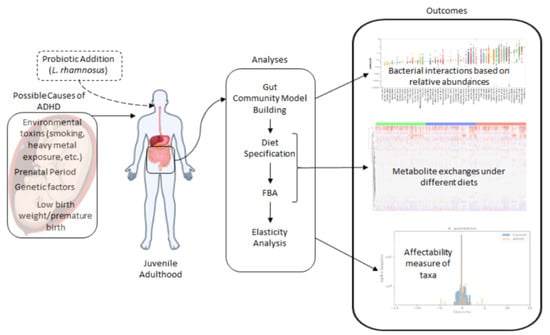
Understanding the ADHD-Gut Axis by Metabolic Network Analysis
Abstract
1. Introduction
| Neurotransmitter Precursors Found in the Human GI Tract | Neurotransmitters | Genus | References |
|---|---|---|---|
| L-Glutamate | GABA | Lactobacillus, Bifidobacterium, Bacteroides, Parabacteroides, Escherichia | [32,33] |
| L-tryptophan | Serotonin | Streptococcus, Escherichia, Enterococcus, Lactococcus, Lactobacillus | [34,35] |
| L-tyrosine, Phenylalanine | Norepinephrine | Escherichia, Bacillus | [35,36] |
| L-tyrosine, Phenylalanine | Dopamine | Escherichia, Bacillus, Lactococcus, Lactobacillus, Streptococcus | [34,35,36] |
| Choline | Acetylcholine | Lactobacillus, Bacillus | [37,38,39,40] |
| L-histidine | Histamine | Lactobacillus, Lactococcus, Streptococcus, Enterococcus | [41,42,43] |
2. Materials and Methods
2.1. Data
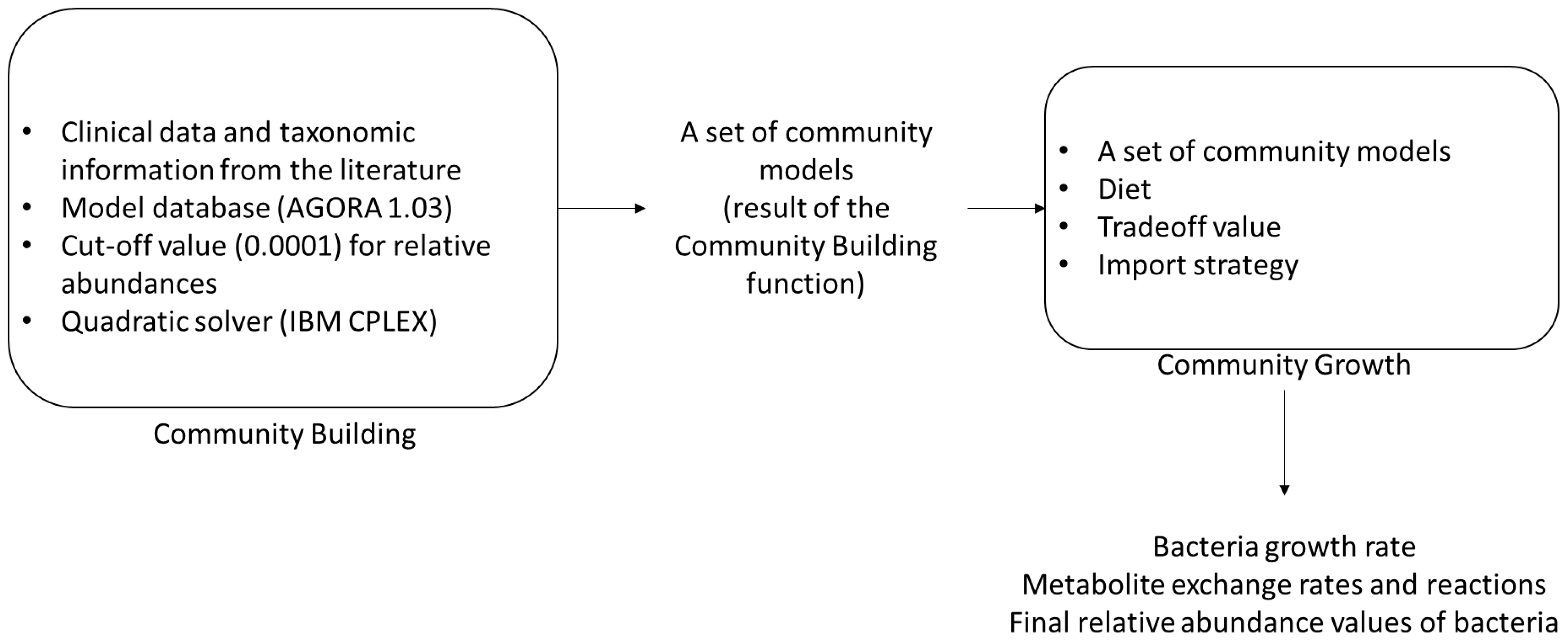
2.2. Reconstruction of Community Model
2.3. Diversity Analyses
2.4. Elasticities
2.5. Probiotic Addition
3. Results
3.1. Abundances
3.2. Diversity
3.3. Production of Short-Chain Fatty Acids and Minor Metabolites (Amino Acids) by Gut Microbiome
3.4. Production of Precursors of Key Neurotransmitters in the Gut
3.5. Elasticities
3.6. Intervention by a Probiotic (L. rhamnosus)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Polanczyk, G.V.; Salum, G.A.; Sugaya, L.S.; Caye, A.; Rohde, L.A. Annual research review: A meta-analysis of the worldwide prevalence of mental disorders in children and adolescents. J. Child Psychol. Psychiatry 2015, 56, 345–365. [Google Scholar] [CrossRef] [PubMed]
- Faraone, S.V.; Biederman, J.; Mick, E. The age-dependent decline of attention deficit hyperactivity disorder: A meta-analysis of follow-up studies. Psychol. Med. 2006, 36, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Barkley, R.A.; Fischer, M.; Edelbrock, C.S.; Smallish, L. The adolescent outcome of hyperactive children diagnosed by research criteria: I. An 8-year prospective follow-up study. J. Am. Acad. Child Adolesc. Psychiatry 1990, 29, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Biederman, J.; Faraone, S.; Milberger, S.; Curtis, S.; Chen, L.; Marrs, A.; Ouellette, C.; Moore, P.; Spencer, T. Predictors of persistence and remission of ADHD into adolescence: Results from a four-year prospective follow-up study. J. Am. Acad. Child Adolesc. Psychiatry 1996, 35, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Rucklidge, J.J. Gender differences in attention-deficit/hyperactivity disorder. Psychiatr. Clin. 2010, 33, 357–373. [Google Scholar] [CrossRef]
- Tripp, G.; Wickens, J.R. Neurobiology of ADHD. Neuropharmacology 2009, 57, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Swanson, J.M.; Kinsbourne, M.; Nigg, J.; Lanphear, B.; Stefanatos, G.A.; Volkow, N.; Taylor, E.; Casey, B.; Castellanos, F.X.; Wadhwa, P.D. Etiologic subtypes of attention-deficit/hyperactivity disorder: Brain imaging, molecular genetic and environmental factors and the dopamine hypothesis. Neuropsychol. Rev. 2007, 17, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Surette, M.; Bercik, P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 2012, 10, 735–742. [Google Scholar] [CrossRef]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Neufeld, K.; Kang, N.; Bienenstock, J.; Foster, J.A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011, 23, 255-e119. [Google Scholar] [CrossRef]
- Lyte, M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: Microbial endocrinology in the design and use of probiotics. Bioessays 2011, 33, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, P.; Kunze, W.A. Voices from within: Gut microbes and the CNS. Cell. Mol. Life Sci. 2013, 70, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.; Gibson, G.; Cummings, J. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 1992, 72, 57–64. [Google Scholar]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA: Acetate CoA-transferase gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014, 8, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. MBio 2014, 5, e00889-14. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Tamboli, C.P.; Neut, C.; Desreumaux, P.; Colombel, J.F. Dysbiosis in inflammatory bowel disease. Gut 2004, 53, 1–4. [Google Scholar] [CrossRef]
- Moos, W.H.; Faller, D.V.; Harpp, D.N.; Kanara, I.; Pernokas, J.; Powers, W.R.; Steliou, K. Microbiota and neurological disorders: A gut feeling. BioResearch Open Access 2016, 5, 137–145. [Google Scholar] [CrossRef]
- Zou, R.; Tian, P.; Xu, M.; Zhu, H.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. Psychobiotics as a novel strategy for alleviating anxiety and depression. J. Funct. Foods 2021, 86, 104718. [Google Scholar] [CrossRef]
- Checa-Ros, A.; Jeréz-Calero, A.; Molina-Carballo, A.; Campoy, C.; Muñoz-Hoyos, A. Current evidence on the role of the gut microbiome in ADHD pathophysiology and therapeutic implications. Nutrients 2021, 13, 249. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Lutgendorff, F.; Akkermans, L.; Soderholm, J.D. The role of microbiota and probiotics in stress-induced gastrointestinal damage. Curr. Mol. Med. 2008, 8, 282–298. [Google Scholar] [CrossRef]
- Szopinska-Tokov, J.; Dam, S.; Naaijen, J.; Konstanti, P.; Rommelse, N.; Belzer, C.; Buitelaar, J.; Franke, B.; Bloemendaal, M.; Aarts, E.; et al. Investigating the gut microbiota composition of individuals with attention-deficit/hyperactivity disorder and association with symptoms. Microorganisms 2020, 8, 406. [Google Scholar] [CrossRef] [PubMed]
- Bull-Larsen, S.; Mohajeri, M.H. The potential influence of the bacterial microbiome on the development and progression of ADHD. Nutrients 2019, 11, 2805. [Google Scholar] [CrossRef]
- MacFabe, D.F. Enteric short-chain fatty acids: Microbial messengers of metabolism, mitochondria, and mind: Implications in autism spectrum disorders. Microb. Ecol. Health Dis. 2015, 26, 28177. [Google Scholar] [CrossRef]
- MacFabe, D.F.; Cain, N.E.; Boon, F.; Ossenkopp, K.P.; Cain, D.P. Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: Relevance to autism spectrum disorder. Behav. Brain Res. 2011, 217, 47–54. [Google Scholar] [CrossRef]
- Wall, R.; Cryan, J.F.; Ross, R.P.; Fitzgerald, G.F.; Dinan, T.G.; Stanton, C. Bacterial neuroactive compounds produced by psychobiotics. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2014; pp. 221–239. [Google Scholar]
- Barrett, E.; Ross, R.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Özogul, F. Effects of specific lactic acid bacteria species on biogenic amine production by foodborne pathogen. Int. J. Food Sci. Technol. 2011, 46, 478–484. [Google Scholar] [CrossRef]
- Shishov, V.; Kirovskaya, T.; Kudrin, V.; Oleskin, A. Amine neuromediators, their precursors, and oxidation products in the culture of Escherichia coli K-12. Appl. Biochem. Microbiol. 2009, 45, 494–497. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Botvinko, I.; Kudrin, V.; Oleskin, A. Detection of neurotransmitter amines in microorganisms with the use of high-performance liquid chromatography. Dokl. Biochem. 2000, 372, 115–117. [Google Scholar]
- Kawashima, K.; Misawa, H.; Moriwaki, Y.; Fujii, Y.X.; Fujii, T.; Horiuchi, Y.; Yamada, T.; Imanaka, T.; Kamekura, M. Ubiquitous expression of acetylcholine and its biological functions in life forms without nervous systems. Life Sci. 2007, 80, 2206–2209. [Google Scholar] [CrossRef]
- Marquardt, P.; Spitznagel, G. Bakterielle acetylcholine bildung in kunstlichen Nahrboden. Arzneimittelforschung 1959, 9, 456–465. [Google Scholar]
- Girvin, G.T.; Stevenson, J. Cell free “choline acetylase” from Lactobacillus plantarum. Can. J. Biochem. Physiol. 1954, 32, 131–146. [Google Scholar] [CrossRef]
- Horiuchi, Y.; Kimura, R.; Kato, N.; Fujii, T.; Seki, M.; Endo, T.; Kato, T.; Kawashima, K. Evolutional study on acetylcholine expression. Life Sci. 2003, 72, 1745–1756. [Google Scholar] [CrossRef]
- Thomas, C.M.; Hong, T.; Van Pijkeren, J.P.; Hemarajata, P.; Trinh, D.V.; Hu, W.; Britton, R.A.; Kalkum, M.; Versalovic, J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 2012, 7, e31951. [Google Scholar] [CrossRef]
- Landete, J.M.; De Las Rivas, B.; Marcobal, A.; Munoz, R. Updated molecular knowledge about histamine biosynthesis by bacteria. Crit. Rev. Food Sci. Nutr. 2008, 48, 697–714. [Google Scholar] [CrossRef]
- Coton, E.; Rollan, G.; Bertrand, A.; Lonvaud-Funel, A. Histamine-producing lactic acid bacteria in wines: Early detection, frequency, and distribution. Am. J. Enol. Vitic. 1998, 49, 199–204. [Google Scholar] [CrossRef]
- Aarts, E.; Ederveen, T.H.A.; Naaijen, J.; Zwiers, M.P.; Boekhorst, J.; Timmerman, H.M.; Smeekens, S.P.; Netea, M.G.; Buitelaar, J.K.; Franke, B.; et al. Gut microbiome in ADHD and its relation to neural reward anticipation. PLoS ONE 2017, 12, e0183509. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, S.; Heinken, A.; Kutt, L.; Ravcheev, D.A.; Bauer, E.; Noronha, A.; Greenhalgh, K.; Jäger, C.; Baginska, J.; Wilmes, P.; et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017, 35, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Gibbons, S.M.; Resendis-Antonio, O.; Chia, N. MICOM: Metagenome-scale modeling to infer metabolic interactions in the gut microbiota. mSystems 2020, 5, e00606-19. [Google Scholar] [CrossRef]
- IBM Corp. IBM ILOG CPLEX Optimization Studio. CPLEX User’s Manual; Version 12; IBM Corp.: Armonk, NY, USA, 1987. [Google Scholar]
- Virtual Metabolic Human Unhealthy Diet. Available online: https://www.vmh.life/#diet/Unhealthy (accessed on 30 September 2010).
- Neal, E.G.; Chaffe, H.; Schwartz, R.H.; Lawson, M.S.; Edwards, N.; Fitzsimmons, G.; Whitney, A.; Cross, J.H. The ketogenic diet for the treatment of childhood epilepsy: A randomised controlled trial. Lancet Neurol. 2008, 7, 500–506. [Google Scholar]
- Virtual Metabolic Human High-Fat, Low Carb Diet. Available online: https://www.vmh.life/#diet/High%20fat,%20low%20carb (accessed on 30 September 2010).
- Virtual Metabolic Human Vegan Diet. Available online: https://www.vmh.life/#diet/Vegan (accessed on 30 September 2010).
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef]
- Pärtty, A.; Kalliomäki, M.; Wacklin, P.; Salminen, S.; Isolauri, E. A possible link between early probiotic intervention and the risk of neuropsychiatric disorders later in childhood: A randomized trial. Pediatr. Res. 2015, 77, 823–828. [Google Scholar] [CrossRef]
- Wang, L.J.; Yang, C.Y.; Chou, W.J.; Lee, M.J.; Chou, M.C.; Kuo, H.C.; Yeh, Y.M.; Lee, S.Y.; Huang, L.H.; Li, S.C. Gut microbiota and dietary patterns in children with attention-deficit/hyperactivity disorder. Eur Child Adolesc Psychiatry 2020, 29, 287–297. [Google Scholar] [CrossRef]
- Prehn-Kristensen, A.; Zimmermann, A.; Tittmann, L.; Lieb, W.; Schreiber, S.; Baving, L.; Fischer, A. Reduced microbiome alpha diversity in young patients with ADHD. PLoS ONE 2018, 13, e0200728. [Google Scholar] [CrossRef]
- Stevens, A.J.; Purcell, R.V.; Darling, K.A.; Eggleston, M.J.F.; Kennedy, M.A.; Rucklidge, J.J. Human gut microbiome changes during a 10 week Randomised Control Trial for micronutrient supplementation in children with attention deficit hyperactivity disorder. Sci. Rep. 2019, 9, 10128. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Ge, W.R.; Zhang, S.; Sun, Y.L.; Wang, B.; Yang, G. Case-control study of the effects of gut microbiota composition on neurotransmitter metabolic pathways in children with attention deficit hyperactivity disorder. Front. Neurosci. 2020, 14, 127. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Han, B.; Ding, M.; Wen, Y.; Ma, M.; Zhang, L.; Qi, X.; Cheng, B.; Li, P.; Kafle, O.P.; et al. Identifying psychiatric disorder-associated gut microbiota using microbiota-related gene set enrichment analysis. Brief. Bioinform. 2020, 21, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Y.; Zhou, Y.Y.; Zhou, G.L.; Li, Y.C.; Yuan, J.; Li, X.H.; Ruan, B. Gut microbiota profiles in treatment-naïve children with attention deficit hyperactivity disorder. Behav. Brain Res. 2018, 347, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Thukral, A.K. A review on measurement of Alpha diversity in biology. Agric. Res. J. 2017, 54, 1–10. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Noronha, A.; Modamio, J.; Jarosz, Y.; Guerard, E.; Sompairac, N.; Preciat, G.; Daníelsdóttir, A.D.; Krecke, M.; Merten, D.; Haraldsdóttir, H.S.; et al. The Virtual Metabolic Human database: Integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019, 47, D614–D624. [Google Scholar] [CrossRef]
- Downes, J.; Munson, M.; Wade, W. Dialister invisus sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2003, 53, 1937–1940. [Google Scholar] [CrossRef]
- Kalenik, A.; Kardaś, K.; Rahnama, A.; Sirojć, K.; Wolańczyk, T. Gut microbiota and probiotic therapy in ADHD: A review of current knowledge. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 110, 110277. [Google Scholar] [CrossRef]
- Mangia, S.; Giove, F.; DiNuzzo, M. Metabolic pathways and activity-dependent modulation of glutamate concentration in the human brain. Neurochem. Res. 2012, 37, 2554–2561. [Google Scholar] [CrossRef]
- De Wit, S.; Standing, H.R.; DeVito, E.E.; Robinson, O.J.; Ridderinkhof, K.R.; Robbins, T.W.; Sahakian, B.J. Reliance on habits at the expense of goal-directed control following dopamine precursor depletion. Psychopharmacology 2012, 219, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Meyers, S. Use of neurotransmitter precursors for treatment of depression. Altern. Med. Rev. 2000, 5, 64–71. [Google Scholar] [PubMed]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef]
- Verlaet, A.A.; Noriega, D.B.; Hermans, N.; Savelkoul, H.F. Nutrition, immunological mechanisms and dietary immunomodulation in ADHD. Eur. Child Adolesc. Psychiatry 2014, 23, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, H.W.; Kephart, W.C.; Holland, A.M.; Mumford, P.; Mobley, C.B.; Lowery, R.P.; Roberts, M.D.; Wilson, J.M.; Kavazis, A.N. A ketogenic diet in rodents elicits improved mitochondrial adaptations in response to resistance exercise training compared to an isocaloric western diet. Front. Physiol. 2016, 7, 533. [Google Scholar] [CrossRef]
- Darand, M.; Hassanizadeh, S.; Martami, F.; Shareghfarid, E.; Hosseinpour-Niazi, S.; Hosseinzadeh, M. A plant-based dietary score and attention deficit/hyperactivity disorder in Iranian children: A case-control study. J. Affect. Disord. 2022, 313, 27–31. [Google Scholar] [CrossRef]
- Rautio, M.; Eerola, E.; Väisänen-Tunkelrott, M.L.; Molitoris, D.; Lawson, P.; Collins, M.D.; Jousimies-Somer, H. Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a new genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and description of Alistipes finegoldii sp. nov., from human sources. Syst. Appl. Microbiol. 2003, 26, 182–188. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clinical microbiology reviews 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Reid, G. When microbe meets human. Clin. Infect. Dis. 2004, 39, 827–830. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [PubMed]
- Karbownik, M.; Reiter, R.J.; Garcia, J.J.; Cabrera, J.; Burkhardt, S.; Osuna, C.; Lewiński, A. Indole-3-propionic acid, a melatonin-related molecule, protects hepatic microsomal membranes from iron-induced oxidative damage: Relevance to cancer reduction. J. Cell. Biochem. 2001, 81, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.K.; Yoo, K.Y.; Li, H.; Park, O.K.; Lee, C.H.; Choi, J.H.; Jeong, Y.G.; Lee, Y.L.; Kim, Y.M.; Kwon, Y.G.; et al. Indole-3-propionic acid attenuates neuronal damage and oxidative stress in the ischemic hippocampus. J. Neurosci. Res. 2009, 87, 2126–2137. [Google Scholar] [CrossRef]
- Chyan, Y.J.; Poeggeler, B.; Omar, R.A.; Chain, D.G.; Frangione, B.; Ghiso, J.; Pappolla, M.A. Potent neuroprotective properties against the Alzheimer β-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J. Biol. Chem. 1999, 274, 21937–21942. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and their molecular communication with the immune system. Front. Microbiol. 2017, 8, 2345. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Z.P.; Liu, C.Q.; Qi, L.; Sheng, Y.; Zou, D.J. Modulation of the gut microbiome: A systematic review of the effect of bariatric surgery. Eur. J. Endocrinol. 2018, 178, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, Y.; Gao, H.; Li, D.; Jiang, R.; Ge, L.; Tong, C.; Xu, K. Associations among dietary omega-3 polyunsaturated fatty acids, the gut microbiota, and intestinal immunity. Mediat. Inflamm. 2021, 2021, 8879227. [Google Scholar] [CrossRef]
- Ezaki, T. Coprococcus. In Bergey’s Manual of Systematics of Archaea and Bacteria; JohnWiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 1–3. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Taras, D.; Simmering, R.; Collins, M.D.; Lawson, P.A.; Blaut, M. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2002, 52, 423–428. [Google Scholar]
- Avgustin, G.; Wallace, R.J.; Flint, H.J. Phenotypic diversity among Ruminai isolates of Prevotella ruminicola: Proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int. J. Syst. Evol. Microbiol. 1997, 47, 284–288. [Google Scholar] [CrossRef]
- Hayashi, H.; Shibata, K.; Sakamoto, M.; Tomita, S.; Benno, Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2007, 57, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Holdeman, L.V.; Moore, W.E.C.; Churn, P.J.; Johnson, J.L. Bacteroides oris and Bacteroides buccae new species from human periodontitis and other human infections. Int. J. Syst. Evol. Microbiol. 1982, 32, 125–131. [Google Scholar] [CrossRef]
- Moore, W.E.C.; Cato, E.P.; Holdeman, L.V. Ruminococcus bromii sp. n. and emendation of the description of Ruminococcus sijpestein. Int. J. Syst. Evol. Microbiol. 1972, 22, 78–80. [Google Scholar] [CrossRef]
- Van Hul, M.; Le Roy, T.; Prifti, E.; Dao, M.C.; Paquot, A.; Zucker, J.D.; Delzenne, N.M.; Muccioli, G.G.; Clément, K.; Cani, P.D. From correlation to causality: The case of Subdoligranulum. Gut Microbes 2020, 12, 1849998. [Google Scholar] [CrossRef]
- Holmstrøm, K.; Collins, M.D.; Møller, T.; Falsen, E.; Lawson, P.A. Subdoligranulum variabile gen. nov., sp. nov. from human feces. Anaerobe 2004, 10, 197–203. [Google Scholar] [CrossRef]
- Gutiérrez-Repiso, C.; Moreno-Indias, I.; Tinahones, F.J. Shifts in gut microbiota and their metabolites induced by bariatric surgery. Impact of factors shaping gut microbiota on bariatric surgery outcomes. Rev. Endocr. Metab. Disord. 2021, 22, 1137–1156. [Google Scholar] [CrossRef]
- Geerlings, S.Y.; Kostopoulos, I.; De Vos, W.M.; Belzer, C. Akkermansia muciniphila in the human gastrointestinal tract: When, where, and how? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef]
- Rainey, F.; Kämpfer, P.; Trujillo, M.; Chun, J.; DeVos, P.; Hedlund, B.; Dedysh, S. Bergey’s manual of systematics of archaea and bacteria. In Association with Bergey’s Manual Trust; Wiley Online Library: Hoboken, NJ, USA, 2015; pp. 1–164. [Google Scholar]
- Cristol, D.A.; Armstrong, J.L.; Whitaker, J.M.; Forsyth, M.H. Feather-degrading bacteria do not affect feathers on captive birds. Auk 2005, 122, 222–230. [Google Scholar] [CrossRef]
- Liu, X.; Yan, H.; Lv, L.; Xu, Q.; Yin, C.; Zhang, K.; Wang, P.; Hu, J. Growth performance and meat quality of broiler chickens supplemented with Bacillus licheniformis in drinking water. Asian-Australas. J. Anim. Sci. 2012, 25, 682. [Google Scholar] [CrossRef]
- Deng, W.; Dong, X.; Tong, J.; Zhang, Q. The probiotic Bacillus licheniformis ameliorates heat stress-induced impairment of egg production, gut morphology, and intestinal mucosal immunity in laying hens. Poult. Sci. 2012, 91, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Muras, A.; Romero, M.; Mayer, C.; Otero, A. Biotechnological applications of Bacillus licheniformis. Crit. Rev. Biotechnol. 2021, 41, 609–627. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Kim, W.S.; Paik, H.D. Bacillus strains as human probiotics: Characterization, safety, microbiome, and probiotic carrier. Food Sci. Biotechnol. 2019, 28, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Sorokulova, I.; Reva, O.; Smirnov, V.; Pinchuk, I.; Lapa, S.; Urdaci, M. Genetic diversity and involvement in bread spoilage of Bacillus strains isolated from flour and ropy bread. Lett. Appl. Microbiol. 2003, 37, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, R.; Chauhan, K.; Seale, R.B.; Deeth, H.C.; Pillidge, C.J.; Powell, I.B.; Craven, H.; Turner, M.S. Genotyping of dairy Bacillus licheniformis isolates by high resolution melt analysis of multiple variable number tandem repeat loci. Food Microbiol. 2013, 34, 344–351. [Google Scholar] [CrossRef]
- Bernalier, A.; Willems, A.; Leclerc, M.; Rochet, V.; Collins, M.D. Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch. Microbiol. 1996, 166, 176–183. [Google Scholar] [CrossRef]
- Intra, J.; Milano, A.; Sarto, C.; Brambilla, P. A rare case of Clostridium paraputrificum bacteremia in a 78-year-old Caucasian man diagnosed with an intestinal neoplasm. Anaerobe 2020, 66, 102292. [Google Scholar] [CrossRef]
- Shinha, T.; Hadi, C. Clostridium paraputrificum bacteremia associated with colonic necrosis in a patient with AIDS. Case Rep. Infect. Dis. 2015, 2015, 312919. [Google Scholar]
- Evvyernie, D.; Yamazaki, S.; Morimoto, K.; Karita, S.; Kimura, T.; Sakka, K.; Ohmiya, K. Identification and characterization of Clostridium paraputrificum M-21, a chitinolytic, mesophilic and hydrogen-producing bacterium. J. Biosci. Bioeng. 2000, 89, 596–601. [Google Scholar] [CrossRef]
- Kageyama, A.; Benno, Y.; Nakase, T. Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 557–565. [Google Scholar] [CrossRef]
- Neumann, A.; Björck, L.; Frick, I.M. Finegoldia magna, an anaerobic gram-positive bacterium of the normal human microbiota, induces inflammation by activating neutrophils. Front. Microbiol. 2020, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, M.E.; Rojtman, A.D.; Frank, E. Finegoldia magna (formerly Peptostreptococcus magnus): An overlooked etiology for toxic shock syndrome? Med. Hypotheses 2012, 79, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; Moore, W.E.C.; Weiss, N.; Collins, M.D. Phenotypic and phylogenetic characterization of some eubacterium-like isolates containing a novel type B wall murein from human feces: Description of Holdemania filiformis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 1997, 47, 1201–1204. [Google Scholar] [CrossRef]
- Spiegel, C.A.; Roberts, M. Mobiluncus gen. nov., Mobiluncus curtisii subsp. curtisii sp. nov., Mobiluncus curtisii subsp. holmesii subsp. nov., and Mobiluncus mulieris sp. nov., curved rods from the human vagina. Int. J. Syst. Evol. Microbiol. 1984, 34, 177–184. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, L.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Roles of intestinal Parabacteroides in human health and diseases. FEMS Microbiol. Lett. 2022, 369, fnac072. [Google Scholar] [CrossRef]
- Sakamoto, M.; Benno, Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 1599–1605. [Google Scholar] [CrossRef] [PubMed]
- Trottier, G.; Srivastava, L.; Walker, C.D. Etiology of infantile autism: A review of recent advances in genetic and neurobiological research. J. Psychiatry Neurosci. 1999, 24, 103. [Google Scholar] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Desbonnet, L.; Garrett, L.; Clarke, G.; Bienenstock, J.; Dinan, T.G. The probiotic Bifidobacteria infantis: An assessment of potential antidepressant properties in the rat. J. Psychiatr. Res. 2008, 43, 164–174. [Google Scholar] [CrossRef]
- Kumperscak, H.G.; Gricar, A.; Ülen, I.; Micetic-Turk, D. A pilot randomized control trial with the probiotic strain Lactobacillus rhamnosus GG (LGG) in ADHD: Children and adolescents report better health-related quality of life. Front. Psychiatry 2020, 11, 181. [Google Scholar] [CrossRef]
- Cenit, M.C.; Nuevo, I.C.; Codoñer-Franch, P.; Dinan, T.G.; Sanz, Y. Gut microbiota and attention deficit hyperactivity disorder: New perspectives for a challenging condition. Eur. Child Adolesc. Psychiatry 2017, 26, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Tengeler, A.C.; Emmerzaal, T.L.; Geenen, B.; Verweij, V.; van Bodegom, M.; Morava, E.; Kiliaan, A.J.; Kozicz, T. Early-adolescent antibiotic exposure results in mitochondrial and behavioral deficits in adult male mice. Sci. Rep. 2021, 11, 12875. [Google Scholar] [CrossRef] [PubMed]

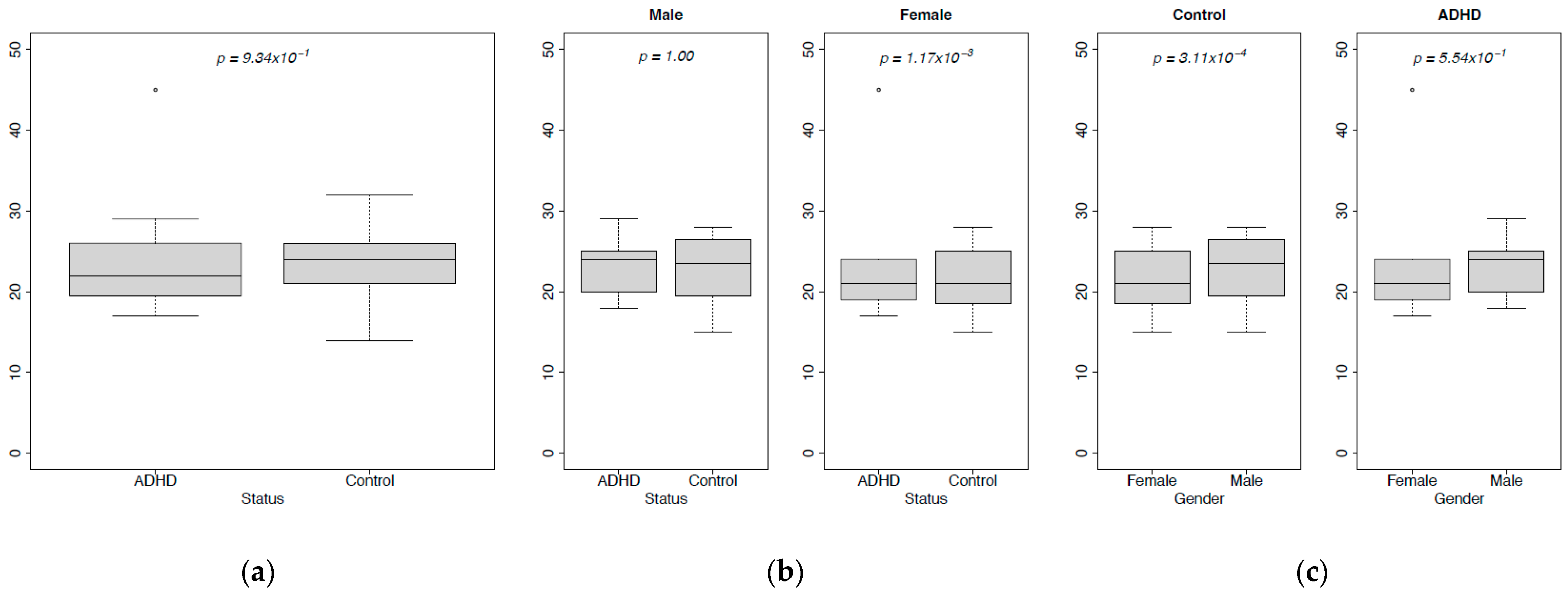

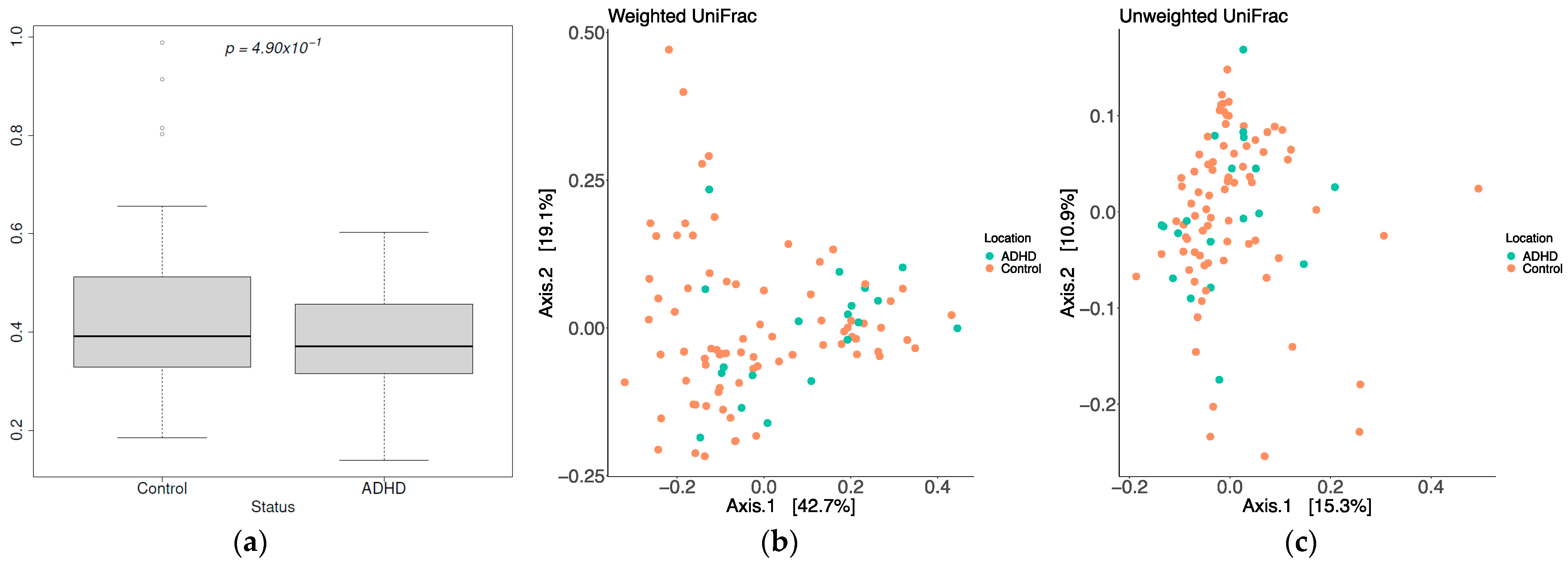
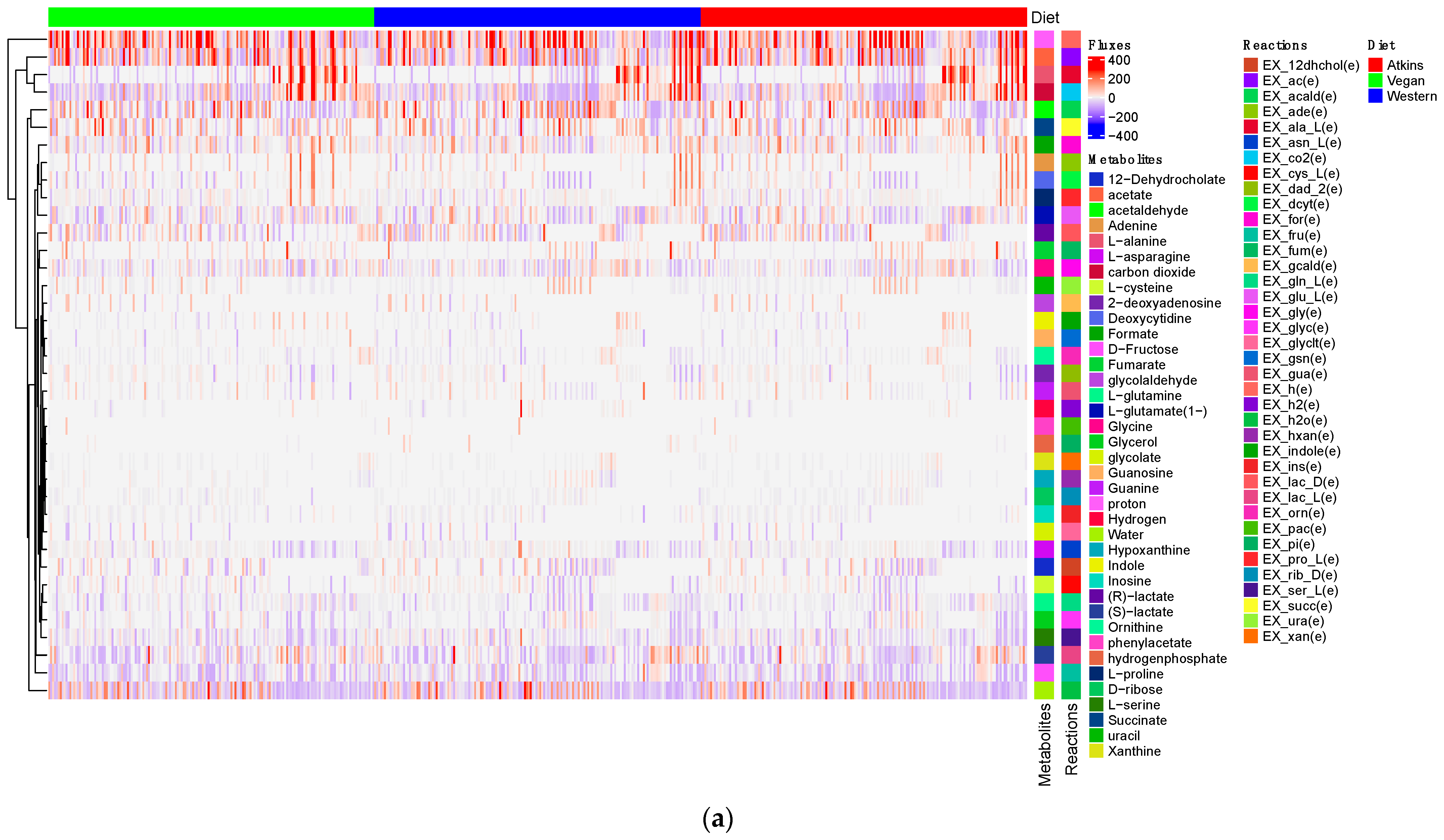
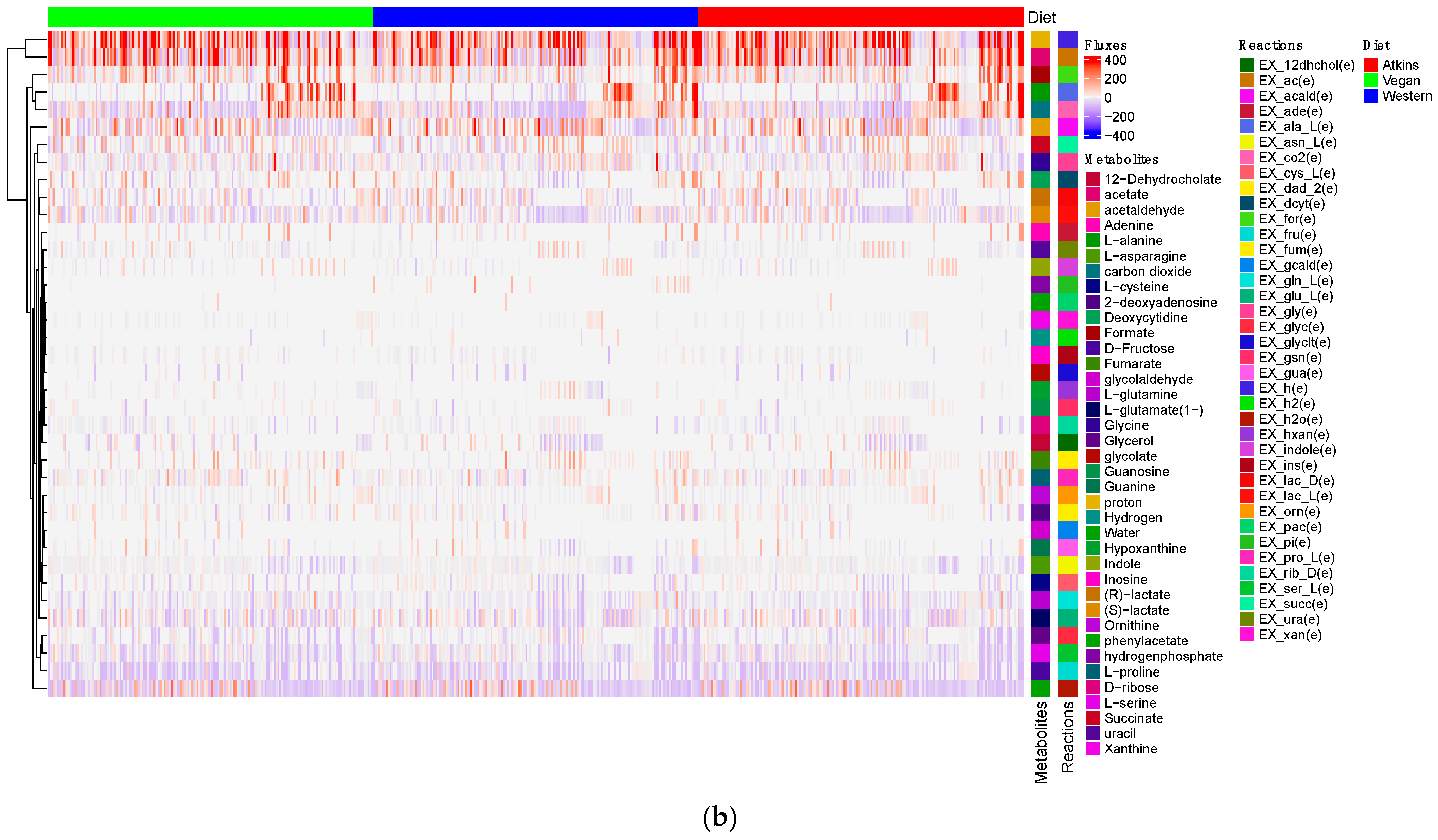
| Sample ID | Age | Gender |
|---|---|---|
| CONTROL01 | 15 | Female |
| ADHD01 | 15 | Female |
| CONTROL02 | 17 | Female |
| ADHD02 | 17 | Female |
| CONTROL03 | 18 | Female |
| ADHD03 | 18 | Female |
| CONTROL04 | 20 | Male |
| ADHD04 | 20 | Male |
| CONTROL05 | 20 | Male |
| ADHD05 | 20 | Male |
| CONTROL06 | 20 | Female |
| ADHD06 | 20 | Female |
| CONTROL07 | 21 | Male |
| ADHD07 | 21 | Male |
| CONTROL08 | 21 | Male |
| ADHD08 | 21 | Male |
| CONTROL09 | 22 | Male |
| ADHD09 | 22 | Male |
| CONTROL10 | 23 | Male |
| ADHD10 | 23 | Male |
| Organism | Pair 1 | Pair 2 | Pair 3 | Pair 4 | Pair 5 | Pair 6 | Pair 7 | Pair 8 | Pair 9 | Pair 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Bifidobacterium adolescentis | Control | ADHD | Control | ADHD | ADHD | Same | ADHD | ADHD | Control | ADHD |
| Collinsella aerofaciens | Control | ADHD | Control | Control | Control | Same | ADHD | Control | Control | ADHD |
| Bifidobacterium angulatum | Control | ADHD | Control | Same | Control | Same | Control | ADHD | ADHD | Control |
| Prevotella buccae | Same | Same | Same | Control | Same | Same | Same | Same | Same | Same |
| Prevotella copri | Control | Control | ADHD | Same | Same | Same | Same | Control | Same | Control |
| Bacteroides coprocola | ADHD | Same | Same | Same | Same | Same | Control | Control | Same | Control |
| Bifidobacterium dentium | Same | Same | ADHD | Same | Same | Same | Same | ADHD | ADHD | ADHD |
| Parabacteroides distasonis | Same | Same | Control | Same | Same | Same | Same | Same | ADHD | Same |
| Enterococcus faecium | ADHD | Same | Same | Same | Same | Same | Same | Same | Same | Same |
| Parabacteroides goldsteinii | Same | Same | Same | Same | Same | Same | Same | Same | ADHD | Same |
| Parabacteroides merdae | Control | Control | ADHD | Control | Control | Control | ADHD | Control | ADHD | Same |
| Bacteroides plebeius | ADHD | Same | Same | Same | Same | Same | Control | Same | Same | ADHD |
| Bifidobacterium pseudocatenulatum | Control | ADHD | Control | ADHD | Control | ADHD | Control | ADHD | ADHD | Control |
| Prevotella ruminicola | ADHD | Same | Same | Same | Same | Same | Same | Control | Control | Same |
| Bacteroides thetaiotaomicron | Same | Same | Same | Same | Same | Same | Same | Same | Same | Same |
| Bacteroides uniformis | Control | Control | ADHD | Control | ADHD | Control | ADHD | ADHD | Control | ADHD |
| Bacteroides vulgatus | Control | Control | ADHD | Control | Same | Control | ADHD | Control | Control | ADHD |
| Paraprevotella xylaniphila | Same | Same | Same | Same | Same | Same | Same | Same | Same | Same |
| Phylum | Family | Genus | Species | Higher in ADHD | Lower in ADHD |
|---|---|---|---|---|---|
| Actinobacteria | [57] | ||||
| Bifidobacteriaceae | Bifidobacterium | [44] | [54] | ||
| B. longum | [57] | ||||
| B. adolescentis | [57] | ||||
| Coriobacteriaceae | Collinsella | [57] | |||
| Bacteroidetes | Bacteroidaceae | [56] | |||
| Bacteroides | [54] | ||||
| B. uniformis | [55] | ||||
| B. ovatus | [55] | ||||
| B. caccae | [58] | ||||
| B. coprocola | [55] | ||||
| Prevotellaceae | [56] | ||||
| Prevotella | [56] | ||||
| Pararevotella | P. xylaniphila | [58] | |||
| Porphyromonadaceae | [56] | ||||
| Parabacteroides | [56] | ||||
| Odoribacteraceae | [58] | ||||
| Odoribacter | O. splanchnicus | [58] | |||
| Firmicutes | [59] | [44] | |||
| Catabacteriaceae | [56] | ||||
| Clostridiaceae | Clostridium | C. histolyticum | [54] | ||
| Enterococcaceae | [58] | ||||
| Enterococcus | [54] | ||||
| Lachnospiraceae | [58] | ||||
| Lactobacillus | [54,55] | ||||
| Ruminococcaceae | [58] | ||||
| Faecalibacterium | [58,60] | ||||
| F. prausnitzii | [58] | ||||
| Ruminococcus | R. gnavus | [58] | |||
| Veillonellaceae | Veillonella | [58] | |||
| V. parvula | [58] | ||||
| Proteobacteria | Neisseriaceae | [56] | |||
| Neisseria | [56] | ||||
| Desulfovibrionaceae | Desulfovibrio | [59] | |||
| Sutterellaceae | Sutterella | S. stercoricanis | [55] | ||
| Fusobacteria | [55] | ||||
| Fusobacteriaceae | Fusobacterium | [55] |
| Organism | Pair 1 | Pair 2 | Pair 3 | Pair 4 | Pair 5 | Pair 6 | Pair 7 | Pair 8 | Pair 9 | Pair 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Alistipes putredinis | ADHD | Control | Control | Control | NA | NA | ADHD | Control | Control | Control |
| Bacteroides coprocola | NA | NA | NA | NA | NA | NA | NA | ADHD | NA | NA |
| Bacteroides uniformis | Control | ADHD | ADHD | ADHD | ADHD | NA | ADHD | Same | Same | Control |
| Bacteroides vulgatus | ADHD | Same | ADHD | Control | NA | Control | ADHD | ADHD | Control | ADHD |
| Bifidobacterium adolescentis | Control | Control | Same | Control | Same | NA | Control | ADHD | Control | Control |
| Bifidobacterium angulatum | ADHD | NA | NA | NA | NA | NA | NA | ADHD | NA | Control |
| Bifidobacterium pseudocatenulatum | ADHD | NA | NA | NA | NA | NA | NA | ADHD | Control | Same |
| Collinsella aerofaciens | Control | Control | Same | ADHD | ADHD | NA | Control | Control | Control | ADHD |
| Coprococcus catus | Same | ADHD | ADHD | ADHD | NA | NA | ADHD | ADHD | NA | Control |
| Coprococcus comes | Control | ADHD | Control | Control | ADHD | Control | ADHD | ADHD | Control | Control |
| Coprococcus eutactus | NA | NA | ADHD | ADHD | NA | NA | NA | Control | NA | NA |
| Dialister invisus | Control | NA | NA | ADHD | NA | NA | NA | NA | NA | Control |
| Dorea formicigenerans | Control | NA | Same | Control | Control | ADHD | ADHD | Control | ADHD | Control |
| Dorea longicatena | Control | ADHD | ADHD | Control | ADHD | Control | Same | Same | ADHD | |
| Holdemania filiformis | NA | NA | NA | ADHD | NA | NA | NA | NA | NA | NA |
| Parabacteroides merdae | Control | ADHD | ADHD | NA | NA | NA | Control | NA | NA | NA |
| Ruminococcus albus | Control | NA | NA | ADHD | NA | NA | NA | ADHD | NA | NA |
| Ruminococcus bromii | Control | ADHD | NA | ADHD | NA | ADHD | ADHD | NA | Control | NA |
| Streptococcus mitis | Control | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Streptococcus parasanguinis | Same | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Streptococcus thermophilus | Control | Same | NA | NA | Same | NA | NA | NA | Control | NA |
| Subdoligranulum variabile | ADHD | ADHD | Control | ADHD | ADHD | ADHD | Control | Same | Control | ADHD |
| Organism | Pair 1 | Pair 2 | Pair 3 | Pair 4 | Pair 5 | Pair 6 | Pair 7 | Pair 8 | Pair 9 | Pair 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Alistipes putredinis | ADHD | ADHD | Control | ADHD | NA | NA | ADHD | Control | Control | ADHD |
| Bacteroides coprocola | NA | NA | NA | NA | NA | NA | NA | Control | NA | NA |
| Bacteroides uniformis | Control | ADHD | Control | ADHD | Control | NA | ADHD | Control | Control | Control |
| Bacteroides vulgatus | ADHD | Control | Control | ADHD | NA | Control | Control | Control | Control | ADHD |
| Bifidobacterium adolescentis | Control | ADHD | Control | Control | ADHD | NA | ADHD | Control | Control | ADHD |
| Bifidobacterium angulatum | Control | NA | NA | NA | NA | NA | NA | Control | NA | ADHD |
| Bifidobacterium pseudocatenulatum | Same | NA | NA | NA | NA | NA | NA | Control | Control | ADHD |
| Collinsella aerofaciens | ADHD | ADHD | ADHD | Same | ADHD | NA | ADHD | Control | ADHD | ADHD |
| Coprococcus catus | Same | Same | Same | Control | NA | NA | ADHD | Control | NA | ADHD |
| Coprococcus comes | ADHD | Control | ADHD | ADHD | ADHD | Control | ADHD | Control | Control | ADHD |
| Coprococcus eutactus | NA | NA | ADHD | Control | NA | NA | NA | Control | NA | NA |
| Dialister invisus | Control | NA | NA | ADHD | NA | NA | NA | NA | NA | Control |
| Dorea formicigenerans | ADHD | NA | Same | Control | ADHD | ADHD | ADHD | Control | Same | Control |
| Dorea longicatena | ADHD | ADHD | Control | Control | ADHD | NA | Control | Control | Control | ADHD |
| Holdemania filiformis | NA | NA | NA | Control | NA | NA | NA | NA | NA | NA |
| Parabacteroides merdae | Same | ADHD | ADHD | NA | NA | NA | ADHD | NA | NA | NA |
| Ruminococcus albus | Control | NA | NA | Control | NA | NA | NA | Control | NA | NA |
| Ruminococcus bromii | Control | ADHD | NA | Control | NA | Control | ADHD | NA | Control | NA |
| Streptococcus mitis | ADHD | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Streptococcus parasanguinis | ADHD | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Streptococcus thermophilus | Control | ADHD | NA | NA | ADHD | NA | NA | NA | Control | NA |
| Subdoligranulum variabile | ADHD | ADHD | ADHD | Same | ADHD | ADHD | ADHD | Control | Control | Control |
| Organism | Pair 1 | Pair 2 | Pair 3 | Pair 4 | Pair 5 | Pair 6 | Pair 7 | Pair 8 | Pair 9 | Pair 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Alistipes putredinis | Control | Control | ADHD | ADHD | NA | NA | Control | Same | ADHD | ADHD |
| Bacteroides coprocola | NA | NA | NA | NA | NA | NA | NA | Control | NA | NA |
| Bacteroides uniformis | Control | Control | ADHD | ADHD | Control | NA | Same | ADHD | ADHD | ADHD |
| Bacteroides vulgatus | Control | Control | ADHD | ADHD | NA | ADHD | Control | ADHD | Control | ADHD |
| Bifidobacterium adolescentis | ADHD | ADHD | ADHD | Control | Control | NA | Control | ADHD | Control | ADHD |
| Bifidobacterium angulatum | Control | NA | NA | NA | NA | NA | NA | ADHD | NA | ADHD |
| Bifidobacterium pseudocatenulatum | Control | NA | NA | NA | NA | NA | NA | Control | Control | Control |
| Collinsella aerofaciens | Same | ADHD | Control | Control | ADHD | NA | Control | Control | Control | Control |
| Coprococcus catus | Control | Control | ADHD | ADHD | NA | NA | Control | ADHD | NA | Control |
| Coprococcus comes | Control | Same | ADHD | ADHD | ADHD | ADHD | Control | Control | Control | Same |
| Coprococcus eutactus | NA | NA | ADHD | Control | NA | NA | NA | Control | NA | Control |
| Dialister invisus | Control | NA | NA | ADHD | NA | NA | NA | NA | NA | NA |
| Dorea formicigenerans | Control | NA | Control | Control | ADHD | ADHD | Control | Control | Control | ADHD |
| Dorea longicatena | Control | Control | Control | Control | Control | NA | Control | Same | Control | Control |
| Holdemania filiformis | NA | NA | NA | ADHD | NA | NA | NA | NA | NA | NA |
| Parabacteroides merdae | Control | ADHD | Control | NA | NA | NA | ADHD | NA | NA | NA |
| Ruminococcus albus | Control | NA | NA | Control | NA | NA | NA | ADHD | NA | NA |
| Ruminococcus bromii | Control | ADHD | NA | Control | NA | Same | Control | Control | NA | |
| Streptococcus mitis | Control | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Streptococcus parasanguinis | Control | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Streptococcus thermophilus | Control | Control | NA | NA | Control | NA | NA | NA | Control | NA |
| Subdoligranulum variabile | ADHD | ADHD | ADHD | Control | ADHD | Control | Control | Control | Same | ADHD |
| Key Metabolite | Western Diet | Atkins’ Diet | Vegan Diet |
|---|---|---|---|
| Acetate | A. putredinis 🠗 C. comes 🠗 C. comes ↑ P. copri 🠗 R. albus 🠗 P. goldsteinii 🠗 P. buccae 🠕 | P. goldsteinii 🠕 R. albus 🠕 | A. putredinis 🠕 P. copri 🠕 R. albus ↑ |
| Formate | C. catus 🠗 C. catus ↓ P. goldsteinii 🠗 P. buccae 🠕 | P. goldsteinii 🠕 | |
| Propionate | P. ruminicola 🠕 | P. goldsteinii 🠕 | P. merdae ↑ P. merdae 🠕 |
| Glutamate | A. Putredinis 🠕 B. licheniformis 🠕 B. vulgatus 🠕 B. hydrogenotrophica 🠕 C. catus 🠕 P. ruminicola 🠕 F. magna ↓ M. curtisii ↓ P. distasonis ↓ P. goldsteinii ↓ F. magna 🠗 P. distasonis 🠗 P. coprii 🠗 | P. ruminicola 🠕 B. hydrogenotrophica ↓ F. magna ↑ | P. goldsteinii ↑ P. copri 🠕 |
| L-Phenylalanine | M. curtisii 🠕 P. distasonis ↓ D. formigeneras | D. formigeneras | D. formigeneras |
| L-Tryptophan | B. uniformis ↓ B. licheniformis | B. licheniformis | B. licheniformis |
| L-Tyrosine | M. curtisii ↑ D. formigeneras | D. formigeneras | D. formigeneras |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taş, E.; Ülgen, K.O. Understanding the ADHD-Gut Axis by Metabolic Network Analysis. Metabolites 2023, 13, 592. https://doi.org/10.3390/metabo13050592
Taş E, Ülgen KO. Understanding the ADHD-Gut Axis by Metabolic Network Analysis. Metabolites. 2023; 13(5):592. https://doi.org/10.3390/metabo13050592
Chicago/Turabian StyleTaş, Ezgi, and Kutlu O. Ülgen. 2023. "Understanding the ADHD-Gut Axis by Metabolic Network Analysis" Metabolites 13, no. 5: 592. https://doi.org/10.3390/metabo13050592
APA StyleTaş, E., & Ülgen, K. O. (2023). Understanding the ADHD-Gut Axis by Metabolic Network Analysis. Metabolites, 13(5), 592. https://doi.org/10.3390/metabo13050592