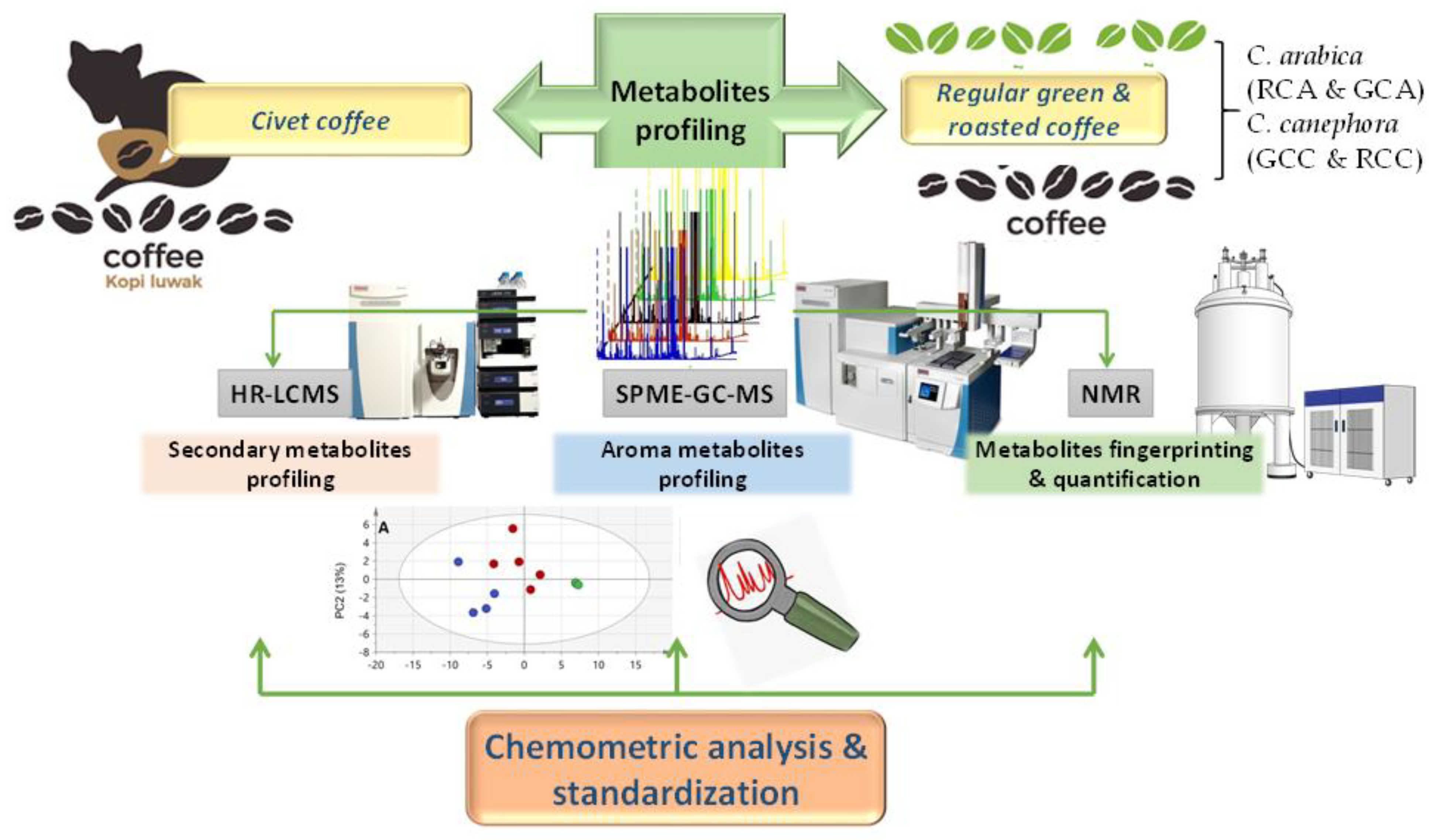
Metabolite Profiling of Premium Civet Luwak Bio-Transformed Coffee Compared with Conventional Coffee Types, as Analyzed Using Chemometric Tools
Abstract
1. Introduction
2. Materials and Methods
2.1. Coffee Specimens, Chemicals, and Extraction
2.2. UPLC-MS Profiling of Secondary Metabolites
2.3. Headspace SPME GC-MS Profiling of Aroma Compounds
2.4. NMR Fingerprinting of Coffee Extracts
2.5. Data Processing and Multivariate Analysis
2.5.1. NMR Quantification of Coffee Metabolites and Dataset Modeling
2.5.2. SPME-GC/MS Dataset Volatiles Identification and Modeling
2.5.3. UPLC-ESI–HRMS Dataset Metabolite Identification and Modeling
3. Results
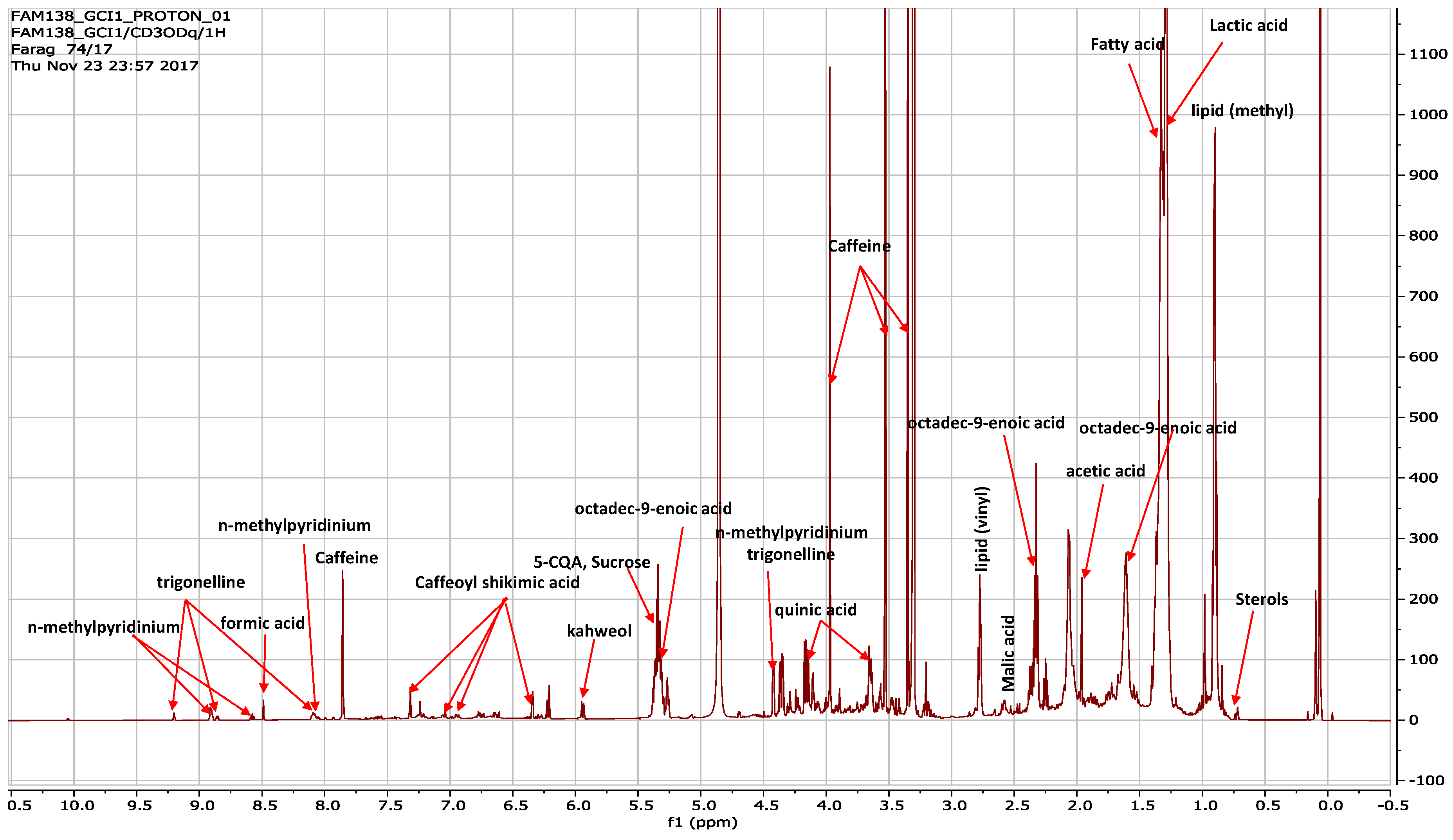
3.1. H-NMR Assignments and the Quantification of Coffee Metabolites
3.1.1. Identification of Coffee Metabolites
3.1.2. NMR Metabolites Quantification
3.2. Metabolite Profiling via UPLC-ESI–HRMS
3.3. SPME/GC-MS Analysis of Luwak Coffee Aroma
4. Discussion
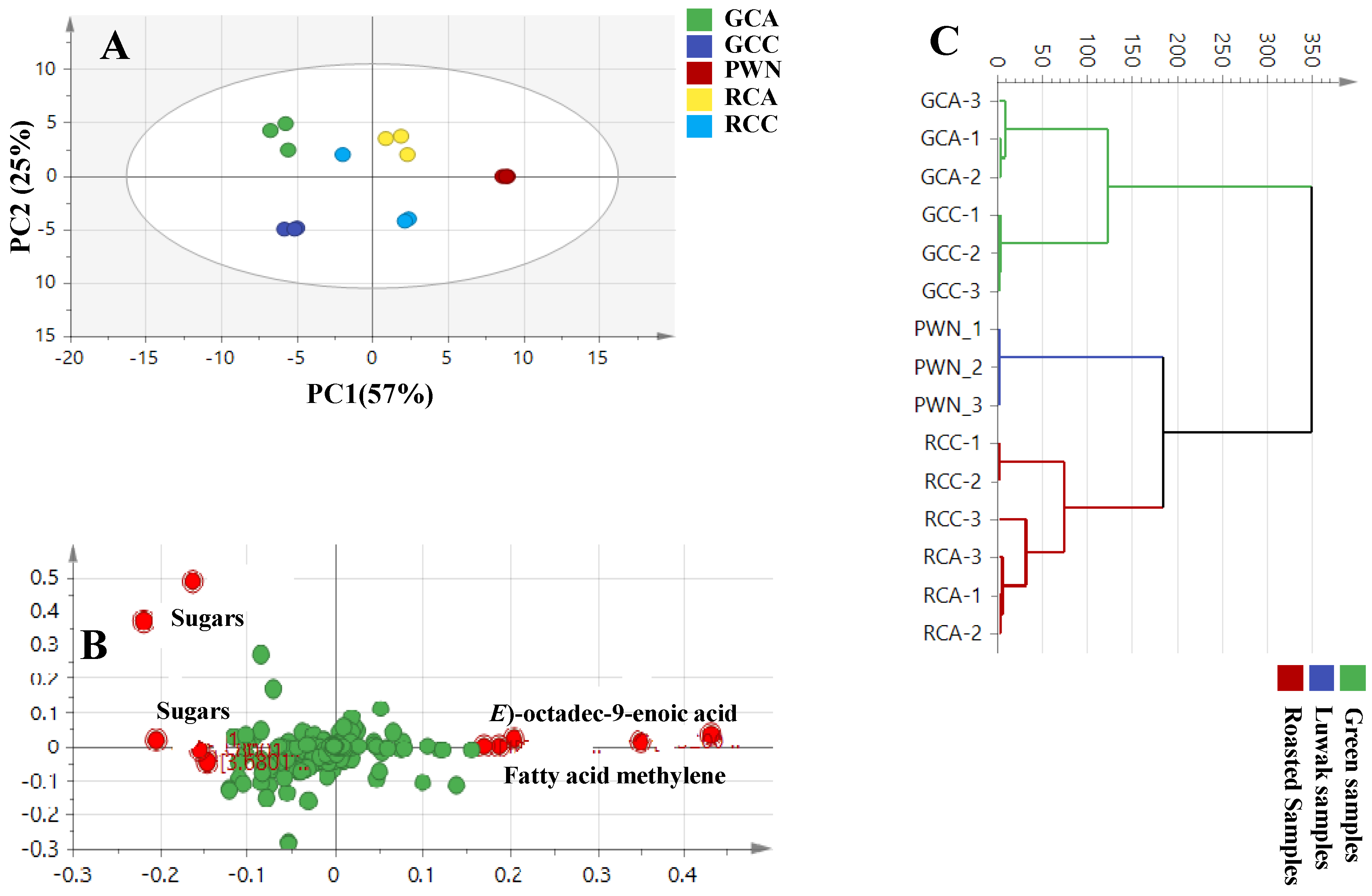
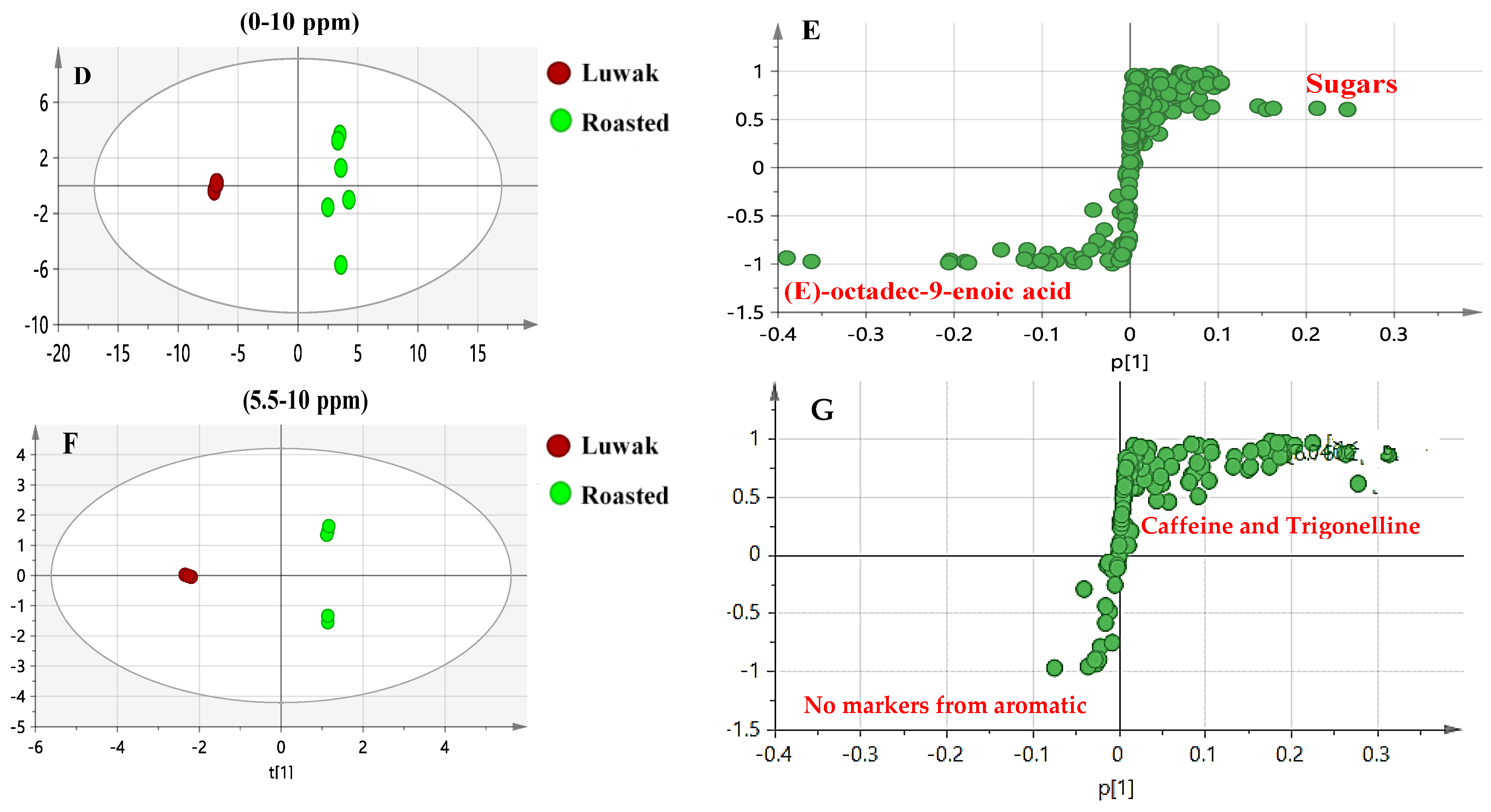
4.1. H-NMR Multivariate Data Analysis of Luwak Coffee and Authenticated Green and Roasted Coffees
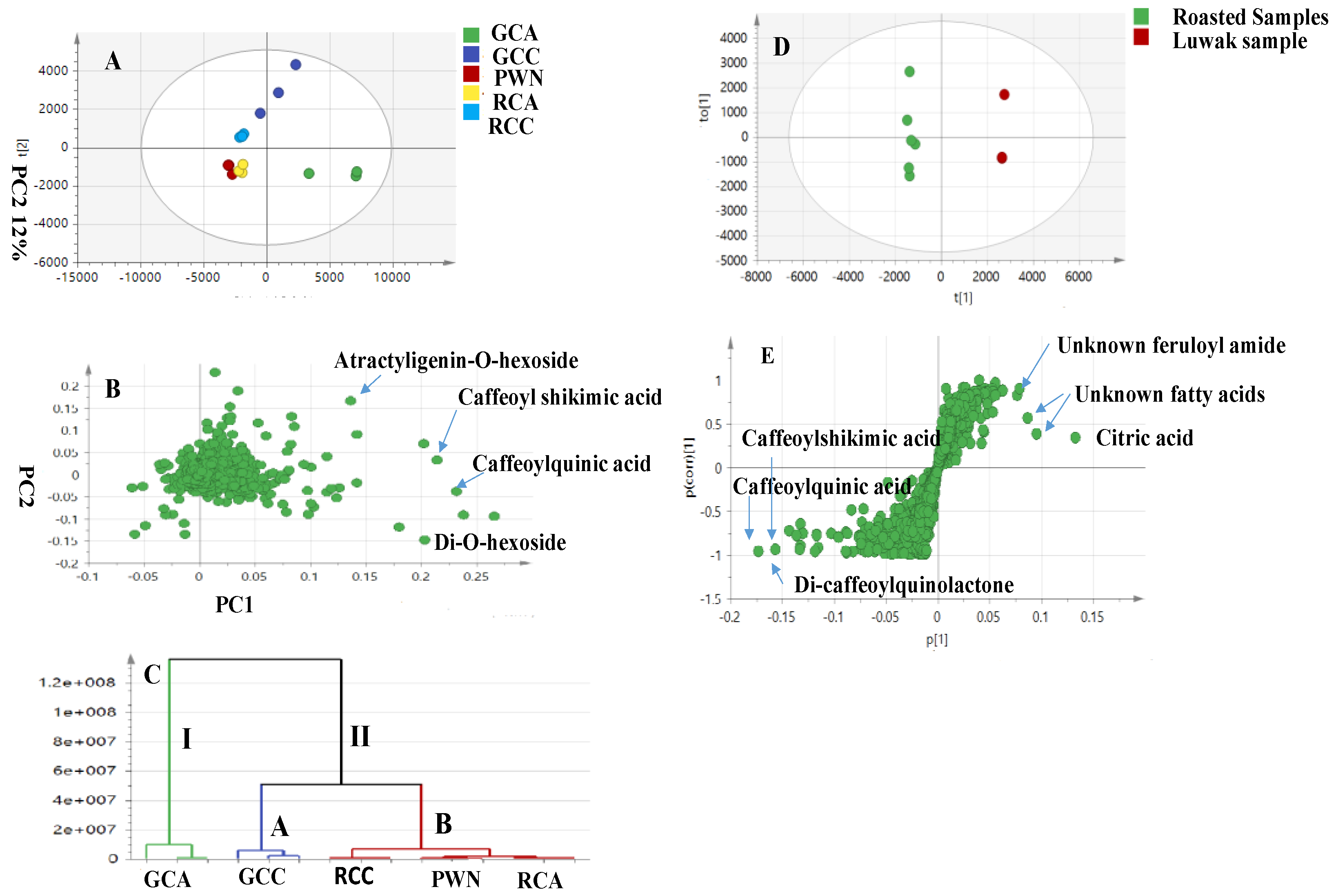
4.2. UPLC–HRMS Multivariate Data Analysis of the Luwak Coffee and Authenticated Coffee Samples
4.3. SPME-GC/MS Multivariate Data Analysis of Luwak Coffee, Roasted Coffee, and Roasted Coffee with Cardamom
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jumhawan, U.; Putri, S.P.; Yusianto; Marwani, E.; Bamba, T.; Fukusaki, E. Selection of discriminant markers for authentication of asian palm civet coffee (kopi luwak): A metabolomics approach. J. Agric. Food Chem. 2013, 61, 7994–8001. [Google Scholar] [CrossRef]
- Jumhawan, U.; Putri, S.; Yusianto; Bamba, T.; Fukusaki, E. Quantification of coffee blends for authentication of asian palm civet coffee (kopi luwak) via metabolomics: A proof of concept. J. Biosci. Bioeng. 2016, 122, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Muzaifa, M.; Hasni, D.; Rahmi, F. What is kopi luwak? A literature review on production, quality and problems. IOP Conf. Ser. Earth and Environ. Sci. 2019, 365, 012041. [Google Scholar]
- Marcone, M. Composition and properties of indonesian palm civet coffee (kopi luwak) and ethiopian civet coffee. Food Res. Int. 2004, 37, 901–912. [Google Scholar] [CrossRef]
- Lachenmeier, D.; Schwarz, S. Digested civet coffee beans (kopi luwak)—an unfortunate trend in specialty coffee caused by mislabeling of coffea liberica? Foods 2021, 10, 1329. [Google Scholar] [CrossRef]
- Iswanto, T.; Shovitri, M.; Altway, A.; Widjaja, T.; Kusumawati, D.I.; Lisdyanti, P. Isolation and identification of caffeine-degrading bacteria from soil, coffee pulp waste and excreted coffee bean in luwak feces. Feces 2019, 20. [Google Scholar] [CrossRef]
- Watanabe, H.; Ng, C.H.; Limviphuvadh, V.; Suzuki, S.; Yamada, T.J.P. Gluconobacter dominates the gut microbiome of the asian palm civet paradoxurus hermaphroditus that produces kopi luwak. luwak 2020, 8, e9579. [Google Scholar] [CrossRef] [PubMed]
- Aditiawati, P.; Astuti, D.I.; Kriswantoro, J.A.; Khanza, S.M.; Kamarisima; Irifune, T.; Amalia, F.; Fukusaki, E.; Putri, S.P. Gc/ms-based metabolic profiling for the evaluation of solid state fermentation to improve quality of arabica coffee beans. Metabolomics 2020, 16, 57. [Google Scholar] [CrossRef] [PubMed]
- Foudah, A.I.; Alam, P.; Abdel-Kader, M.S.; Shakeel, F.; Alqasoumi, S.I.; Salkini, A.M.; Yusufoglu, H.S. High-performance thin-layer chromatographic determination of trigonelline content in various extracts and different varieties of some commercial coffees available in the saudi arabian market. JPC–J. Planar Chromatogr.–Modern TLC 2020, 33, 43–50. [Google Scholar] [CrossRef]
- Foudah, A.I.; Shakeel, F.; Salkini, M.A.; Alshehri, S.; Ghoneim, M.M.; Alam, P. A green high-performance thin-layer chromatography method for the determination of caffeine in commercial energy drinks and formulations. Materials 2022, 15, 2965. [Google Scholar] [CrossRef]
- Alam, P.; Shakeel, F.; Ali, A.; Alqarni, M.H.; Foudah, A.I.; Aljarba, T.M.; Alkholifi, F.K.; Alshehri, S.; Ghoneim, M.M.; Ali, A. Simultaneous determination of caffeine and paracetamol in commercial formulations using greener normal-phase and reversed-phase hptlc methods: A contrast of validation parameters. Molecules 2022, 27, 405. [Google Scholar] [CrossRef]
- Jumhawan, U.; Putri, S.P.; Bamba, T.; Fukusaki, E. Application of gas chromatography/flame ionization detector-based metabolite fingerprinting for authentication of asian palm civet coffee (kopi luwak). J. biosci. bioeng. 2015, 120, 555–561. [Google Scholar] [CrossRef]
- El-Hawary, E.A.; Zayed, A.; Laub, A.; Modolo, L.V.; Wessjohann, L.; Farag, M.A. How does lc/ms compare to uv in coffee authentication and determination of antioxidant effects? Brazilian and middle eastern coffee as case studies. Antioxidants 2022, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Abdelwareth, A.; Zayed, A.; Farag, M.A. Chemometrics-based aroma profiling for revealing origin, roasting indices, and brewing method in coffee seeds and its commercial blends in the middle east. Food Chem. 2021, 349, 129162. [Google Scholar] [CrossRef]
- Porzel, A.; Farag, M.A.; Mülbradt, J.; Wessjohann, L.A. Metabolite profiling and fingerprinting of hypericum species: A comparison of ms and nmr metabolomics. Metabolomics 2014, 10, 574–588. [Google Scholar] [CrossRef]
- Farag, M.A.; Porzel, A.; Mahrous, E.A.; El-Massry, M.M.; Wessjohann, L.A. Integrated comparative metabolite profiling via ms and nmr techniques for senna drug quality control analysis. Anal. Bioanal. Chem. 2015, 407, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Rasheed, D.M.; Kamal, I.M. Volatiles and primary metabolites profiling in two hibiscus sabdariffa (roselle) cultivars via headspace spme-gc-ms and chemometrics. Food Res. Int. 2015, 78, 327–335. [Google Scholar] [CrossRef]
- Farag, M.A.; Hegazi, N.; Dokhalahy, E.; Khattab, A.R. Chemometrics based gc-ms aroma profiling for revealing freshness, origin and roasting indices in saffron spice and its adulteration. Food Chem. 2020, 331, 127358. [Google Scholar] [CrossRef] [PubMed]
- Zayed, A.; Abdelwareth, A.; Mohamed, T.A.; Fahmy, H.A.; Porzel, A.; Wessjohann, L.A.; Farag, M.A. Dissecting coffee seeds metabolome in context of genotype, roasting degree, and blending in the middle east using nmr and gc/ms techniques. Food Chem. 2022, 373, 131452. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Porzel, A.; Wessjohann, L.A. Unraveling the active hypoglycemic agent trigonelline in balanites aegyptiaca date fruit using metabolite fingerprinting by nmr. J. Pharm. Biomed. Anal. 2015, 115, 383–387. [Google Scholar] [CrossRef] [PubMed]
- del Campo, G.; Berregi, I.; Caracena, R.; Zuriarrain, J. Quantitative determination of caffeine, formic acid, trigonelline and 5-(hydroxymethyl) furfural in soluble coffees by 1h nmr spectrometry. Talanta 2010, 81, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Riedel, A.; Hochkogler, C.M.; Lang, R.; Bytof, G.; Lantz, I.; Hofmann, T.; Somoza, V. N-methylpyridinium, a degradation product of trigonelline upon coffee roasting, stimulates respiratory activity and promotes glucose utilization in hepg2 cells. Food Funct. 2014, 5, 454–462. [Google Scholar]
- Kotyczka, C.; Boettler, U.; Lang, R.; Stiebitz, H.; Bytof, G.; Lantz, I.; Hofmann, T.; Marko, D.; Somoza, V. Dark roast coffee is more effective than light roast coffee in reducing body weight, and in restoring red blood cell vitamin e and glutathione concentrations in healthy volunteers. Mol. Nutr. Food Res. 2011, 55, 1582–1586. [Google Scholar]
- Belton, P.S.; Colquhoun, I.J.; Kemsley, E.K.; Delgadillo, I.; Roma, P.; Dennis, M.J.; Sharman, M.; Holmes, E.; Nicholson, J.K.; Spraul, M. Application of chemometrics to the 1h nmr spectra of apple juices: Discrimination between apple varieties. Food Chem. 1998, 61, 207–213. [Google Scholar] [CrossRef]
- Girelli, C.R.; Angilè, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR Metabolite Fingerprinting Analysis Reveals a Disease Biomarker and a Field Treatment Response in Xylella fastidiosa subsp. pauca-Infected Olive Trees. Plants 2019, 8, 115. [Google Scholar] [CrossRef]
- Wei, F.; Furihata, K.; Hu, F.; Miyakawa, T.; Tanokura, M. Complex mixture analysis of organic compounds in green coffee bean extract by two-dimensional nmr spectroscopy. Magn. Reson. Chem. 2010, 48, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Hall, L. The conformations of cyclic compounds in solution. I. Shikimic acid. J. Org. Chem. 1964, 29, 297–299. [Google Scholar] [CrossRef]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High resolution nmr spectroscopy as a structural and analytical tool for unsaturated lipids in solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef] [PubMed]
- Luisa, P.F.; Flavio, M.B.; Fabiana, C.R.; Gerson, S.G.; Jose, H.d.S.T.; Marcelo, R.M. Fatty acid profiles and parameters of quality of specialty coffees produced in different brazilian regions. Afr. J. Agric. Res. 2015, 10, 3484–3493. [Google Scholar] [CrossRef]
- Yang, S.; Hao, Y.; Wang, J.; Wang, H.; Zheng, Y.; Tian, H.; Liu, Y.; Sun, B. Selective catalytic dehydration of furfuryl alcohol to 2, 2′-difurfuryl ether using a polyoxometalate catalyst. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Karunanithi, G.; Varadappan, A.M.S. Exploring the effectiveness of novel coffea arabica leaf pigment as a natural antioxidant additive for date seed biodiesel. Fuel 2022, 324, 124561. [Google Scholar] [CrossRef]
- Fang, N.; Yu, S.; Prior, R.L. Lc/ms/ms characterization of phenolic constituents in dried plums. J. Agric. Food Chem. 2002, 50, 3579–3585. [Google Scholar] [CrossRef] [PubMed]
- Asamenew, G.; Kim, H.W.; Lee, M.K.; Lee, S.H.; Lee, S.; Cha, Y.S.; Lee, S.H.; Yoo, S.M.; Kim, J.B. Comprehensive characterization of hydroxycinnamoyl derivatives in green and roasted coffee beans: A new group of methyl hydroxycinnamoyl quinate. Food Chem. X 2019, 2, 100033. [Google Scholar] [CrossRef] [PubMed]
- Bianco, G.; Abate, S.; Labella, C.; Cataldi, T.R. Identification and fragmentation pathways of caffeine metabolites in urine samples via liquid chromatography with positive electrospray ionization coupled to a hybrid quadrupole linear ion trap (ltq) and fourier transform ion cyclotron resonance mass spectrometry and tandem mass spectrometry. Rapid Commun. Mass Spectro. RCM 2009, 23, 1065–1074. [Google Scholar]
- Lee, L.W.; Cheong, M.W.; Curran, P.; Yu, B.; Liu, S.Q. Modulation of coffee aroma via the fermentation of green coffee beans with rhizopus oligosporus: Ii. Effects of different roast levels. Food Chem. 2016, 211, 925–936. [Google Scholar] [CrossRef]
- Setoyama, D.; Iwasa, K.; Seta, H.; Shimizu, H.; Fujimura, Y.; Miura, D.; Wariishi, H.; Nagai, C.; Nakahara, K. High-throughput metabolic profiling of diverse green coffea arabica beans identified tryptophan as a universal discrimination factor for immature beans. PLoS ONE 2013, 8, e70098. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.T.; Walker, M.J. Critical review of analytical and bioanalytical verification of the authenticity of coffee. J. AOAC Int. 2020, 103, 283–294. [Google Scholar] [CrossRef]
- Medina, S.; Pereira, J.A.; Silva, P.; Perestrelo, R.; Câmara, J.S. Food fingerprints—A valuable tool to monitor food authenticity and safety. Food Chem. 2019, 278, 144–162. [Google Scholar] [CrossRef]
- Putri, S.P.; Ikram, M.M.M.; Sato, A.; Dahlan, H.A.; Rahmawati, D.; Ohto, Y.; Fukusaki, E. Application of gas chromatography-mass spectrometry-based metabolomics in food science and technology. J. Biosci. Bioeng. 2022, 133, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Lee, S.; Bang, E.; Lee, S.; Rhee, J.-K.; Na, Y.-C. Comparative evaluation of flavor compounds in fermented green and roasted coffee beans by solid phase microextraction-gas chromatography/mass spectrometry. Flavour Fragr. J. 2019, 34, 365–376. [Google Scholar] [CrossRef]
- Lestari, W.; Hasballah, K.; Listiawan, M.; Sofia, S. Identification of antioxidant components of gayo arabica coffee cascara using the gc-ms method. IOP Conf. Ser. Earth and Environ. Sci. 2022, 956, 012011. [Google Scholar] [CrossRef]

| No | Metabolite | Assignment | 1H (Multiplicity) | HSQC | HMBC |
|---|---|---|---|---|---|
| 1 | (E)-octadec-9-enoic acid | H-9 | 5.33 | 129 | C-7 (27.9) |
| H-10 | 5.33 | 129 | C-12 (27.9) | ||
| H-2 | 2.35 | 34.8 | C-1 (174.6), C-3 (26.4) | ||
| H-8, H-11 | 2.3 | 34.8 | |||
| H-3 | 1.6 | 25.9 | C-1 (174.6) | ||
| 2 | Trigonelline | H-2 | 9.20 s | 147.5 | |
| H-4 | 8.89 d | 146.8 | C-5 (129.2), C-2 (146.8) | ||
| H-5 | 8.09 t | 129.2 | C-3 (143.9) | ||
| H-6 | 8.86 d | 146.8 | |||
| CH3 | 4.43 s | 48.8 | C-2 (147.5) | ||
| 3 | Caffeoyl shikimic acid | CH2-2axial | 2.09 | 40.4 | |
| CH2-2 equatorial | 2 | 40.4 | |||
| H-5 | 5.26 | 70.5 | C-4 (74.8) | ||
| H-8′ | 6.34 | 109.5 | |||
| H-7′ | 7.56 | 146.9 | |||
| H-2′ | 7.04 | 115 | |||
| H-5′ | 6.77 | 116.4 | |||
| H-6′ | 6.95 | 122.8 | |||
| 4 | Caffeine | H-8 | 3.35 | 28.1 | C-1 (156.8), C-2 (153.3) |
| H-7 | 3.53 | 30 | C-2 (153.3), C-3 (149.8) | ||
| H-6 | 3.97 | 33.7 | C-4 (108.8), C-5 (144.0) | ||
| H-5 | 7.86 | 144.1 | C-2 (115.0) | ||
| 5 | Kahweol | H-1 | 5.95 d (10.2) | 139.6 | C-5 (45.7), C-9 (49.7), C-10 (42.9) |
| H-2 | 6.21 d (10.2) | 116.2 | C-10 (42.9) | ||
| H-3 | --- | 151.5 | |||
| H-4 | --- | 122.9 | |||
| H-5 | 2.57 | 45.7 | |||
| H-9 | 1.59 | 49.7 | |||
| H-10 | --- | 42.9 | |||
| H-18 | 6.21 (10.2) | 109 | C-3 (151.5), C-4 (122.9) | ||
| H-19 | 7.23 | 141.8 | C-3 (151.5), C-4 (122.9) | ||
| H-20 | 0.97 | 15.9 | C-1 (156.8), C-5 (45.7), C-9 (49.7), C-10 (42.9) | ||
| 6 | Caffeic acid | H-2 | 7.06 d | 115 | |
| H-5 | 6.77 d | 116.3 | |||
| H-6 | 6.98 dd | 124.3 | |||
| H-7 | 7.57 d | 146.8 | |||
| H-8 | 6.28 d | 117.1 | |||
| 7 | N-methylpyridinium | H-2, H-6 | 9.90 d | 146.8 | |
| H-3, H-5 | 8.05 t | 128.4 | |||
| H-4 | 8.57 t | 146.5 | C-2 (146.8), C-6 (146.8) | ||
| CH3 | 4.42 s | 48.8 | C-2 (146.8), C-6 (146.8) | ||
| 8 | Myo-inositol | H | 3.41 dd | 73.1 | 73.1 |
| 9 | Xanthine | H-8 | 7.93 s | 132.9 | |
| 10 | Difurfuryl ether | H-3 | 6.26 | 115.4 | |
| H-4 | 6.29 | 115.3 | |||
| H-5 | 7.57 | 14.1 | |||
| 11 | fumaric acid | H | 6.61 | 132.6 | C=O (173.5) |
| 12 | Caffeine | H-8 | 3.35 | 28.1 | C-1 (156.7), C-2 (153.3) |
| H-7 | 3.53 | 30 | C-2 (153.3), C-3 (149.8) | ||
| H-6 | 3.97 | 33.7 | C-4 (108.8), C-5 (144.0) | ||
| H-5 | 7.86 | 144.1 | C-2 (108.8) | ||
| 13 | Lactic acid | CH3 | 1.34 | 23.5 | |
| CH | 4 | 68.2 | |||
| 14 | Sucrose | H-1 | 5.37 | 93.8 | C-5 (74.5), C-1′ (105.3) |
| H-2 | 3.41 | 73.1 | C-3 (74.4) | ||
| H-3 | 3.68 | 74.4 | |||
| H-4 | 3.34 | 71.4 | C-3 (74.4), C-5 (74.5) | ||
| H-5 | 3.81 | 74.5 | |||
| H-6 | 3.68 | 61.7 | |||
| H-2′ | 4.08 | 79.3 | |||
| H-3′ | 3.95 | 74.1 | C-1′ (105.3), C-5′ (64.5), C-2′ (79.3) | ||
| H-4′ | 3.75 | 83.8 | |||
| H-5′ | 3.7 | 64.5 | |||
| H-6′ | 3.75 | 63.2 |
| No | RT (min) | Compound Name | Chemical Class | [M-H]- | Molecular Formula | Mass Error | MS/MS Fragments | References |
|---|---|---|---|---|---|---|---|---|
| L1 | 0.27 | Malic acid | Organic acid | 133.01411 | C4H5O5− | −2.36 | n.d | [13] |
| L2 | 0.22 | O-Malonyl-hexopyranoside | Sugar | 471.07425 | C19H19O14− | 4.44 | n.d | [13] |
| L3 | 0.32 | Di-O-hexoside | Sugar | 341.10815 | C12H21O11− | −2.32 | 179,161 | [13] |
| L4 | 0.37 | Citric acid | Organic acid | 191.01926 | C6H7O7− | −2.43 | 111,173 | [13] |
| L5 | 0.38 | Quinic acid | Organic acid | 191.05556 | C7H11O6− | −2.86 | 173, 111 | [13] |
| L6 | 0.38 | O-Caffeoylquinic acid | Phenolic acid | 353.08664 | C16H17O9− | −3.37 | 191,179,135 | [13] |
| L7 | 3.2 | Caffeoylshikimic acid | Phenolic acid | 335.07614 | C16H15O8− | −3.21 | 179,161,135 | [13] |
| L8 | 6.1 | Dicaffeoylquinic acid | Phenolic acid | 515.11804 | C25H23O12− | −2.82 | 353,335 | [13] |
| L9 | 6.3 | Feruloylquinic acid | Phenolic acid | 367.10223 | C17H19O9− | −3.33 | 161,193,135 | [13] |
| L10 | 6.6 | P-Coumaroyl quinic acid | Phenolic acid | 337.09174 | C16H17O8− | −3.41 | 191,163 | [13] |
| L11 | 7.01 | Atractyligenin-O-hexoside | Diterpene | 481.24234 | C25H37O9− | −4.08 | 301 | [13] |
| L12 | 7.11 | Carboxtatractyligenin-O-hexoside | Diterpene | 525.23193 | C26H37O11− | −4.18 | 396,203 | [13] |
| L13 | 7.7 | Trihydroxy-kauranoic acid | Diterpene | 351.21664 | C20H31O5− | −3.12 | 289,321 | [13] |
| L14 | 7.7 | Caffeoyl-feuloylquinic acid | Phenolic acid | 529.13293 | C26H25O12− | −2.35 | 367,353 | [13] |
| L15 | 7.9 | Desoxycarboxyatractyligenin-O-hexoside | Diterpene | 771.34113 | C37H55O17− | −4.31 | 727 | [13] |
| L16 | 7.9 | Desoxyatractyligenin-O-hexoside | Diterpene | 727.35138 | C36H55O15− | −4.42 | 643,625 | [13] |
| L17 | 8.1 | Caffeoyl-N-tryptophan | Amino acid | 365.11272 | C20H17N2O5− | −2.08 | 135,229 | [13] |
| L18 | 8.3 | Unknown chlorogenic acid | Phenolic acid | 437.14359 | C21H25O10− | −3.95 | 173,275 | [13] |
| L19 | 8.47 | Di-caffeoylquinolactone | Phenolic acid | 497.10716 | C25H22O11− | −2.25 | 335 | [13] |
| L20 | 8.56 | Unknown fatty acid | Fatty acid | 538.23956 | C25H48O11N− | −4.25 | 311,198,180 | [13] |
| L21 | 8.8 | Isovaleryl-atractyligenin-O-hexoside | Diterpene | 565.29987 | C30H45O10− | −3.45 | 481,463,303 | [13] |
| L22 | 12.1 | Unknown fatty acid | Fatty acid | 311.12355 | C20H13O3− | −3.12 | 183 | [13] |
| L23 | 12.5 | Unknown fatty acid | Fatty acid | 325.18341 | C14H29O8− | −9.61 | 183 | [13] |
| L24 | 13.7 | Unknown fatty acid | Fatty acid | 339.19907 | C15H31O8− | −9.91 | 183 | [13] |
| Category | Metabolite | RI | RT | Luwak Samples | Roasted Coffee with Cardamom (R&C) | Roasted C. arabica (RCA) |
|---|---|---|---|---|---|---|
| Alcohol | 1-Octanol | 1101.5 | 10.3042 | 0.29 ± 0.251 | ||
| 2,7-dimethyl-4-Octene-2,7-diol | 1152.7 | 11.0333 | 0.2 ± 0.09 | |||
| Cineole | 1007.2 | 8.8292 | 0.35 ± 0.433 | 1.98 ± 1.405 | ||
| Furfuryl alcohol | 842.7 | 5.7867 | 12.1 ± 2.204 | 5.5 ± 7.774 | ||
| Nerolidol | 1519.7 | 15.7508 | 3.71 ± 4.156 | |||
| Maltol | 1115.9 | 10.5092 | 2.23 ± 2.078 | |||
| Aldehyde | 2-Methoxy-4-methylbenzaldehyde | 1132.7 | 10.7492 | 0.41 ± 0.361 | ||
| Cinnamic aldehyde | 1264.8 | 12.5617 | 1.44 ± 2.038 | 14.54 ± 0.247 | ||
| Aliphatic hydrocarbon | Dodecane | 1163.5 | 11.1875 | 0.17 ± 0.161 | ||
| Hexadecane | 1545.4 | 16.1208 | 0.18 ± 0.184 | 0.01 ± 0.017 | ||
| Pentadecane | 1450.6 | 14.8658 | 0.44 ± 0.372 | |||
| Tetradecane | 1356.1 | 13.7125 | 0.49 ± 0.406 | |||
| Aromatic hydrocarbon | Naphthalene | 1169.6 | 11.275 | 4.73 ± 1.833 | ||
| Styrene | 867.9 | 6.31 | 0.09 ± 0.093 | |||
| Ester | Decyl acetate | 1369.9 | 13.8808 | 0.94 ± 0.957 | 0.13 ± 0.186 | |
| Diisobutyl phthalate | 1812.3 | 20.935 | 0.09 ± 0.083 | 0.29 ± 0.332 | ||
| Ethylene diacetate | 851.8 | 5.9767 | 0.88 ± 0.802 | |||
| Linalyl acetate | 1216.4 | 11.9242 | 12.16 ± 10.269 | 2.32 ± 0.707 | ||
| Myrcenylacetate | 1285.3 | 12.8317 | 0.33 ± 0.262 | |||
| Oxalic acid, allyl isobutyl ester | 1037.2 | 9.3 | 0.07 ± 0.063 | |||
| Terpinyl acetate | 1316.9 | 13.2325 | 35.53 ± 49.328 | 13.05 ± 18.453 | ||
| Ether/oxide | Anethole | 1267.7 | 12.5992 | 3.98 ± 5.211 | ||
| Biphenyl oxide | 1382.4 | 14.035 | 0.12 ± 0.116 | |||
| Cineol | 1007.2 | 8.8292 | 0.35 ± 0.433 | 1.98 ± 1.405 | ||
| Dicyclobutylidene oxide | 1083.4 | 10.0233 | 0.61 ± 0.291 | |||
| Eugenol | 1334.8 | 13.4517 | 3.44 ± 3.801 | 5.84 ± 0.965 | ||
| Furan / pyrrole | 2-Furanmethanol | 842.7 | 5.7867 | 6.05 ± 1.102 | 2.75 ± 3.887 | |
| Acetylfuran | 891.6 | 6.8008 | 0.65 ± 0.048 | |||
| Furfural | 820.3 | 5.3225 | 4.18 ± 1.87 | 4.55 ± 6.44 | ||
| Furfuryl 3-methylbutanoate | 1190.2 | 11.5683 | 2.06 ± 1.164 | |||
| Furfuryl acetate | 969.5 | 8.185 | 6.47 ± 4.393 | |||
| 2-Furfuryl-5-methylfuran | 1149.8 | 10.9933 | 3.42 ± 0.901 | |||
| 2-Furfurylfuran | 1057.5 | 9.6175 | 0.73 ± 0.694 | |||
| 2-Pentylfuran | 963.1 | 8.0742 | 1.04 ± 0.909 | |||
| 5-Methyl furfural | 947.4 | 7.8008 | 2.56 ± 0.609 | 0.44 ± 0.626 | ||
| Difurfuryl ether | 1274.8 | 12.6925 | 8.96 ± 6.84 | |||
| 1-Furfurylpyrrole | 1157.4 | 11.1008 | 1.44 ± 0.383 | |||
| 2-Formyl-1-methylpyrrole | 990.1 | 8.5442 | 0.65 ± 0.609 | |||
| 2-Formyl-4,5-dimethyl-pyrrole | 1141.3 | 10.8717 | 0.45 ± 0.425 | |||
| N-Furfurylpyrrole | 1157.4 | 11.1008 | 1.44 ± 0.383 | |||
| Ketone | 2,2-Dimethylbutanone | 943.3 | 7.73 | 0.13 ± 0.132 | ||
| 3,3-dimethyl-2-butanone | 1550.9 | 16.2008 | 0.4 ± 0.257 | |||
| p-Anisylacetone | 1374.1 | 13.9325 | 0.2 ± 0.285 | 29.78 ± 40.553 | ||
| Monoterpene hydrocarbon | alpha-Terpineol | 1175.5 | 11.3583 | 5.69 ± 5.262 | ||
| Camphor | 1130.3 | 10.7142 | 0.23 ± 0.127 | |||
| Isoterpinolene | 1061.3 | 9.6775 | 0.6 ± 0.103 | |||
| Terpin-4-ol | 1161 | 11.1517 | 0.95 ± 0.754 | |||
| Unknown monoterpene | 948.2 | 7.815 | 0.18 ± 0.105 | |||
| Others | Butyryl lactone | 895.6 | 6.8833 | 2.74 ± 2.129 | ||
| Phenolics | 4-Ethylguaiacol | 1256 | 12.445 | 9.59 ± 4.57 | ||
| 4-Vinylguaiacol | 1302.3 | 13.0533 | 1.78 ± 1.197 | 1.28 ± 1.812 | ||
| o-Guaiacol | 1070.7 | 9.8242 | 1.52 ± 0.078 | |||
| Pyrazines | 1-(6-Methyl-2-pyrazinyl)-1-ethanone | 1097.4 | 10.2417 | 0.28 ± 0.262 | ||
| 2,3-dimethylpyrazine | 899.9 | 6.9725 | 0.1 ± 0.099 | |||
| 2,6-dimethylpyrazine | 894.8 | 6.8667 | 1.62 ± 1.205 | |||
| 2-Acetyl-3-methylpyrazine | 1097.4 | 10.2417 | 0.21 ± 0.27 | |||
| 2-Ethyl-3-methylpyrazine | 978.6 | 8.345 | 1.05 ± 1.181 | |||
| 2-Methyl-3,5-diethylpyrazine | 1130.5 | 10.7175 | 0.56 ± 0.115 | |||
| 2-Methyl-5-propenyl-pyrazine | 1181.4 | 11.4433 | 0.68 ± 0.248 | |||
| 5-Methyl-2,3-diethylpyrazine | 1126.1 | 10.655 | 0.25 ± 0.063 | |||
| 5-Methyl-5H-cyclopenta [b]pyrazine | 1123.5 | 10.6183 | 0.23 ± 0.235 | |||
| Methylpyrazine | 842.7 | 5.7867 | 6.05 ± 1.102 | 2.75 ± 3.887 | ||
| Pyrazine, 2-ethyl-3-methyl- | 978.6 | 8.345 | 1.62 ± 1.404 | |||
| 2-Methyl-5-(1-propenyl)-, (E)-pyrazine | 1181.4 | 11.4433 | 0.68 ± 0.248 | |||
| Sesquiterpene hydrocarbon | alpha-Farnesene | 1461.1 | 14.9942 | 1.02 ± 1.11 | ||
| beta-Curcumene | 1471.7 | 15.1225 | 3.68 ± 0.702 | |||
| beta-Eudesmene | 1463.6 | 15.0242 | 1.19 ± 0.518 | |||
| Calamenene | 1493.1 | 15.3833 | 0.17 ± 0.151 | |||
| beta-Caryophyllene | 1395.9 | 14.1992 | 2.09 ± 1.751 | |||
| Curcumene | 1444.6 | 14.7933 | 7.28 ± 0.257 | |||
| Germacrene | 1484.4 | 15.2775 | 6.25 ± 5.915 | |||
| alpha-Humulene | 1430.2 | 14.6175 | 0.45 ± 0.31 | |||
| alpha-Bergamotene | 1400.2 | 14.2525 | 2.94 ± 3.195 | |||
| beta-Farnesene | 1411.8 | 14.3933 | 0.63 ± 0.386 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farag, M.A.; Mohamed, T.A.; El-Hawary, E.A.; Abdelwareth, A. Metabolite Profiling of Premium Civet Luwak Bio-Transformed Coffee Compared with Conventional Coffee Types, as Analyzed Using Chemometric Tools. Metabolites 2023, 13, 173. https://doi.org/10.3390/metabo13020173
Farag MA, Mohamed TA, El-Hawary EA, Abdelwareth A. Metabolite Profiling of Premium Civet Luwak Bio-Transformed Coffee Compared with Conventional Coffee Types, as Analyzed Using Chemometric Tools. Metabolites. 2023; 13(2):173. https://doi.org/10.3390/metabo13020173
Chicago/Turabian StyleFarag, Mohamed A., Tarik A. Mohamed, Enas A. El-Hawary, and Amr Abdelwareth. 2023. "Metabolite Profiling of Premium Civet Luwak Bio-Transformed Coffee Compared with Conventional Coffee Types, as Analyzed Using Chemometric Tools" Metabolites 13, no. 2: 173. https://doi.org/10.3390/metabo13020173
APA StyleFarag, M. A., Mohamed, T. A., El-Hawary, E. A., & Abdelwareth, A. (2023). Metabolite Profiling of Premium Civet Luwak Bio-Transformed Coffee Compared with Conventional Coffee Types, as Analyzed Using Chemometric Tools. Metabolites, 13(2), 173. https://doi.org/10.3390/metabo13020173










