The Effects of Pregnancy on Amino Acid Levels and Nitrogen Disposition
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Metabolomics Analysis
2.3. Confirmational Liquid Chromatography Mass Spectrometry
2.4. Statistical Analysis
2.5. Pathways Analysis
3. Results
3.1. Characteristics of Subjects
3.2. Multivariate Analysis
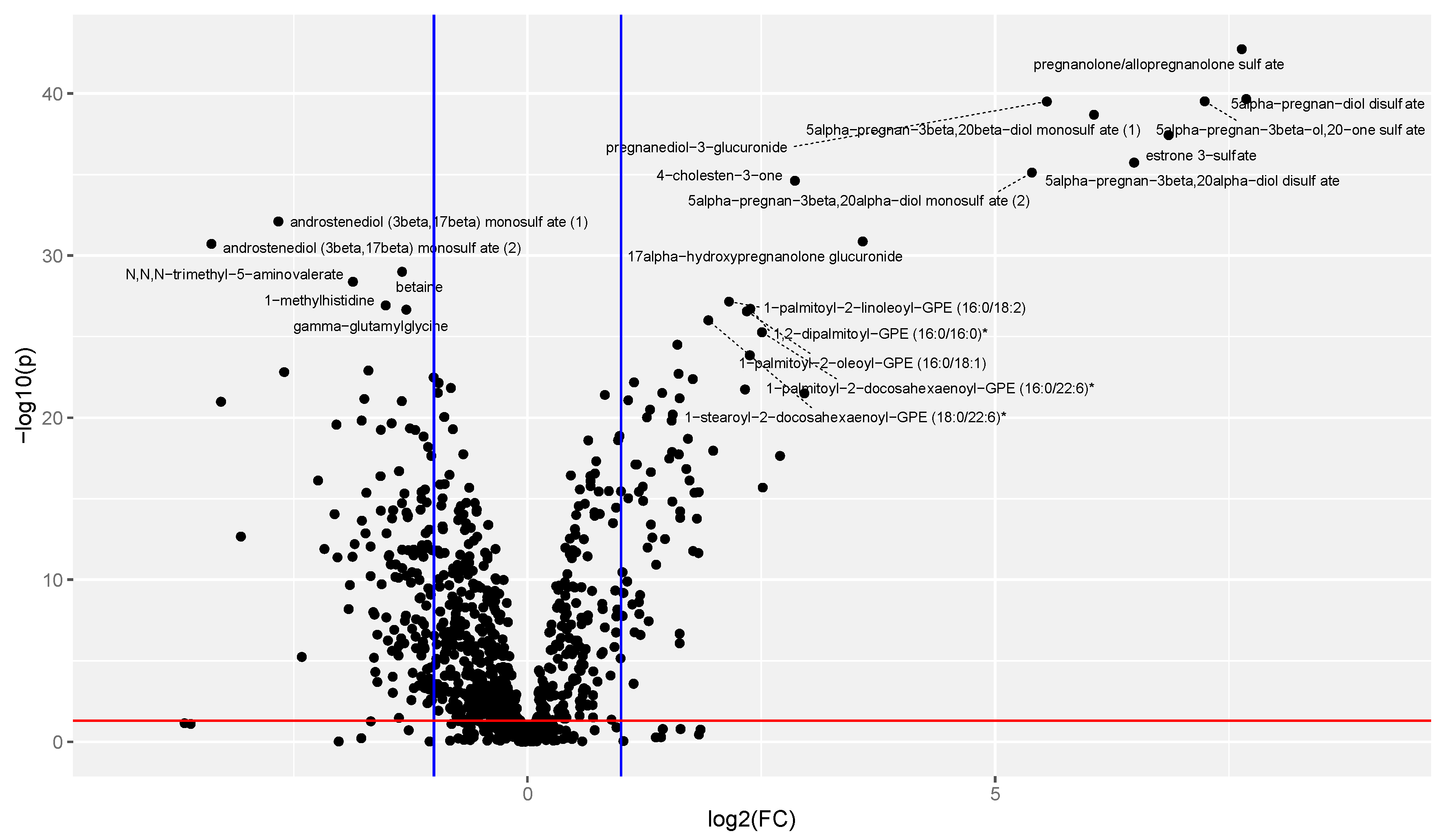
3.3. Univariate Analysis
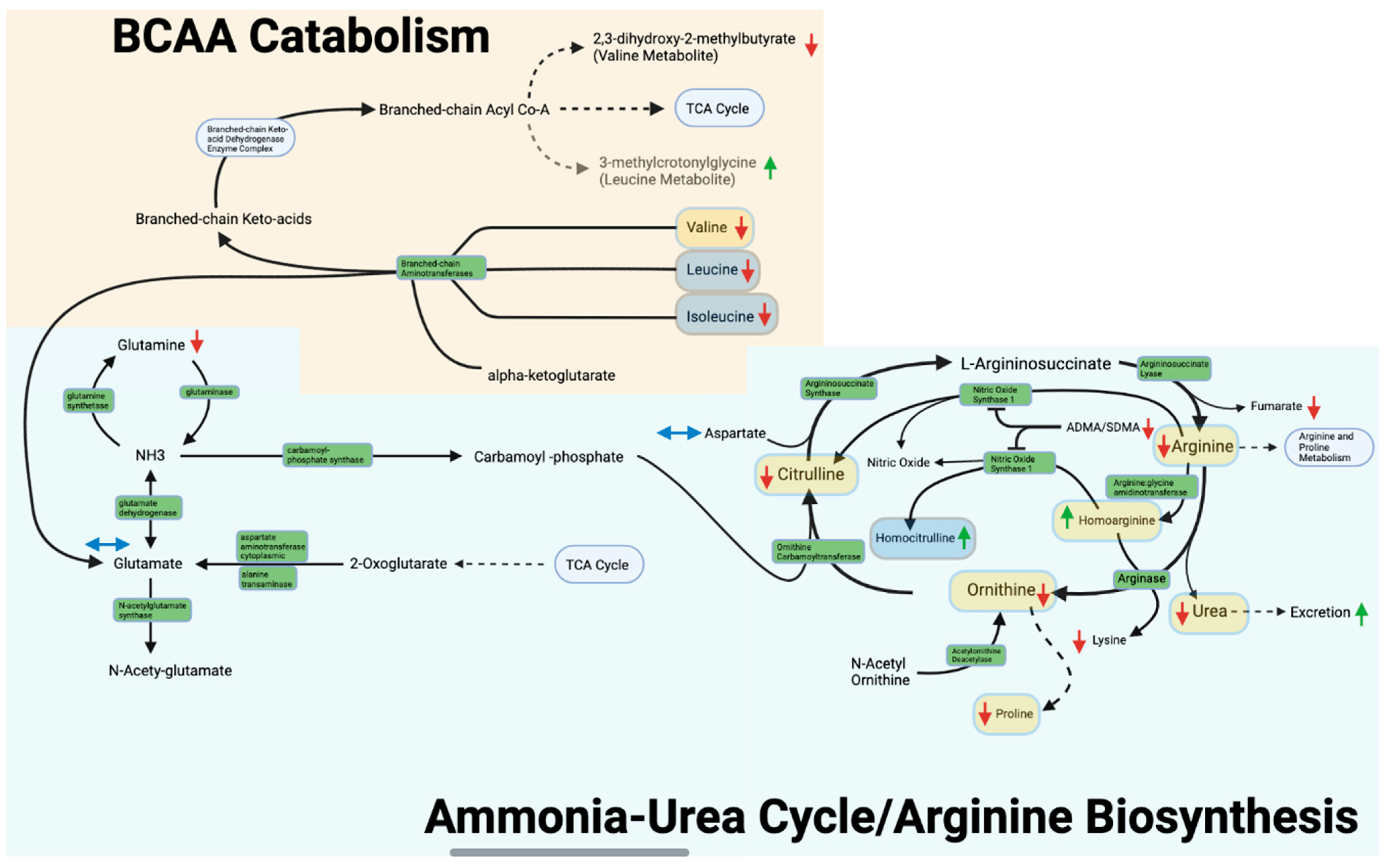
3.4. Arginine Biosynthesis and Ammonia-Urea Cycle
3.5. Valine, Leucine, and Isoleucine Metabolism
4. Discussion
4.1. Overview
4.2. Arginine Biosynthesis/Nitrogen Disposition
4.3. Valine, Leucine, and Isoleucine Metabolism
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soma-Pillay, P.; Nelson-Piercy, C.; Tolppanen, H.; Mebazaa, A. Physiological changes in pregnancy. Cardiovasc. J. Afr. 2016, 27, 89–94. [Google Scholar] [CrossRef]
- Wu, G. Amino Acids: Biochemistry and Nutrition, 1st ed.; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar] [CrossRef]
- Di Giulio, A.M.; Carelli, S.; Castoldi, R.; Gorio, A.; Taricco, E.; Cetin, I. Plasma amino acid concentrations throughout normal pregnancy and early stages of intrauterine growth restricted pregnancy. J. Matern. Neonatal Med. 2004, 15, 356–362. [Google Scholar] [CrossRef]
- Schoengold, D.M.; Defiore, R.H.; Parlett, R.C. Free amino acids in plasma throughout pregnancy. Am. J. Obstet. Gynecol. 1978, 131, 490–499. [Google Scholar] [CrossRef]
- Kalhan, S.C. Protein metabolism in pregnancy. Am. J. Clin. Nutr. 2000, 71, 1249S–1255S. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Tserng, K.-Y.; Gilfillan, C.; Dierker, L.J. Metabolism of urea and glucose in normal and diabetic pregnancy. Metabolism 1982, 31, 824–833. [Google Scholar] [CrossRef]
- Holden, D.P.; Fickling, S.A.; Whitley, G.S.; Nussey, S.S. Plasma concentrations of asymmetric dimethylarginine, a natural inhibitor of nitric oxide synthase, in normal pregnancy and preeclampsia. Am. J. Obstet. Gynecol. 1998, 178, 551–556. [Google Scholar] [CrossRef]
- Brosnan, J.T.; Brosnan, M.E. Branched-Chain Amino Acids: Enzyme and Substrate Regulation. J. Nutr. 2006, 136 (Suppl. S1), S207–S211. [Google Scholar] [CrossRef]
- Hutson, S.M.; Sweatt, A.J.; Lanoue, K.F. Branched-chain amino acid metabolism: Implications for establishing safe intakes. J. Nutr. 2005, 135 (Suppl. S1), 1557S–1564S. [Google Scholar] [CrossRef]
- Hutson, S.M.; Fenstermacher, D.; Mahar, C. Role of mitochondrial transamination in branched chain amino acid metabolism. J. Biol. Chem. 1988, 263, 3618–3625. [Google Scholar] [CrossRef]
- Mogami, H.; Yura, S.; Itoh, H.; Kawamura, M.; Fujii, T.; Suzuki, A.; Aoe, S.; Ogawa, Y.; Sagawa, N.; Konishi, I.; et al. Isocaloric high-protein diet as well as branched-chain amino acids supplemented diet partially alleviates adverse consequences of maternal undernutrition on fetal growth. Growth Horm. IGF Res. 2009, 19, 478–485. [Google Scholar] [CrossRef]
- Shum, S.; Bs, A.Y.; Fay, E.; Moreni, S.; Mao, J.; Czuba, L.; Wang, C.; Ms, N.I.; Hebert, M.F. Infant Dextromethorphan and Dextrorphan Exposure via Breast Milk From Mothers Who Are CYP2D6 Extensive Metabolizers. J. Clin. Pharmacol. 2021, 62, 747–755. [Google Scholar] [CrossRef]
- Collet, T.H.; Sonoyama, T.; Henning, E.; Keogh, J.M.; Ingram, B.; Kelway, S.; Guo, L.; Farooqi, I.S. A Metabolomic Signature of Acute Caloric Restriction. J. Clin. Endocrinol. Metab. 2017, 102, 4486–4495. [Google Scholar] [CrossRef]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. Qvalue: Q-Value Estimation for False Discovery Rate Control. R Package Version 2.28.0. 2022. Available online: https://github.com/jdstorey/qvalue (accessed on 14 December 2022).
- Xia, J.; Wishart, D.S. Metabolomic Data Processing, Analysis, and Interpretation Using MetaboAnalyst. Curr. Protoc. Bioinform. 2011, 34, 14.10.1–14.10.48. [Google Scholar] [CrossRef]
- Han, L.W.; Shi, Y.; Paquette, A.; Wang, L.; Bammler, T.K.; Mao, Q. Key hepatic metabolic pathways are altered in germ-free mice during pregnancy. PLoS ONE 2021, 16, e0248351. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Zheng, W.; Lan, Z.; Huang, Z.; Yang, Q.; Liu, C.; Gao, R.; Zhang, Y. Clinical, biochemical and molecular analysis of two infants with familial chylomicronemia syndrome. Lipids Health Dis. 2016, 15, 88. [Google Scholar] [CrossRef]
- Handelman, S.; Romero, R.; Tarca, A.L.; Pacora, P.; Ingram, B.; Maymon, E.; Chaiworapongsa, T.; Hassan, S.S.; Erez, O. The plasma metabolome of women in early pregnancy differs from that of non-pregnant women. PLoS ONE 2019, 14, e0224682. [Google Scholar] [CrossRef]
- Zhou, T.; Du, S.; Sun, D.; Li, X.; Heianza, Y.; Hu, G.; Sun, L.; Pei, X.; Shang, X.; Qi, L. Prevalence and Trends in Gestational Diabetes Mellitus Among Women in the United States, 2006–2017: A Population-Based Study. Front. Endocrinol. 2022, 13, 868094. [Google Scholar] [CrossRef]
- Cheung, K.; Lafayette, R.A. Renal physiology of pregnancy. Adv. Chronic. Kidney Dis. 2013, 20, 209–214. [Google Scholar] [CrossRef]
- Valtonen, P.; Laitinen, T.; Lyyra-Laitinen, T.; Raitakari, O.T.; Juonala, M.; Viikari, J.S.; Heiskanen, N.; Vanninen, E.; Punnonen, K.; Heinonen, S. Serum L-Homoarginine Concentration is Elevated During Normal Pregnancy and is Related to Flow-Mediated Vasodilatation. Circ. J. 2008, 72, 1879–1884. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Rossi, K.Q.; Gruca, L.L.; Super, D.M.; Savin, S.M. Relation between transamination of branched-chain amino acids and urea synthesis: Evidence from human pregnancy. Am. J. Physiol. Metab. 1998, 275, E423–E431. [Google Scholar] [CrossRef]
- Denne, S.C.; Patel, D.; Kalhan, S.C. Leucine kinetics and fuel utilization during a brief fast in human pregnancy. Metabolism 1991, 40, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Haüssinger, D. Nitrogen metabolism in liver: Structural and functional organization and physiological relevance. Biochem. J. 1990, 267, 281–290. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Keane, K.N.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef]
- Weiner, I.D.; Verlander, J.W. Renal Ammonia Metabolism and Transport. Compr. Physiol. 2013, 3, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Elango, R.; Ball, R.O. Protein and Amino Acid Requirements during Pregnancy. Adv. Nutr. Int. Rev. J. 2016, 7, 839S–844S. [Google Scholar] [CrossRef] [PubMed]
- Pearson, D.L.; Dawling, S.; Walsh, W.F.; Haines, J.L.; Christman, B.W.; Bazyk, A.; Scott, N.; Summar, M.L. Neonatal Pulmonary Hypertension: Urea-Cycle Intermediates, Nitric Oxide Production, and Carbamoyl-Phosphate Synthetase Function. N. Engl. J. Med. 2001, 344, 1832–1838. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Datta, S.; Johnson, G.A.; Li, P.; Satterfield, M.C.; Spencer, T. Proline metabolism in the conceptus: Implications for fetal growth and development. Amino Acids 2008, 35, 691–702. [Google Scholar] [CrossRef]
- Lopez-Jaramillo, P.; Arenas, W.D.; Garcia, R.G.; Rincon, M.Y.; López, M. Review: The role of the L-arginine-nitric oxide pathway in preeclampsia. Ther. Adv. Cardiovasc. Dis. 2008, 2, 261–275. [Google Scholar] [CrossRef]
- Maul, H.; Longo, M.; Saade, G.; Garfield, R. Nitric Oxide and its Role During Pregnancy: From Ovulation to Delivery. Curr. Pharm. Des. 2003, 9, 359–380. [Google Scholar] [CrossRef]
- Summar, M.L.; Gainer, J.V.; Pretorius, M.; Malave, H.; Harris, S.; Hall, L.D.; Weisberg, A.; Vaughan, D.E.; Christman, B.W.; Brown, N.J. Relationship between Carbamoyl-Phosphate Synthetase Genotype and Systemic Vascular Function. Hypertension 2004, 43, 186–191. [Google Scholar] [CrossRef]
- Deng, A.; Engels, K.; Baylis, C. Impact of nitric oxide deficiency on blood pressure and glomerular hemodynamic adaptations to pregnancy in the rat. Kidney Int. 1996, 50, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Hrabák, A.; Bajor, T.; Temesi, A. Comparison of Substrate and Inhibitor Specificity of Arginase and Nitricm Oxide (NO) Synthase for Arginine Analogs and Related Compounds in Murine and Rat Macrophages. Biochem. Biophys. Res. Commun. 1994, 198, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Cynober, L.A. Plasma amino acid levels with a note on membrane transport: Characteristics, regulation, and metabolic significance. Nutrition 2002, 18, 761–766. [Google Scholar] [CrossRef]
- Cetin, I.; Ronzoni, S.; Marconi, A.M.; Perugino, G.; Corbetta, C.; Battaglia, F.C.; Pardi, G. Maternal concentrations and fetal-maternal concentration differences of plasma amino acids in normal and intrauterine growth-restricted pregnancies. Am. J. Obstet. Gynecol. 1996, 174, 1575–1583. [Google Scholar] [CrossRef]
- Teodoro, G.F.R.; Vianna, D.; Torres-Leal, F.L.; Pantaleão, L.C.; Matos-Neto, E.M.; Donato, J., Jr.; Tirapegui, J. Leucine Is Essential for Attenuating Fetal Growth Restriction Caused by a Protein-Restricted Diet in Rats. J. Nutr. 2012, 142, 924–930. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Syngelaki, A.; Mandal, R.; Graham, S.F.; Akolekar, R.; Han, B.; Bjondahl, T.C.; Dong, E.; Bauer, S.; Alpay-Savasan, Z.; et al. Metabolomic determination of pathogenesis of late-onset preeclampsia. J. Matern. Neonatal Med. 2017, 30, 658–664. [Google Scholar] [CrossRef]
- Gh, B.F.N.M. Application of metabolomics to preeclampsia diagnosis. Syst. Biol. Reprod. Med. 2018, 64, 324–339. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Syngelaki, A.; Akolekar, R.; Mandal, R.; Bjondahl, T.C.; Han, B.; Dong, E.; Bauer, S.; Alpay-Savasan, Z.; Graham, S.; et al. Validation of metabolomic models for prediction of early-onset preeclampsia. Am. J. Obstet. Gynecol. 2015, 213, 530.e1–530.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, M.; Li, J.; Bi, Y.; Li, M.; Yang, J. Association of Circulating Branched-Chain Amino Acids with Gestational Diabetes Mellitus: A Meta-Analysis. Int. J. Endocrinol. Metab. 2019, 17, e85413. [Google Scholar] [CrossRef]
- Baumgartner, M.R.; Dantas, M.; Suormala, T.; Almashanu, S.; Giunta, C.; Friebel, D.; Gebhardt, B.; Fowler, B.; Hoffmann, G.F.; Baumgartner, E.; et al. Isolated 3-Methylcrotonyl-CoA Carboxylase Deficiency: Evidence for an Allele-Specific Dominant Negative Effect and Responsiveness to Biotin Therapy. Am. J. Hum. Genet. 2004, 75, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimons, P.E.; Alston, C.L.; Bonnen, P.E.; Hughes, J.; Crushell, E.; Geraghty, M.T.; Tetreault, M.; O’Reilly, P.; Twomey, E.; Sheikh, Y.; et al. Clinical, biochemical, and genetic features of four patients with short-chain enoyl-CoA hydratase (ECHS1) deficiency. Am. J. Med Genet. Part A 2018, 176, 1115–1127. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Pregnant (n = 47) | Postpartum (n = 47) | p-Value |
|---|---|---|---|
| Gestational Age (weeks) or Time Postpartum (weeks) | 27.0 ± 1.3 | 15.0 ± 2.1 | NA |
| Height (cm) | 163.6 ± 16.8 | NA | |
| Weight (kg) | 71.6 ± 10.6 | 67.4 ± 9.3 | 1 × 10−5 |
| IBW (kg) | 58.2 ±6.4 | NA | |
| BMI (kg/m2) | 25.7 ± 3.2 | NA | |
| Albumin (g/dL) | 3.6 ± 0.2 | 4.6 ± 0.2 | 9 × 10−28 |
| Bilirubin (mg/dL) | 0.5 ± 0.2 | 0.7 ± 0.4 | 7 × 10−7 |
| Serum Creatinine (mg/dL) | 0.5 ± 0.1 | 0.7 ± 0.1 | 9 × 10−21 |
| BUN (mg/dL) | 7.8 ± 1.9 | 14.0 ± 4.0 | 1 × 10−14 |
| CrCl (mL/min) | 194.3 ± 81.4 | 130.9 ± 25.6 | 3 × 10−6 |
| Mean Protein Intake (g/day) | 81.3 ± 26.7 | 73.9 ± 22.4 | 0.13 |
| Pathway | p-Value | FDR | Holm Adjusted p-Value | ||
|---|---|---|---|---|---|
| Valine, Leucine, and Isoleucine Metabolism Caffeine/Xanthine Metabolism Arginine Biosynthesis | 2 × 10−5 4 × 10−5 5 × 10−5 | 1 × 10−3 1 × 10−3 1 × 10−3 | 2 × 10−3 4 × 10−3 4 × 10−3 | ||
| Arginine Biosynthesis Metabolite | Pregnant | Postpartum | Pregnancy/ Postpartum | p-Value | q-Value |
| ADMA/SDMA | 0.92 ± 0.16 | 1.15 ± 0.22 | 0.80 | 1 × 10−8 | 4 × 10−9 |
| Arginine | 0.80 ± 0.17 | 1.25 ± 0.23 | 0.63 | 2 × 10−15 | 2 × 10−15 |
| Aspartate | 1.23 ± 0.78 | 1.07 ± 0.40 | 1.15 | 6 × 10−1 | 7 × 10−2 |
| Citrulline | 0.82 ± 0.26 | 1.30 ± 0.35 | 0.63 | 9 × 10−14 | 6 × 10−14 |
| Fumarate | 0.90 ± 0.19 | 1.15 ± 0.26 | 0.79 | 8 × 10−11 | 4 × 10−11 |
| Glutamate | 1.08 ± 0.72 | 1.06 ± 0.43 | 1.02 | 3 × 10−1 | 3 × 10−2 |
| Glutamine | 0.93 ± 0.13 | 1.11 ± 0.16 | 0.84 | 1 × 10−10 | 5 × 10−11 |
| Homoarginine | 1.88 ± 1.04 | 0.75 ± 0.31 | 2.49 | 2 × 10−17 | 3 × 10−17 |
| Homocitrulline | 1.22 ± 0.55 | 0.95 ± 0.51 | 1.29 | 1 × 10−3 | 2 × 10−4 |
| N-acetylglutamate | 0.96 ± 0.20 | 1.13 ± 0.32 | 0.85 | 3 × 10−4 | 5 × 10−5 |
| N-acetylarginine | 0.78 ± 0.36 | 1.55 ± 0.66 | 0.50 | 3 × 10−23 | 1 × 10−22 |
| Ornithine | 0.68 ± 0.19 | 1.39 ± 0.32 | 0.49 | 3 × 10−18 | 3 ×10−18 |
| Proline | 0.86 ± 0.25 | 1.16 ± 0.34 | 0.74 | 3 × 10−8 | 1 × 10−8 |
| Urea | 0.80 ± 0.27 | 1.53 ± 0.46 | 0.52 | 1 × 10−16 | 2 × 10−16 |
| 3-amino-2-piperidone | 0.79 ± 0.16 | 1.37 ± 0.37 | 0.58 | 5 × 10−20 | 1 × 10−19 |
| BCAA Metabolism Metabolite | Pregnant | Postpartum | Pregnancy/ Postpartum | p-Value | q-Value |
| Isoleucine | 1.00 ± 0.30 | 1.09 ± 0.23 | 0.92 | 3 × 10−2 | 5 × 10−3 |
| Leucine | 0.97 ± 0.32 | 1.11 ± 0.24 | 0.87 | 1 × 10−3 | 2 × 10−4 |
| Threonine | 1.25 ± 0.26 | 0.76 ± 0.21 | 1.64 | 3 × 10−14 | 3 × 10−14 |
| Valine | 0.92 ± 0.23 | 1.09 ± 0.21 | 0.84 | 8 × 10−5 | 2 × 10−5 |
| 3-methylcrotonylglycine | 0.96 ± 0.49 | 0.81 ± 0.43 | 1.22 | 5 × 10−2 | 4 × 10−3 |
| 2,3-dihydroxy-2-methylbutyrate | 0.66 ± 0.21 | 1.43 ± 0.41 | 0.46 | 3 × 10−17 | 5 × 10−17 |
| Metabolite | Pregnant (nM) | Postpartum (nM) | Pregnancy/ Postpartum | Bonferroni Corrected p-Value |
|---|---|---|---|---|
| Arginine | 844 ± 208 | 1042 ± 239 | 0.82 | 9 × 10−6 |
| Citrulline | 0.15 ± 0.07 | 0.28 ± 0.12 | 0.54 | 2 × 10−12 |
| Homoarginine | 0.06 ± 0.04 | 0.02 ± 0.01 | 2.41 | 2 × 10−7 |
| Homocitrulline | 0.005 ± 0.003 | 0.004 ± 0.002 | 1.18 | 5 × 10−1 |
| Isoleucine | 280 ± 129 | 312 ± 82 | 0.90 | 1 |
| Leucine | 448 ± 206 | 524 ± 153 | 0.86 | 5 × 10−1 |
| Ornithine | 0.35 ± 0.17 | 0.78 ± 0.37 | 0.45 | 2 × 10−11 |
| Proline | 626 ± 21 | 871 ± 285 | 0.72 | 7 × 10−5 |
| Threonine | 800 ± 177 | 525 ± 126 | 1.49 | 2 × 10−9 |
| Valine | 821 ± 272 | 1030 ± 220 | 0.80 | 2 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enthoven, L.F.; Shi, Y.; Fay, E.E.; Moreni, S.; Mao, J.; Honeyman, E.M.; Smith, C.K.; Whittington, D.; Brockerhoff, S.E.; Isoherranen, N.; et al. The Effects of Pregnancy on Amino Acid Levels and Nitrogen Disposition. Metabolites 2023, 13, 242. https://doi.org/10.3390/metabo13020242
Enthoven LF, Shi Y, Fay EE, Moreni S, Mao J, Honeyman EM, Smith CK, Whittington D, Brockerhoff SE, Isoherranen N, et al. The Effects of Pregnancy on Amino Acid Levels and Nitrogen Disposition. Metabolites. 2023; 13(2):242. https://doi.org/10.3390/metabo13020242
Chicago/Turabian StyleEnthoven, Luke F., Yuanyuan Shi, Emily E. Fay, Sue Moreni, Jennie Mao, Emma M. Honeyman, Chase K. Smith, Dale Whittington, Susan E. Brockerhoff, Nina Isoherranen, and et al. 2023. "The Effects of Pregnancy on Amino Acid Levels and Nitrogen Disposition" Metabolites 13, no. 2: 242. https://doi.org/10.3390/metabo13020242
APA StyleEnthoven, L. F., Shi, Y., Fay, E. E., Moreni, S., Mao, J., Honeyman, E. M., Smith, C. K., Whittington, D., Brockerhoff, S. E., Isoherranen, N., Totah, R. A., & Hebert, M. F. (2023). The Effects of Pregnancy on Amino Acid Levels and Nitrogen Disposition. Metabolites, 13(2), 242. https://doi.org/10.3390/metabo13020242







