O3 | - | Unknown | - | 4 |
| 18 | 7.225 | 487.343 | C31H42N4O | - | Unknown | - | 4 |
| 19 | 7.339 | 719.306 | C39H40N7O7 | - | Unknown | - | 4 |
| MS (m/z) [M+H]+ | Molecular Formula | Parent Ion | Cosine Score | Compound Name |
|---|---|---|---|---|
| 307.081 | C15H14O7 | [M+H]+ | 0.74 | Epigallocatechin |
| 291.086 | C15H14O6 | [M+H]+ | 0.88 | Epicatechin |
| 459.092 | C22H18O11 | [M+H]+ | 0.78 | Epigallocatechin gallate |
| 496.203 | C20H30O13 | [M+NH4]+ | 0.83 | Kelampayoside A |
| 306.2064 | C15H14O7 | [M+H]+ | 0.70 | Leucocyanidin |
| 322.2379 | C15H14O8 | [M+H]+ | 0.75 | Leucodelphinidin |
| 320.2220 | C15H12O8 | [M+H]+ | 0.81 | Dihydromyricetin |
| 357.133 | C20H20O6 | [M+H]+ | 0.79 | Conidendrin |
| 259.097 | C16H18O3 | [M+H]+ | 0.76 | 3,5-O-Dimethyl Resveratrol |
| 243.102 | C15H14O3 | [M+H]+ | 0.72 | Methyl Resveratrol |
| 229.086 | C14H12O3 | [M+H]+ | 0.97 | Resveratrol |
| 152.107 | C9H13NO | [M+H]+ | 0.94 | 2-Amino-1-phenylpropan-1-ol |
| 256.133 | C16H17NO2 | [M+H]+ | 0.93 | N-Benzoyl-L-phenylalaninol |
| 652.115 | C27H22O18 | [M+NH4]+ | 0.96 | Corilagin |
| 255.065 | C15H10O4 | [M+H]+ | 0.81 | 4’,7-Dihydroxyflavone |
| 438.176 | C21H24O9 | [M+NH4]+ | 0.77 | Isorhapontin |
| 285.075 | C16H12O5 | [M+H]+ | 0.80 | Biochanin A |
| Inhibition-Zone Diameter (IZD, mm) | |||||
|---|---|---|---|---|---|
| Sample | Bacterial Strains | ||||
| E. coli ATCC 25922 | E. coli ATCC 8739 | K.p MDR | S. enterica | E. coli ESBL | |
| Bt EtOH Ce | 15 ± 0.00 | 16 ± 0.47 | 20 ± 0.00 | 14 ± 0.47 | 22 ± 0.82 |
| Bt EtOH F1 | 12 ± 0.00 | 13 ± 0.00 | 10 ± 0.00 | Na | 9.33 ± 0.47 |
| Bt EtOH F2 | 13.33 ± 0.94 | 14.67 ± 0.47 | 21.33 ± 0.47 | 16 ± 0.82 | 19.67 ± 0.47 |
| Bt EtOH F3 | 11 ± 0.82 | 11.33 ± 0.94 | 10.33 ± 0.47 | Na | 11.33 ± 0.94 |
| Bt EtOH F4 | 20 ± 0.82 | 12.33 ± 0.47 | 14.33 ± 0.47 | 18 ± 0.47 | 11.67 ± 0.47 |
| Bt EtOH F5 | 9 ± 0.00 | 10.33 ± 0.47 | 10.67 ± 0.47 | 11 ± 0.00 | 10 ± 0.82 |
| Bt EtOH F6 | 10 ± 0.00 | Na | 9 ± 0.00 | 10.67 ± 0.47 | Na |
| Bt EtOH F7 | 9 ± 0.00 | Na | Na | 10 ± 0.00 | Na |
| Ticarcillin | 19.33 ± 0.47 | 21.33 ± 0.47 | 20 ± 0.00 | 20 ± 0.00 | Na |
| Gentamicin | 30 ± 0.00 | 23 ± 0.00 | 21 ± 0.00 | 22.33 ± 0.47 | 22 ± 0.00 |
| Tetracycline | Na | Na | 12 ± 0.00 | Na | Na |
| 1% DMSO | - | - | - | - | - |
| MICs and MBCs (mg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | E. coli ATCC 25922 | E. coli ATCC 8739 | K.p MDR | S. enterica | E. coli ESBL | |||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| Bt EtOH Ce | 0.62 | 2.5 | 1.25 | 5 | 1.25 | 5 | 1.25 | 5 | 1.25 | 5 |
| Bt EtOH F1 | 1.25 | >5 | 1.25 | 5 | 0.31 | 2.5 | Nt | Nt | 2.5 | >5 |
| Bt EtOH F2 | 1.25 | 2.5 | 0.62 | 1.25 | 1.25 | 5 | 1.25 | 2.5 | 1.25 | 5 |
| Bt EtOH F3 | 2.5 | 5 | 1.25 | 5 | 1.25 | 5 | >5 | >5 | 2.5 | 5 |
| Bt EtOH F4 | 2.5 | 5 | 1.25 | 2.5 | 1.25 | 2.5 | 2.5 | 5 | 1.25 | 2.5 |
| Bt EtOH F5 | 5 | >5 | 2.5 | >5 | 0.62 | 5 | 5 | >5 | 5 | >5 |
| Bt EtOH F6 | 1.25 | >5 | Nt | Nt | 5 | >5 | 2.5 | 5 | Nt | Nt |
| Bt EtOH F7 | 5 | >5 | Nt | Nt | Nt | Nt | 2.5 | >5 | Nt | Nt |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Essono Mintsa, M.; Kumulungui, B.S.; Obiang, C.S.; Dussert, E.; Choque, E.; Herfurth, D.; Ravallec, R.; Ondo, J.-P.; Mesnard, F. Cytotoxicity and Identification of Antibacterial Compounds from Baillonella toxisperma Bark Using a LC-MS/MS and Molecular Networking Approach. Metabolites 2023, 13, 599. https://doi.org/10.3390/metabo13050599
Essono Mintsa M, Kumulungui BS, Obiang CS, Dussert E, Choque E, Herfurth D, Ravallec R, Ondo J-P, Mesnard F. Cytotoxicity and Identification of Antibacterial Compounds from Baillonella toxisperma Bark Using a LC-MS/MS and Molecular Networking Approach. Metabolites. 2023; 13(5):599. https://doi.org/10.3390/metabo13050599
Chicago/Turabian StyleEssono Mintsa, Morel, Brice Serge Kumulungui, Cédric Sima Obiang, Elodie Dussert, Elodie Choque, Damien Herfurth, Rozenn Ravallec, Joseph-Privat Ondo, and François Mesnard. 2023. "Cytotoxicity and Identification of Antibacterial Compounds from Baillonella toxisperma Bark Using a LC-MS/MS and Molecular Networking Approach" Metabolites 13, no. 5: 599. https://doi.org/10.3390/metabo13050599
APA StyleEssono Mintsa, M., Kumulungui, B. S., Obiang, C. S., Dussert, E., Choque, E., Herfurth, D., Ravallec, R., Ondo, J.-P., & Mesnard, F. (2023). Cytotoxicity and Identification of Antibacterial Compounds from Baillonella toxisperma Bark Using a LC-MS/MS and Molecular Networking Approach. Metabolites, 13(5), 599. https://doi.org/10.3390/metabo13050599





