Regulation of Human Endogenous Metabolites by Drug Transporters and Drug Metabolizing Enzymes: An Analysis of Targeted SNP-Metabolite Associations
Abstract
1. Introduction
2. Materials and Methods
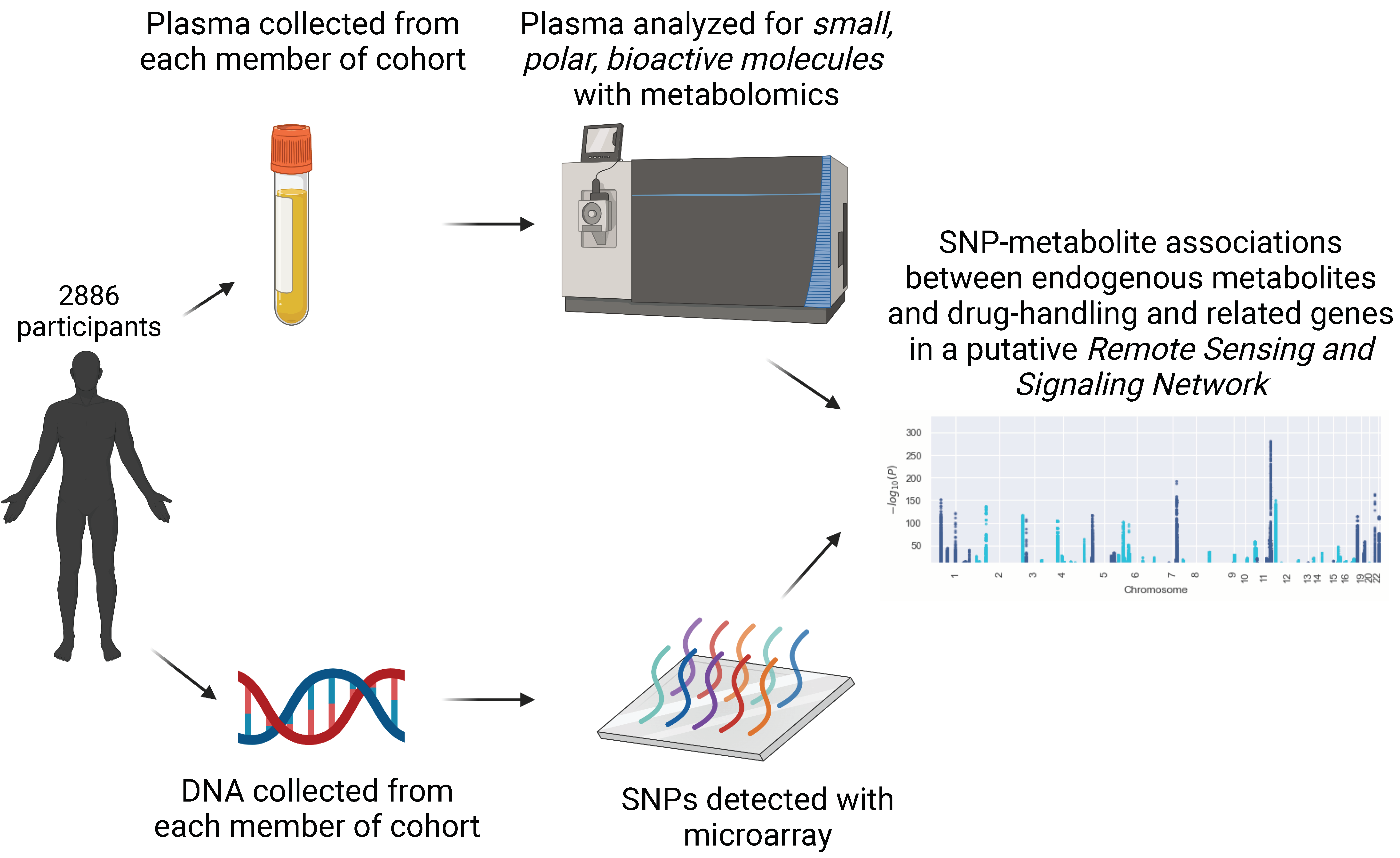
2.1. Sample Population
2.2. Gene List
2.3. SNP Identification
2.4. Imputation
2.5. Metabolomics Analysis
2.6. Statistical Analysis
2.7. Genomic Loci
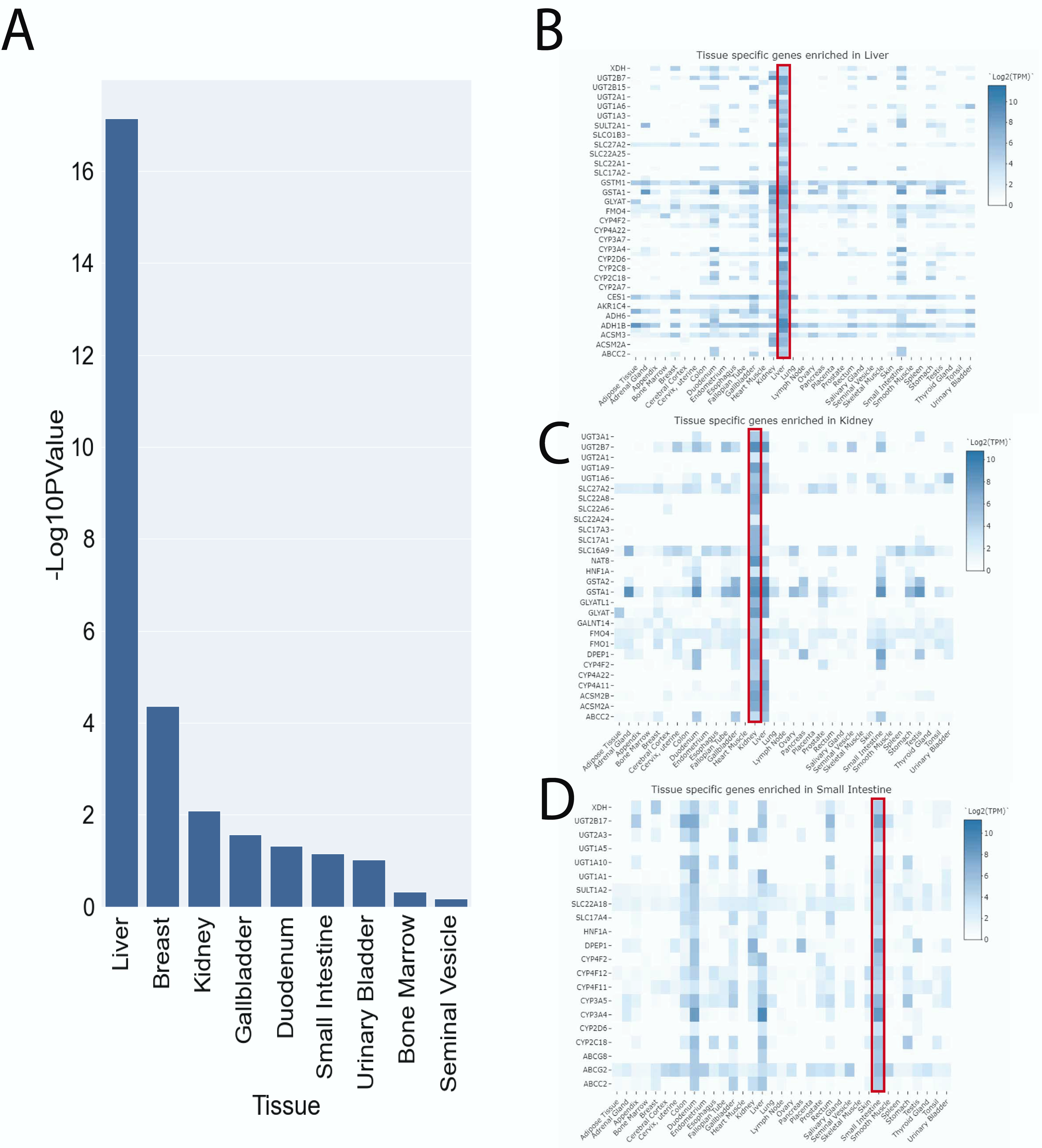
2.8. Tissue-Specific Enrichment
2.9. Disease and Pharmaceutical Variant Associations
3. Results
3.1. 77 Genomic Loci Are Linked to Circulating Levels of Small, Polar Bioactive Molecules
3.2. Tissue-Specific Enrichment of Genes with SNPs Shows Overrepresentation of Liver Genes
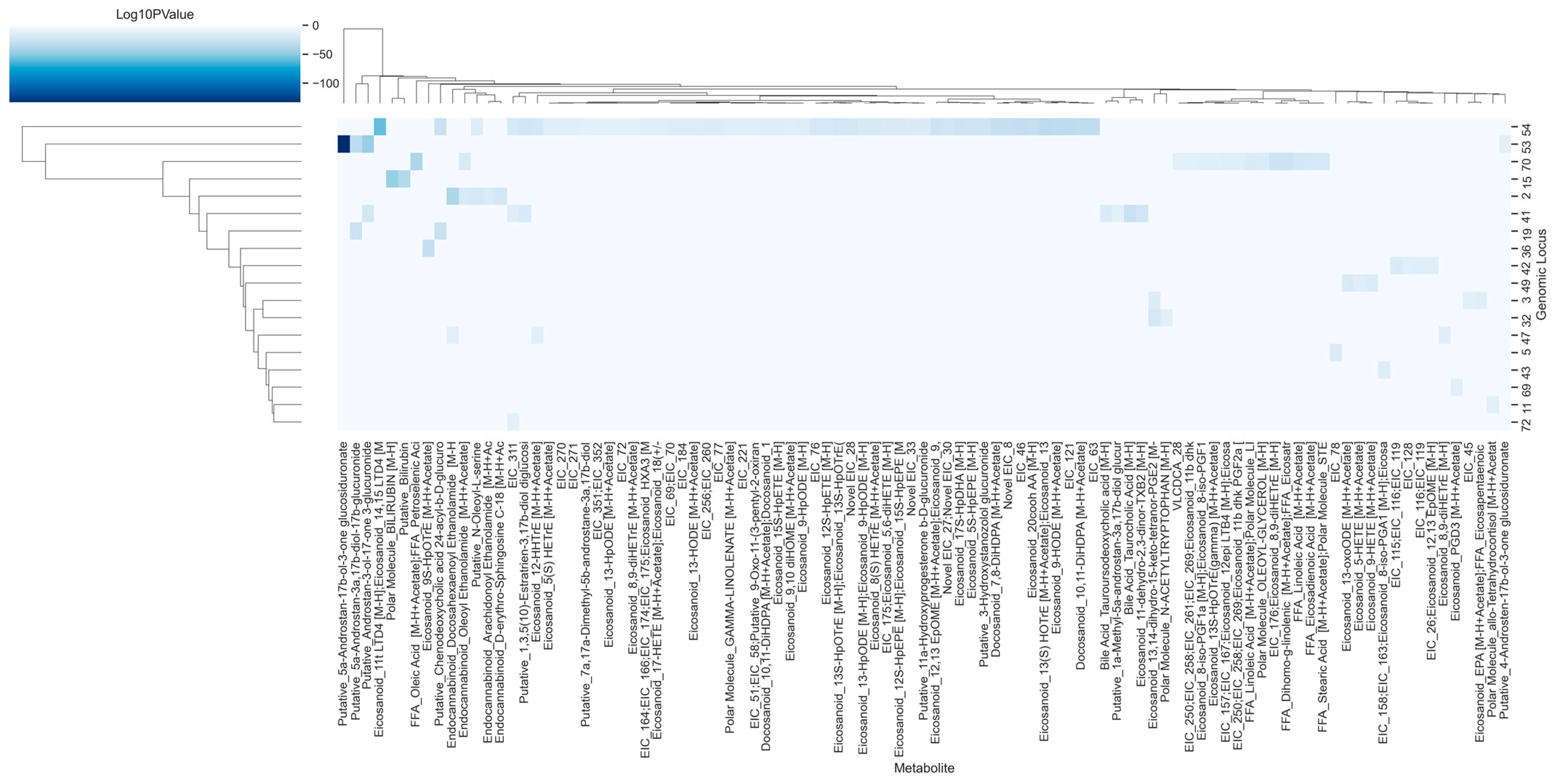
3.3. Unidentified Metabolites Are Potentially Regulated by Distinct Genomic Loci
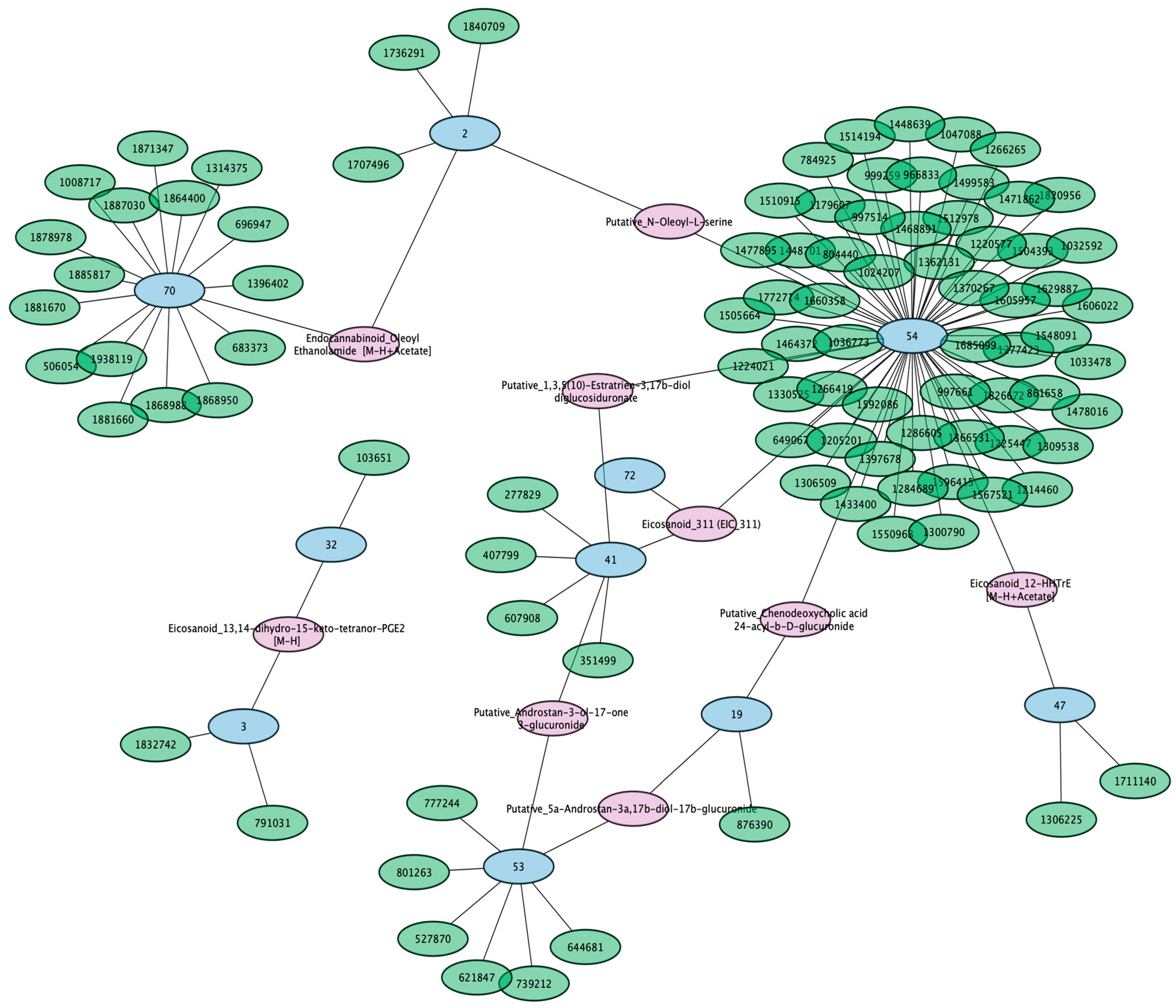
3.4. Circulating Eicosanoids, Fatty Acids, and Bile Acids Are Impacted by SNPs in 18 Genomic Loci
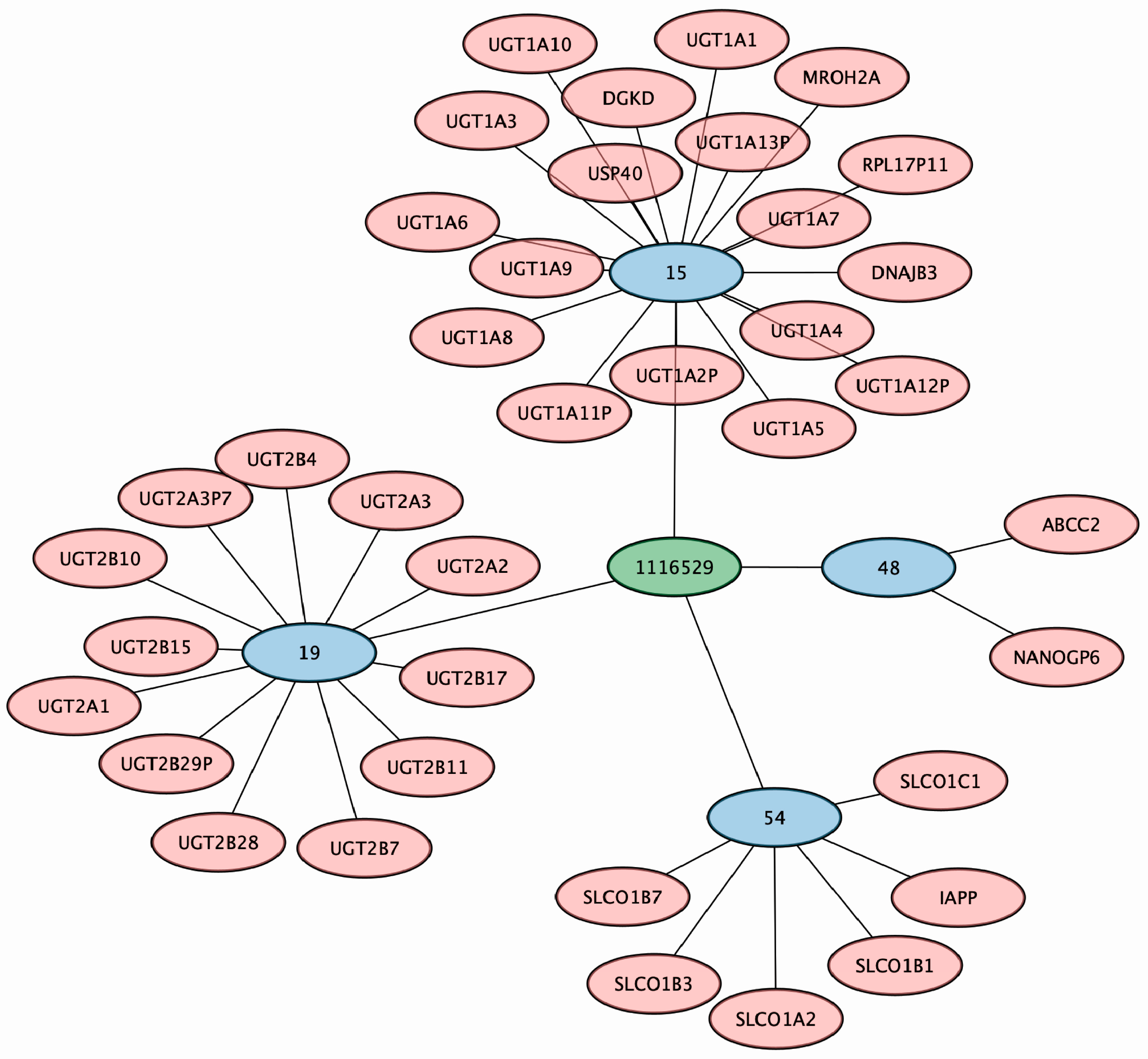
3.5. A Putative Eicosanoid Is Independently Associated with SNPs in Phase I and II Drug Metabolism and Transporter Genes
3.6. Conjugated Sex Steroids Are Strongly Associated with SLC22 Genes
3.7. SNPs in Drug Transporter and DME Genes Are Pleiotropic and Linked to Multiple Identified Metabolites
3.8. Implicated SNPs in Endogenous Metabolism Have Been Reported to Impact Drug Handling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Kooperberg, C.; LeBlanc, M.; Obenchain, V. Risk Prediction Using Genome-Wide Association Studies. Genet. Epidemiol. 2010, 34, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bosse, Y.; Pare, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Breen, G.; Li, Q.; Roth, B.L.; O’Donnell, P.; Didriksen, M.; Dolmetsch, R.; O’Reilly, P.F.; Gaspar, H.A.; Manji, H.; Huebel, C.; et al. Translating genome-wide association findings into new therapeutics for psychiatry. Nat. Neurosci. 2016, 19, 1392–1396. [Google Scholar] [CrossRef]
- van der Sijde, M.R.; Ng, A.; Fu, J.Y. Systems genetics: From GWAS to disease pathways. Bba-Mol. Basis. Dis. 2014, 1842, 1903–1909. [Google Scholar] [CrossRef] [PubMed]
- Rhee, E.P.; Ho, J.E.; Chen, M.H.; Shen, D.X.; Cheng, S.; Larson, M.G.; Ghorbani, A.; Shi, X.; Helenius, I.T.; O’Donnell, C.J.; et al. A Genome-wide Association Study of the Human Metabolome in a Community-Based Cohort. Cell Metab. 2013, 18, 130–143. [Google Scholar] [CrossRef]
- Suhre, K.; Arnold, M.; Bhagwat, A.M.; Cotton, R.J.; Engelke, R.; Raffler, J.; Sarwath, H.; Thareja, G.; Wahl, A.; DeLisle, R.K.; et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017, 8, 14357. [Google Scholar] [CrossRef]
- Raffler, J.; Friedrich, N.; Arnold, M.; Kacprowski, T.; Rueedi, R.; Altmaier, E.; Bergmann, S.; Budde, K.; Gieger, C.; Homuth, G.; et al. Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality. PLoS Genet. 2015, 11, e1005487. [Google Scholar] [CrossRef]
- Shin, S.Y.; Fauman, E.B.; Petersen, A.K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef]
- Suhre, K.; Shin, S.Y.; Petersen, A.K.; Mohney, R.P.; Meredith, D.; Wagele, B.; Altmaier, E.; Deloukas, P.; Erdmann, J.; Grundberg, E.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54-U60. [Google Scholar] [CrossRef]
- Rosenthal, S.B.; Bush, K.T.; Nigam, S.K. A Network of SLC and ABC Transporter and DME Genes Involved in Remote Sensing and Signaling in the Gut-Liver-Kidney Axis. Sci. Rep. 2019, 9, 11879. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.J.; Li, Y.Q. Interplay of Drug-Metabolizing Enzymes and Transporters in Drug Absorption and Disposition. Curr. Drug Metab. 2014, 15, 915–941. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.K.; Bush, K.T.; Martovetsky, G.; Ahn, S.Y.; Liu, H.C.; Richard, E.; Bhatnagar, V.; Wu, W. The organic anion transporter (OAT) family: A systems biology perspective. Physiol. Rev. 2015, 95, 83–123. [Google Scholar] [CrossRef] [PubMed]
- Yee, S.W.; Giacomini, K.M. Emerging Roles of the Human Solute Carrier 22 Family. Drug Metab. Dispos. 2021, 50, 1193–1210. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.K.; Granados, J.C. OAT, OATP, and MRP Drug Transporters and the Remote Sensing and Signaling Theory. Annu. Rev. Pharmacol. Toxicol. 2022, 63. [Google Scholar] [CrossRef]
- Li, J.; Bluth, M.H. Pharmacogenomics of drug metabolizing enzymes and transporters: Implications for cancer therapy. Pharmacogen. Pers. Med. 2011, 4, 11–33. [Google Scholar] [CrossRef]
- Ahmed, S.; Zhou, Z.; Zhou, J.; Chen, S.Q. Pharmacogenomics of Drug Metabolizing Enzymes and Transporters: Relevance to Precision Medicine. Genom. Proteom. Bioinform. 2016, 14, 298–313. [Google Scholar] [CrossRef]
- Nigam, S.K.; Granados, J.C. A Biological Basis for Pharmacokinetics: The Remote Sensing and Signaling Theory. Clin. Pharmacol. Ther. 2022, 112, 456–460. [Google Scholar] [CrossRef]
- Nigam, S.K. What do drug transporters really do? Nat. Rev. Drug Discov. 2015, 14, 29–44. [Google Scholar] [CrossRef]
- Smith, C.J.; Sinnott-Armstrong, N.; Cichonska, A.; Julkunen, H.; Fauman, E.B.; Wurtz, P.; Pritchard, J.K. Integrative analysis of metabolite GWAS illuminates the molecular basis of pleiotropy and genetic correlation. Elife 2022, 11, e79348. [Google Scholar] [CrossRef]
- Yin, X.; Chan, L.S.; Bose, D.; Jackson, A.U.; VandeHaar, P.; Locke, A.E.; Fuchsberger, C.; Stringham, H.M.; Welch, R.; Yu, K.; et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat. Commun. 2022, 13, 1644. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, P.; Li, Y.; Sekula, P.; Raffler, J.; Grundner-Culemann, F.; Pietzner, M.; Cheng, Y.; Wuttke, M.; Steinbrenner, I.; Schultheiss, U.T.; et al. Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nat. Genet. 2020, 52, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Granados, J.C.; Bhatnagar, V.; Nigam, S.K. Blockade of organic anion transport in humans after treatment with the drug probenecid leads to major metabolic alterations in plasma and urine. Clin. Pharmacol. Ther. 2022, 112, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Granados, J.C.; Nigam, A.K.; Bush, K.T.; Jamshidi, N.; Nigam, S.K. A key role for the transporter OAT1 in systemic lipid metabolism. J. Biol. Chem. 2021, 100603. [Google Scholar] [CrossRef]
- Granados, J.C.; Richelle, A.; Gutierrez, J.M.; Zhang, P.; Zhang, X.; Bhatnagar, V.; Lewis, N.E.; Nigam, S.K. Coordinate regulation of systemic and kidney tryptophan metabolism by the drug transporters OAT1 and OAT3. J. Biol. Chem. 2021, 100575. [Google Scholar] [CrossRef]
- Liu, H.C.; Jamshidi, N.; Chen, Y.C.; Eraly, S.A.; Cho, S.Y.; Bhatnagar, V.; Wu, W.; Bush, K.T.; Abagyan, R.; Palsson, B.O.; et al. An Organic Anion Transporter 1 (OAT1)-centered Metabolic Network. J. Biol. Chem. 2016, 291, 19474–19486. [Google Scholar] [CrossRef]
- Ahn, S.Y.; Jamshidi, N.; Mo, M.L.; Wu, W.; Eraly, S.A.; Dnyanmote, A.; Bush, K.T.; Gallegos, T.F.; Sweet, D.H.; Palsson, B.O.; et al. Linkage of Organic Anion Transporter-1 to Metabolic Pathways through Integrated “Omics”-driven Network and Functional Analysis. J. Biol. Chem. 2011, 286, 31522–31531. [Google Scholar] [CrossRef]
- Jansen, J.; Jansen, K.; Neven, E.; Poesen, R.; Othman, A.; van Mil, A.; Sluijter, J.; Sastre Torano, J.; Zaal, E.A.; Berkers, C.R.; et al. Remote sensing and signaling in kidney proximal tubules stimulates gut microbiome-derived organic anion secretion. Proc. Natl. Acad. Sci. USA 2019, 116, 16105–16110. [Google Scholar] [CrossRef]
- Bhatnagar, V.; Richard, E.L.; Wu, W.; Nievergelt, C.M.; Lipkowitz, M.S.; Jeff, J.; Maihofer, A.X.; Nigam, S.K. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: Potential role of remote sensing and signaling. Clin. Kidney J. 2016, 9, 444–453. [Google Scholar] [CrossRef]
- Sever, R.; Glass, C.K. Signaling by Nuclear Receptors. Csh. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef]
- Lowenstein, J.; Nigam, S.K. Uremic Toxins in Organ Crosstalk. Front. Med. 2021, 8, 592602. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.K.; Bush, K.T.; Bhatnagar, V.; Poloyac, S.M.; Momper, J.D. The Systems Biology of Drug Metabolizing Enzymes and Transporters: Relevance to Quantitative Systems Pharmacology. Clin. Pharmacol. Ther. 2020, 108, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, A.; Welch, M.A.; van Vlijmen, H.; Korzekwa, K.; Swaan, P.W.; Matsson, P. Molecular Modeling of Drug-Transporter Interactions-An International Transporter Consortium Perspective. Clin. Pharmacol. Ther. 2018, 104, 818–835. [Google Scholar] [CrossRef]
- Lagerborg, K.A.; Watrous, J.D.; Cheng, S.; Jain, M. High-Throughput Measure of Bioactive Lipids Using Non-targeted Mass Spectrometry. Methods Mol. Biol. 2019, 1862, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Watrous, J.D.; Niiranen, T.J.; Lagerborg, K.A.; Henglin, M.; Xu, Y.J.; Rong, J.; Sharma, S.; Vasan, R.S.; Larson, M.G.; Armando, A.; et al. Directed Non-targeted Mass Spectrometry and Chemical Networking for Discovery of Eicosanoids and Related Oxylipins. Cell Chem. Biol. 2019, 26, 433–442.e434. [Google Scholar] [CrossRef]
- Tsao, C.W.; Vasan, R.S. Cohort Profile: The Framingham Heart Study (FHS): Overview of milestones in cardiovascular epidemiology. Int. J. Epidemiol. 2015, 44, 1800–1813. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Loh, P.R.; Tucker, G.; Bulik-Sullivan, B.K.; Vilhjalmsson, B.J.; Finucane, H.K.; Salem, R.M.; Chasman, D.I.; Ridker, P.M.; Neale, B.M.; Berger, B.; et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015, 47, 284–290. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Fuma: Functional Mapping and Annotation of Genetic Associations. European Neuropsychopharmacol. 2019, 29, S789–S790. [Google Scholar] [CrossRef]
- Khramtsova, E.A.; Stranger, B.E. Assocplots: A Python package for static and interactive visualization of multiple-group GWAS results. Bioinformatics 2017, 33, 432–434. [Google Scholar] [CrossRef]
- Jain, A.; Tuteja, G. TissueEnrich: Tissue-specific gene enrichment analysis. Bioinformatics 2019, 35, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Whirl-Carrillo, M.; McDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; Klein, T.E. Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef]
- Sladek, F.M. What are nuclear receptor ligands? Mol. Cell Endocrinol. 2011, 334, 3–13. [Google Scholar] [CrossRef]
- Wacker, D.; Stevens, R.C.; Roth, B.L. How Ligands Illuminate GPCR Molecular Pharmacology. Cell 2017, 170, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Joshi, A.D.; Andersson, C.; Buch, S.; Stender, S.; Noordam, R.; Weng, L.C.; Weeke, P.E.; Auer, P.L.; Boehm, B.; Chen, C.; et al. Four Susceptibility Loci for Gallstone Disease Identified in a Meta-analysis of Genome-Wide Association Studies. Gastroenterology 2016, 151, 351–363.e328. [Google Scholar] [CrossRef]
- Yee, S.W.; Giacomini, M.M.; Hsueh, C.H.; Weitz, D.; Liang, X.; Goswami, S.; Kinchen, J.M.; Coelho, A.; Zur, A.A.; Mertsch, K.; et al. Metabolomic and Genome-wide Association Studies Reveal Potential Endogenous Biomarkers for OATP1B1. Clin. Pharmacol. Ther. 2016, 100, 524–536. [Google Scholar] [CrossRef]
- Engelhart, D.C.; Granados, J.C.; Shi, D.; Saier Jr, M.H., Jr.; Baker, M.E.; Abagyan, R.; Nigam, S.K. Systems Biology Analysis Reveals Eight SLC22 Transporter Subgroups, Including OATs, OCTs, and OCTNs. Int. J. Mol. Sci. 2020, 21, 1791. [Google Scholar] [CrossRef]
- Yee, S.; Stecula, A.; Chien, H.; Zou, L.; Feofanova, E.; van Borselen, M.; Artegoitia, V.; Newman, J.; Cheung, K.; Yousri, N.; et al. Unraveling the Functional Role of Orphan Transporter Slc22a24 as an Anion Transporter Selective for Steroid Conjugates. Clin. Pharmacol. Ther. 2020, 107, S118. [Google Scholar]
- Galetin, A.; Ito, K.; Hallifax, D.; Houston, J.B. CYP3A4 substrate selection and substitution in the prediction of potential drug-drug interactions. J. Pharmacol. Exp. Ther. 2005, 314, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Strobel, H.W. Cytochrome P4504F subfamily: At the crossroads of eicosanoid and drug metabolism. Pharmacol. Therapeut. 2006, 112, 589–611. [Google Scholar] [CrossRef]
- Roth, M.; Obaidat, A.; Hagenbuch, B. OATPs, OATs and OCTs: The organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. Br. J. Pharmacol. 2012, 165, 1260–1287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Azad, P.; Engelhart, D.C.; Haddad, G.G.; Nigam, S.K. SLC22 Transporters in the Fly Renal System Regulate Response to Oxidative Stress In Vivo. Int. J. Mol. Sci. 2021, 22, 13407. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, D.C.; Azad, P.; Ali, S.; Granados, J.C.; Haddad, G.G.; Nigam, S.K. Drosophila SLC22 Orthologs Related to OATs, OCTs, and OCTNs Regulate Development and Responsiveness to Oxidative Stress. Int. J. Mol. Sci. 2020, 21, 2002. [Google Scholar] [CrossRef]
- Bush, K.T.; Wu, W.; Lun, C.; Nigam, S.K. The drug transporter OAT3 (SLC22A8) and endogenous metabolite communication via the gut-liver-kidney axis. J. Biol. Chem. 2017, 292, 15789–15803. [Google Scholar] [CrossRef]
- Volpe, D.A.; Balimane, P.V. Application of in vitro CYP and transporter assays to predict clinical drug-drug interactions. Bioanalysis 2018, 10, 619–623. [Google Scholar] [CrossRef]
- Demel, M.A.; Schwaha, R.; Kramer, O.; Ettmayer, P.; Haaksma, E.E.; Ecker, G.F. In silico prediction of substrate properties for ABC-multidrug transporters. Expert Opin. Drug Metab. Toxicol. 2008, 4, 1167–1180. [Google Scholar] [CrossRef]
- Hohne, M.; Schatzle, S.; Jochens, H.; Robins, K.; Bornscheuer, U.T. Rational assignment of key motifs for function guides in silico enzyme identification. Nat. Chem. Biol. 2010, 6, 807–813. [Google Scholar] [CrossRef]
- Yu, M.S.; Lee, H.M.; Park, A.; Park, C.; Ceong, H.; Rhee, K.H.; Na, D. In silico prediction of potential chemical reactions mediated by human enzymes. BMC Bioinform. 2018, 19, 207. [Google Scholar] [CrossRef]
- Guo, Y.Z. Be Cautious with Crystal Structures of Membrane Proteins or Complexes Prepared in Detergents. Crystals 2020, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Raj, D.; Tomar, B.; Lahiri, A.; Mulay, S.R. The gut-liver-kidney axis: Novel regulator of fatty liver associated chronic kidney disease. Pharmacol. Res. 2020, 152, 104617. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, M. Drug transport in intestine, liver and kidney. Arch. Toxicol. 1987, 60, 37–42. [Google Scholar] [CrossRef]
- Nigam, S.K.; Bhatnagar, V. The systems biology of uric acid transporters: The role of remote sensing and signaling. Curr. Opin. Neph. Hypertens 2018, 27, 305–313. [Google Scholar] [CrossRef]
- Pochini, L.; Galluccio, M.; Scalise, M.; Console, L.; Indiveri, C. OCTN: A Small Transporter Subfamily with Great Relevance to Human Pathophysiology, Drug Discovery, and Diagnostics. SLAS Discov. 2019, 24, 89–110. [Google Scholar] [CrossRef]
- Bosma, P.J.; Seppen, J.; Goldhoorn, B.; Bakker, C.; Elferink, R.P.J.O.; Chowdhury, J.R.; Chowdhury, N.R.; Jansen, P.L.M. Bilirubin Udp-Glucuronosyltransferase-1 Is the Only Relevant Bilirubin Glucuronidating Isoform in Man. J. Biol. Chem. 1994, 269, 17960–17964. [Google Scholar] [CrossRef]
- Theken, K.N.; Deng, Y.M.; Kannon, M.A.; Miller, T.M.; Poloyac, S.M.; Lee, C.R. Activation of the Acute Inflammatory Response Alters Cytochrome P450 Expression and Eicosanoid Metabolism. Drug Metab. Dispos. 2011, 39, 22–29. [Google Scholar] [CrossRef]
- Sehgal, N.; Agarwal, V.; Valli, R.K.; Joshi, S.D.; Antonovic, L.; Strobel, H.W.; Ravindranath, V. Cytochrome P4504f, a potential therapeutic target limiting neuroinflammation. Biochem. Pharmacol. 2011, 82, 53–64. [Google Scholar] [CrossRef]
- Edson, K.Z.; Rettie, A.E. CYP4 Enzymes As Potential Drug Targets: Focus on Enzyme Multiplicity, Inducers and Inhibitors, and Therapeutic Modulation of 20-Hydroxyeicosatetraenoic Acid (20-HETE) Synthase and Fatty Acid omega-Hydroxylase Activities. Curr. Top. Med. Chem. 2013, 13, 1429–1440. [Google Scholar] [CrossRef]
- Hardwick, J.P.; Eckman, K.; Lee, Y.K.; Abdelmegeed, M.A.; Esterle, A.; Chilian, W.M.; Chiang, J.Y.; Song, B.J. Eicosanoids in Metabolic Syndrome. Adv. Pharmacol. 2013, 66, 157–266. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A. SLCO1B1 Polymorphisms and Statin-Induced Myopathy. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, C.; Mangravite, L.; Klein, T.; Altman, R. PharmGKB very important pharmacogene: SLCO1B1. Pharmacogenet. Genom. 2010, 20, 211–216. [Google Scholar] [CrossRef]
- Dainese, E.; Oddi, S.; Simonetti, M.; Sabatucci, A.; Angelucci, C.B.; Ballone, A.; Dufrusine, B.; Fezza, F.; De Fabritiis, G.; Maccarrone, M. The endocannabinoid hydrolase FAAH is an allosteric enzyme. Sci. Rep. 2020, 10, 2292. [Google Scholar] [CrossRef]
- Zheng, W.; Hussein, S.; Yang, J.; Huang, J.; Zhang, F.; Hernandez-Anzaldo, S.; Fernandez-Patron, C.; Cao, Y.; Zeng, H.B.; Tang, J.F.; et al. A novel PKD2L1 C-terminal domain critical for trimerization and channel function. Sci. Rep. 2015, 5, 9460. [Google Scholar] [CrossRef]
- Lau, H.H.; Ng, N.H.J.; Loo, L.S.W.; Jasmen, J.B.; Teo, A.K.K. The molecular functions of hepatocyte nuclear factors - In and beyond the liver. J. Hepatol. 2018, 68, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Martovetsky, G.; Tee, J.B.; Nigam, S.K. Hepatocyte nuclear factors 4alpha and 1alpha regulate kidney developmental expression of drug-metabolizing enzymes and drug transporters. Mol. Pharmacol. 2013, 84, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Snowman, A.M.; Eliasson, M.J.L.; Cohen, N.A.; Snyder, S.H. CAPON: A protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron 1998, 20, 115–124. [Google Scholar] [CrossRef]
- Daly, A.K. Pharmacogenetics: A general review on progress to date. Brit. Med. Bull. 2017, 124, 65–79. [Google Scholar] [CrossRef]
- Pirmohamed, M. Pharmacogenomics: Relevance to Personalized Medicine. Drug Metab. Pharmacok. 2017, 32, S10. [Google Scholar] [CrossRef]
- Wu, W.; Dnyanmote, A.V.; Nigam, S.K. Remote Communication through Solute Carriers and ATP Binding Cassette Drug Transporter Pathways: An Update on the Remote Sensing and Signaling Hypothesis. Mol. Pharmacol. 2011, 79, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.Y.; Nigam, S.K. Toward a Systems Level Understanding of Organic Anion and Other Multispecific Drug Transporters: A Remote Sensing and Signaling Hypothesis. Mol. Pharmacol. 2009, 76, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.K.; Wu, W.; Bush, K.T.; Hoenig, M.P.; Blantz, R.C.; Bhatnagar, V. Handling of Drugs, Metabolites, and Uremic Toxins by Kidney Proximal Tubule Drug Transporters. Clin. J. Am. Soc. Nephrol. 2015, 10, 2039–2049. [Google Scholar] [CrossRef] [PubMed]




| Category | Value |
|---|---|
| Participants | 2886 |
| Men | 1315 |
| Women | 1571 (54.4%) |
| Age | 66 ± 9 years |
| Body Mass Index (BMI) | 28.3 ± 5.4 kg/m2 |
| Genomic Locus 1 | Genomic Locus 2 | Genomic Locus 3 | Genomic Locus 4 | mtb | MZ | RT |
|---|---|---|---|---|---|---|
| 4 | 28 | 29 | 54 | 1,380,594 | 284.2233 | 4.248833 |
| 15 | 19 | 48 | 54 | 1,116,529 | 607.3553 | 3.428667 |
| 28 | 29 | 46 | 54 | 1,272,586 | 282.2076 | 3.878833 |
| 28 | 29 | 31 | 46 | 1,291,919 | 282.2085 | 3.959 |
| 28 | 29 | 46 | 54 | 1,592,026 | 310.2399 | 5.155334 |
| Identity | Genomic Locus 1 | Genomic Locus 2 | MZ | RT |
|---|---|---|---|---|
| Eicosanoid_13,14-dihydro-15-keto-tetranor-PGE2 [M-H] | 3 | 32 | 297.1744 | 1.816083 |
| Eicosanoid_12-HHTrE [M-H+Acetate] | 47 | 54 | 339.2178 | 3.669167 |
| Eicosanoid_12-HHTrE [M-H+Acetate] | 47 | 54 | 339.2197 | 3.766292 |
| Putative_N-Oleoyl-L-serine | 2 | 54 | 368.2847 | 6.353979 |
| Putative_N-Oleoyl-L-serine | 2 | 54 | 368.286 | 6.253 |
| Endocannabinoid_Oleoyl Ethanolamide [M-H+Acetate] | 2 | 70 | 384.3096 | 6.401 |
| Endocannabinoid_Oleoyl Ethanolamide [M-H+Acetate] | 2 | 70 | 384.3174 | 6.479625 |
| Putative_Androstan-3-ol-17-one 3-glucuronide | 41 | 53 | 465.2491 | 2.152167 |
| Putative_Androstan-3-ol-17-one 3-glucuronide | 41 | 53 | 465.2492 | 2.0535 |
| Putative_Androstan-3-ol-17-one 3-glucuronide | 41 | 53 | 465.2498 | 2.2015 |
| Putative_5a-Androstan-3a,17b-diol-17b-glucuronide | 19 | 53 | 467.2574 | 2.035 |
| Putative_5a-Androstan-3a,17b-diol-17b-glucuronide | 19 | 53 | 467.2634 | 1.89625 |
| Putative_Chenodeoxycholic acid 24-acyl-b-D-glucuronide | 19 | 54 | 567.3177 | 2.890625 |
| Putative_Chenodeoxycholic acid 24-acyl-b-D-glucuronide | 19 | 54 | 567.3182 | 2.76575 |
| Putative_1,3,5(10)-Estratrien-3,17b-diol diglucosiduronate | 41 | 54 | 623.3406 | 2.713333 |
| Putative_1,3,5(10)-Estratrien-3,17b-diol diglucosiduronate | 41 | 54 | 623.342 | 2.58075 |
| Putative_1,3,5(10)-Estratrien-3,17b-diol diglucosiduronate | 41 | 54 | 623.3441 | 2.540667 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Granados, J.C.; Watrous, J.D.; Long, T.; Rosenthal, S.B.; Cheng, S.; Jain, M.; Nigam, S.K. Regulation of Human Endogenous Metabolites by Drug Transporters and Drug Metabolizing Enzymes: An Analysis of Targeted SNP-Metabolite Associations. Metabolites 2023, 13, 171. https://doi.org/10.3390/metabo13020171
Granados JC, Watrous JD, Long T, Rosenthal SB, Cheng S, Jain M, Nigam SK. Regulation of Human Endogenous Metabolites by Drug Transporters and Drug Metabolizing Enzymes: An Analysis of Targeted SNP-Metabolite Associations. Metabolites. 2023; 13(2):171. https://doi.org/10.3390/metabo13020171
Chicago/Turabian StyleGranados, Jeffry C., Jeramie D. Watrous, Tao Long, Sara Brin Rosenthal, Susan Cheng, Mohit Jain, and Sanjay K. Nigam. 2023. "Regulation of Human Endogenous Metabolites by Drug Transporters and Drug Metabolizing Enzymes: An Analysis of Targeted SNP-Metabolite Associations" Metabolites 13, no. 2: 171. https://doi.org/10.3390/metabo13020171
APA StyleGranados, J. C., Watrous, J. D., Long, T., Rosenthal, S. B., Cheng, S., Jain, M., & Nigam, S. K. (2023). Regulation of Human Endogenous Metabolites by Drug Transporters and Drug Metabolizing Enzymes: An Analysis of Targeted SNP-Metabolite Associations. Metabolites, 13(2), 171. https://doi.org/10.3390/metabo13020171






