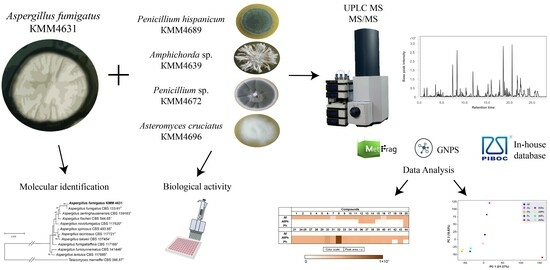
The Metabolite Profiling of Aspergillus fumigatus KMM4631 and Its Co-Cultures with Other Marine Fungi
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Fungal Strains
2.3. DNA Extraction and Amplification
2.4. Phylogenetic Analysis
2.5. Cultivation of Fungi
2.6. Extraction and HPLC MS Analysis
2.6.1. Extraction of Fungal Cultures
2.6.2. HPLC MS Analysis of Fungal Extracts
2.6.3. UHPLC-Q-TOF Data Analysis
2.7. Principal Component Analysis (PCA)
2.8. Bioassays
2.8.1. Urease Inhibition Assay
2.8.2. DPPH Radical Scavenger Assay
2.8.3. Cell Culture
2.8.4. Cell Viability Assay
2.8.5. Statistical Data Evaluation
3. Results
3.1. Molecular Identification of the Fungal Strain
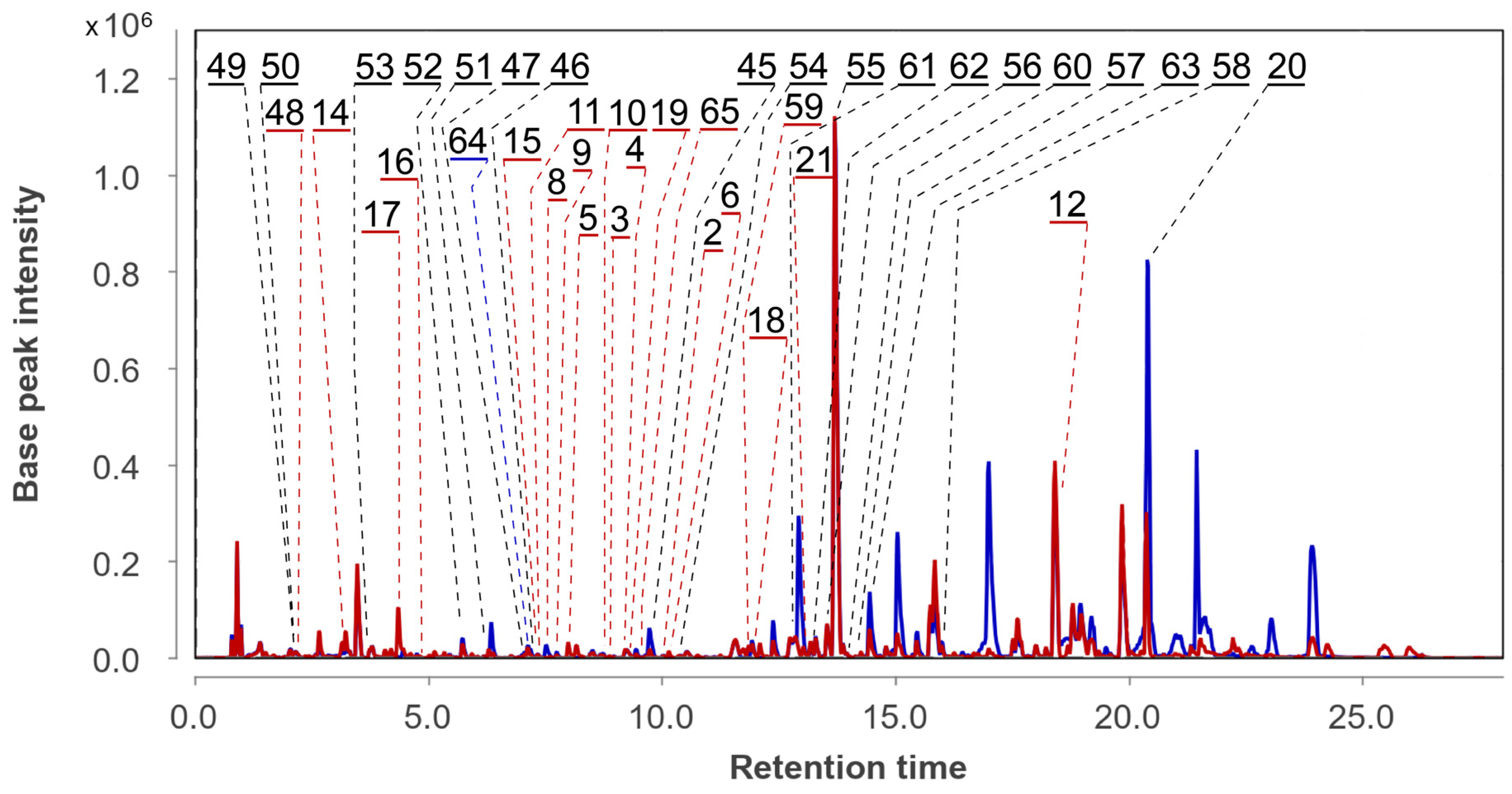
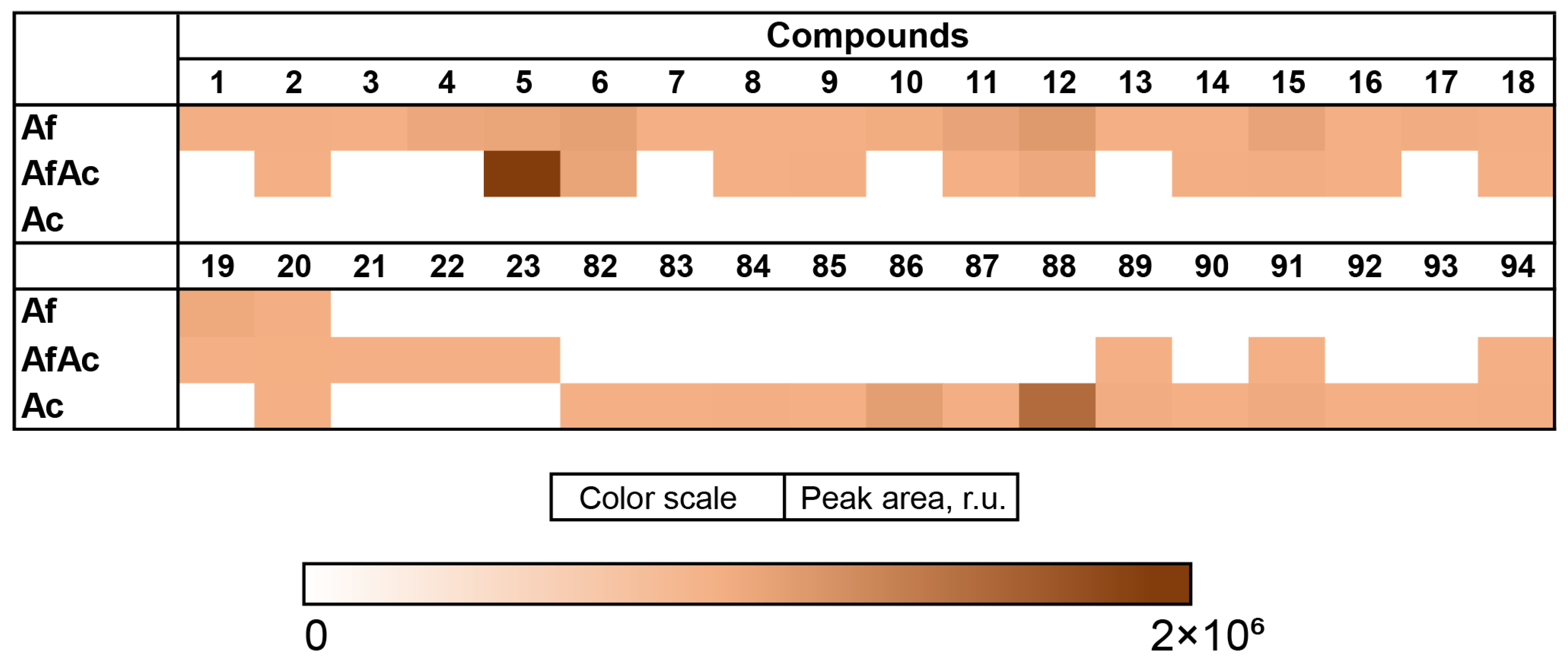
3.2. Aspergillus fumigatus KMM 4631 Monoculture Metabolites
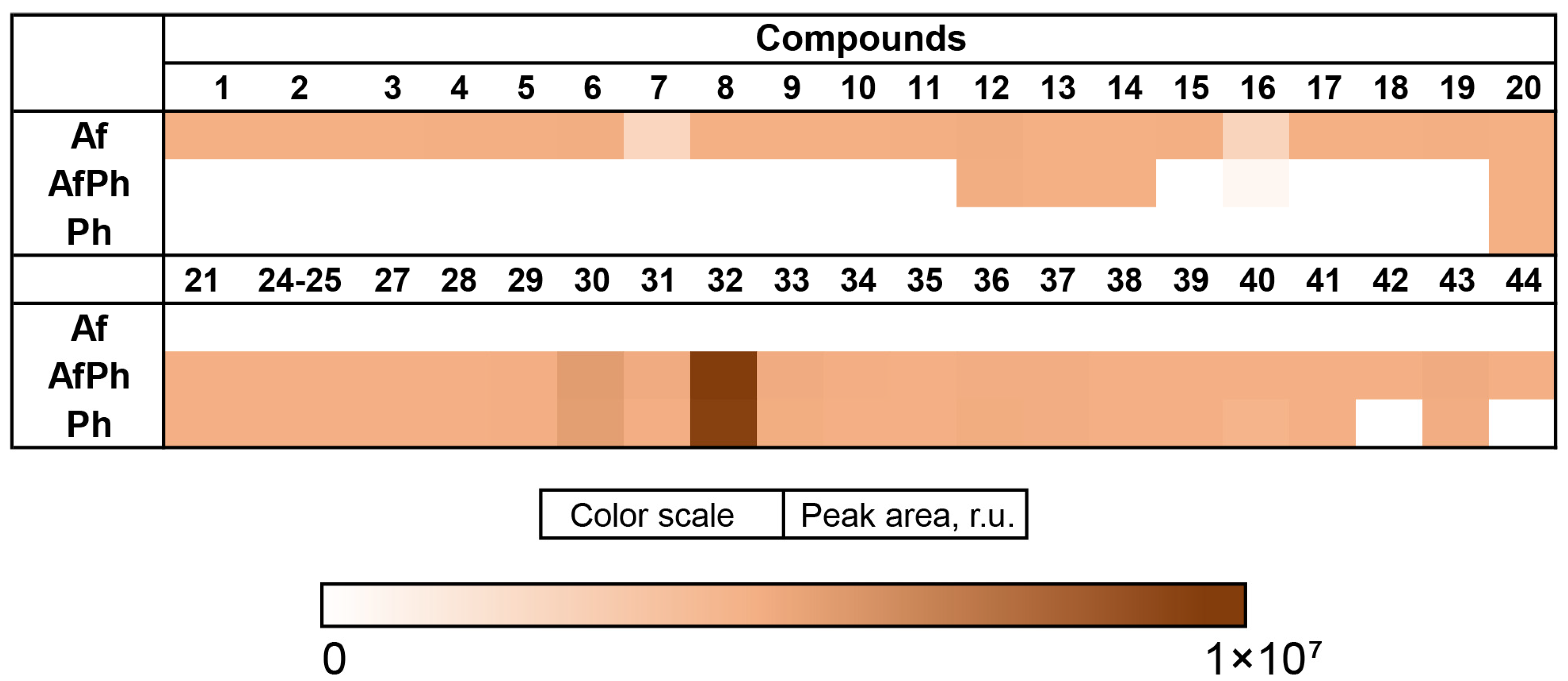
3.3. Aspergillus fumigatus KMM 4631 and Penicilliun hispanicum KMM 4689 Co-Culture Metabolites
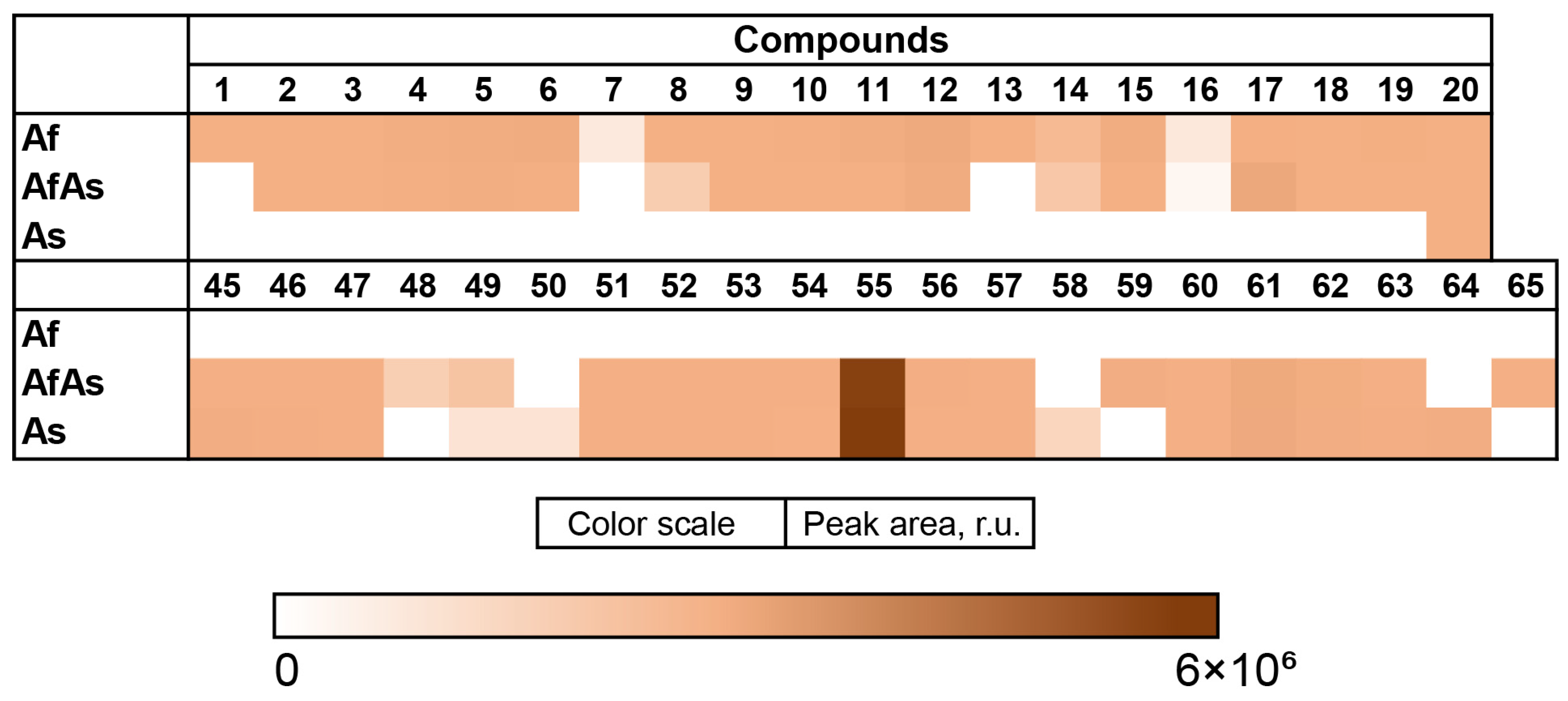
3.4. Aspergillus fumigatus KMM 4631 and Amphichorda sp. KMM 4639 Co-Culture Metabolites
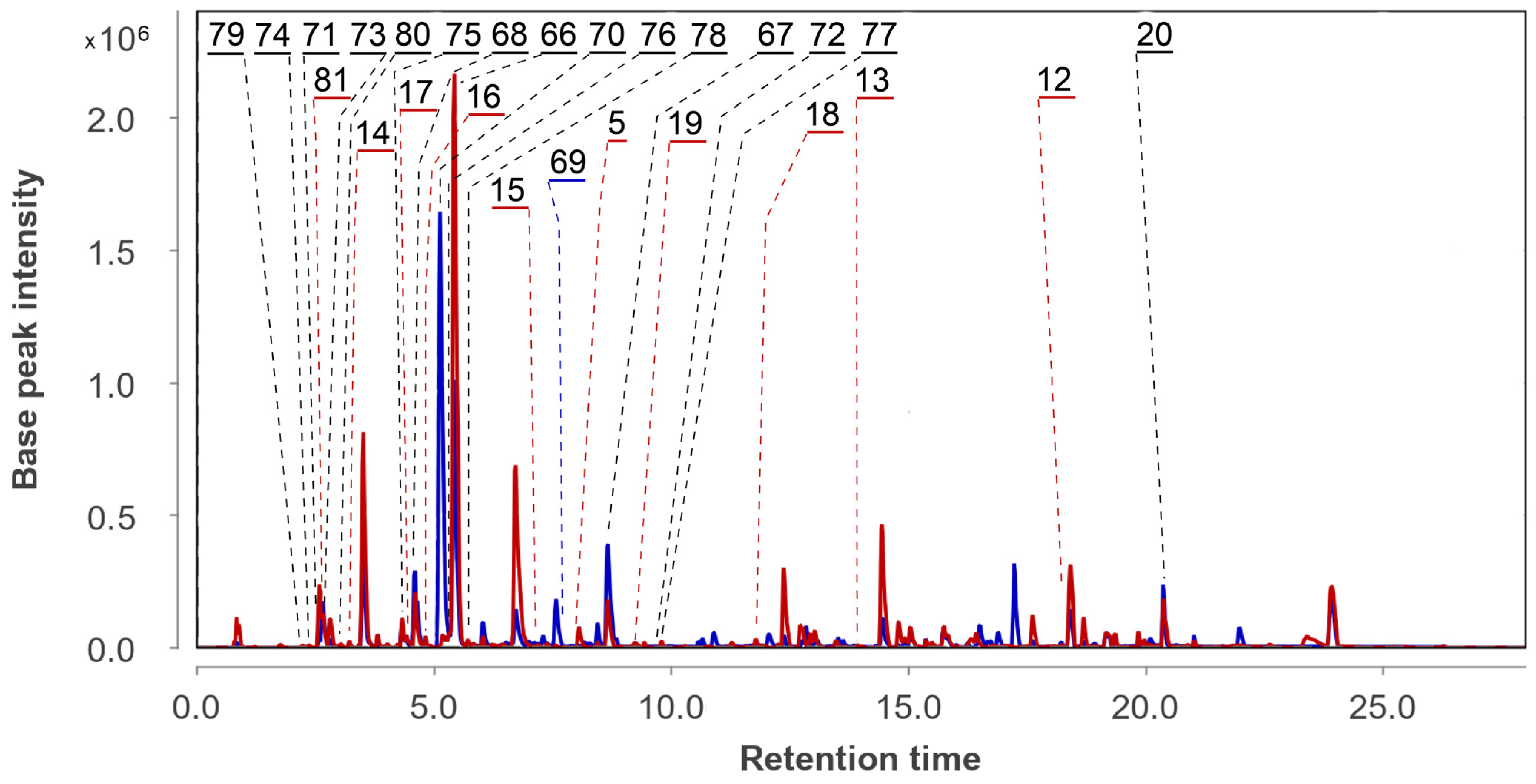
3.5. Aspergillus fumigatus KMM 4631 and Penicillium sp. KMM 4672 Co-Culture Metabolites
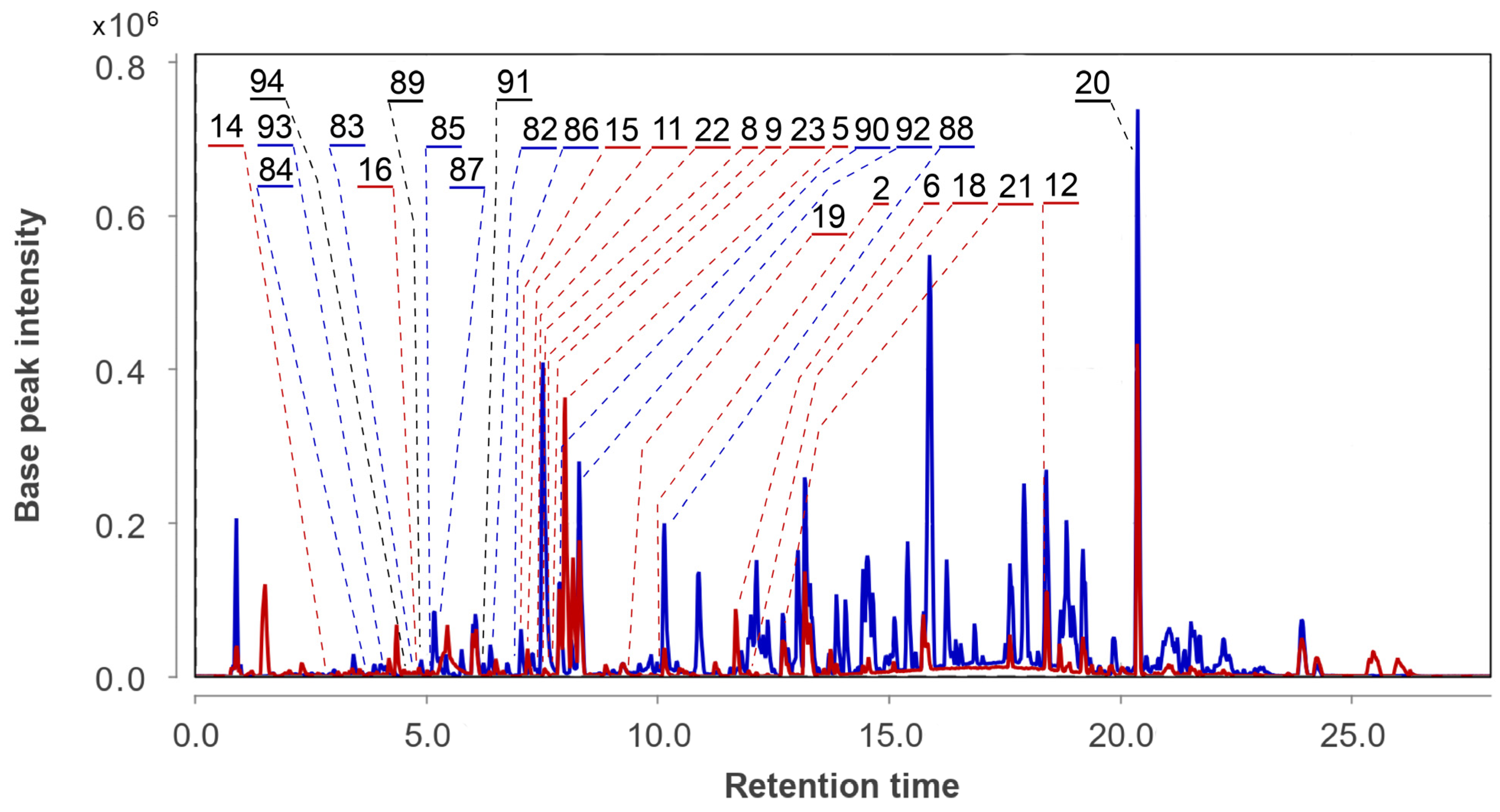
3.6. Aspergillus fumigatus KMM 4631 and Asteromyces cruciatus KMM 4696 Co-Culture Metabolites
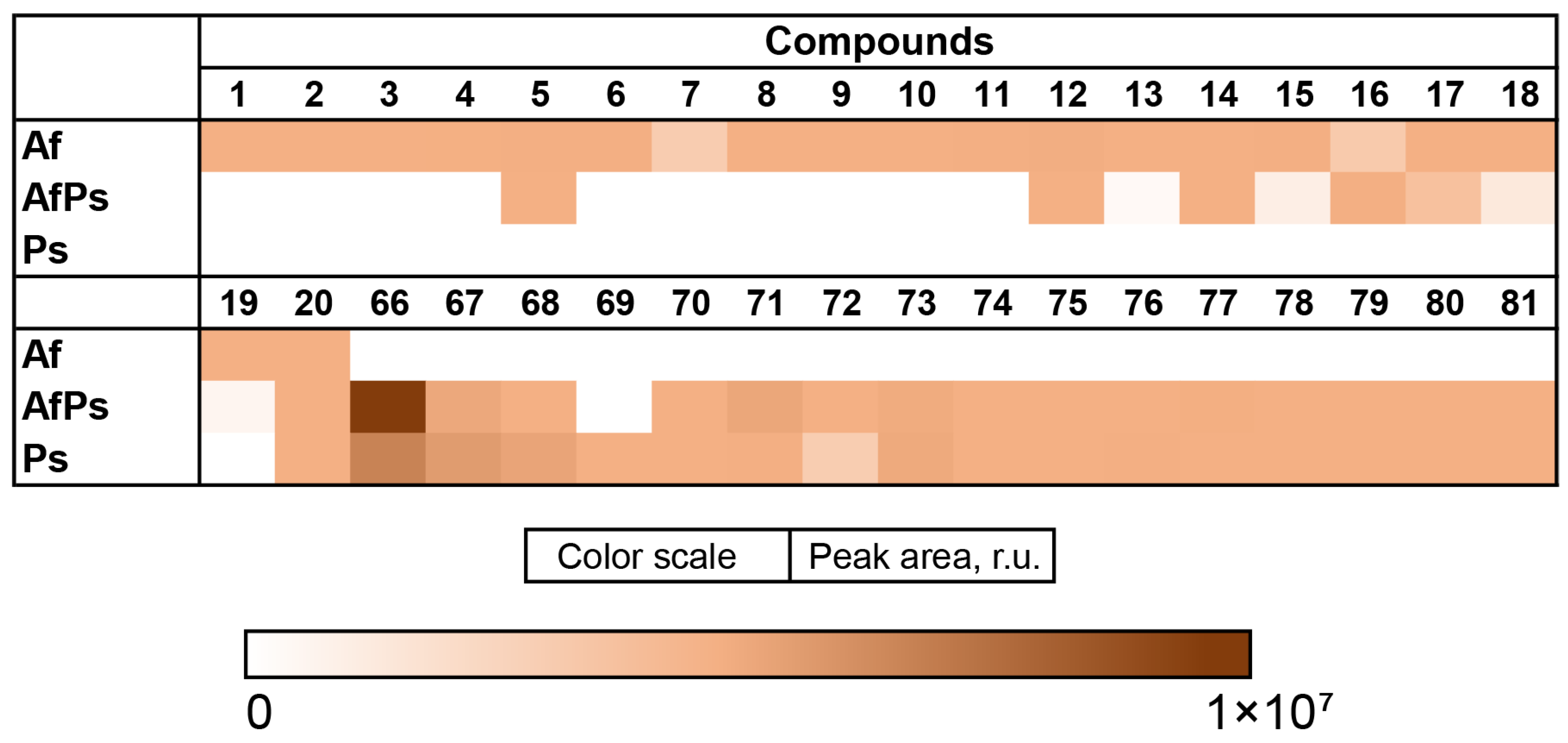
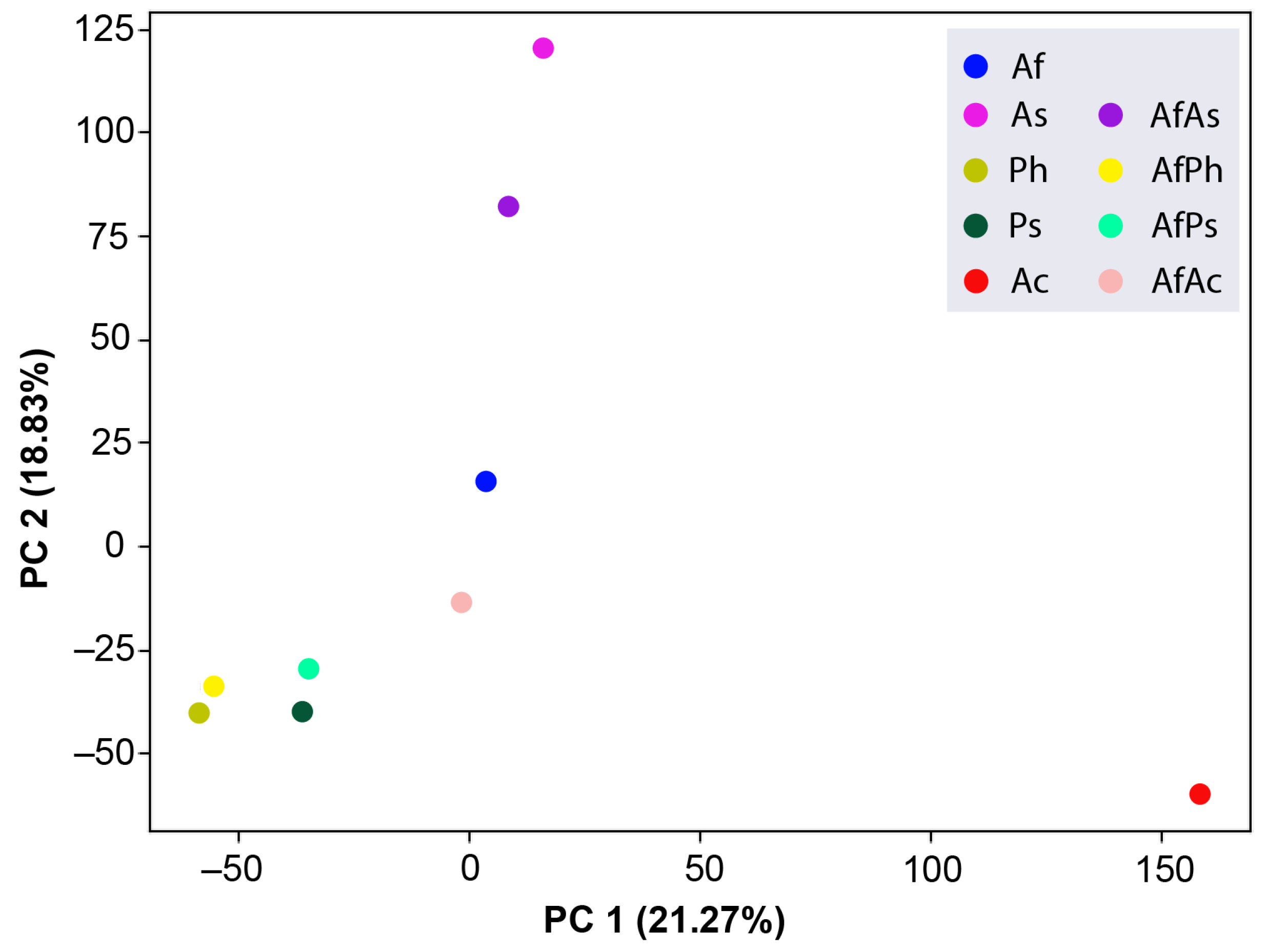
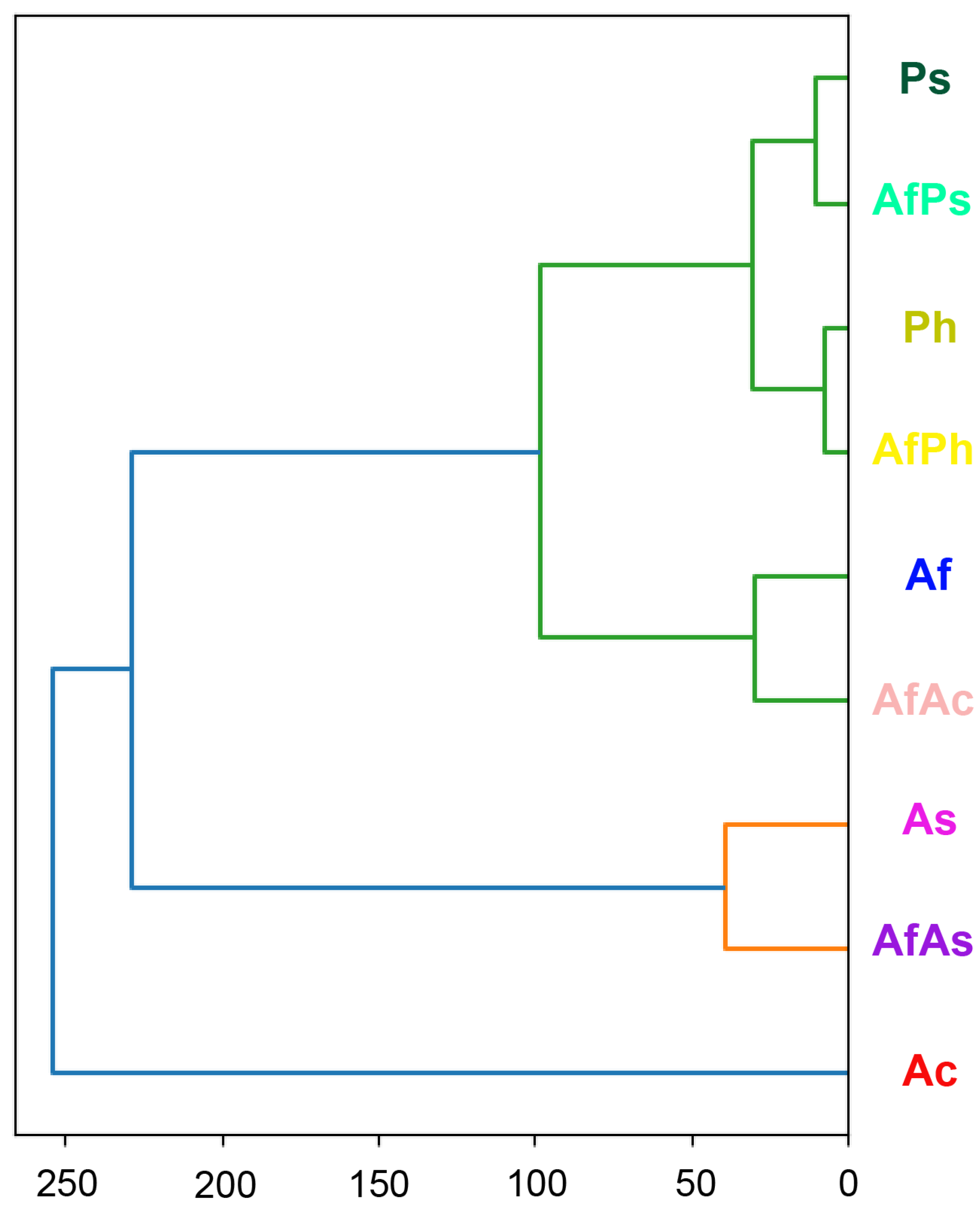
3.7. PCA Analysis of the HPLC MS Chromatograms of Fungal Extracts
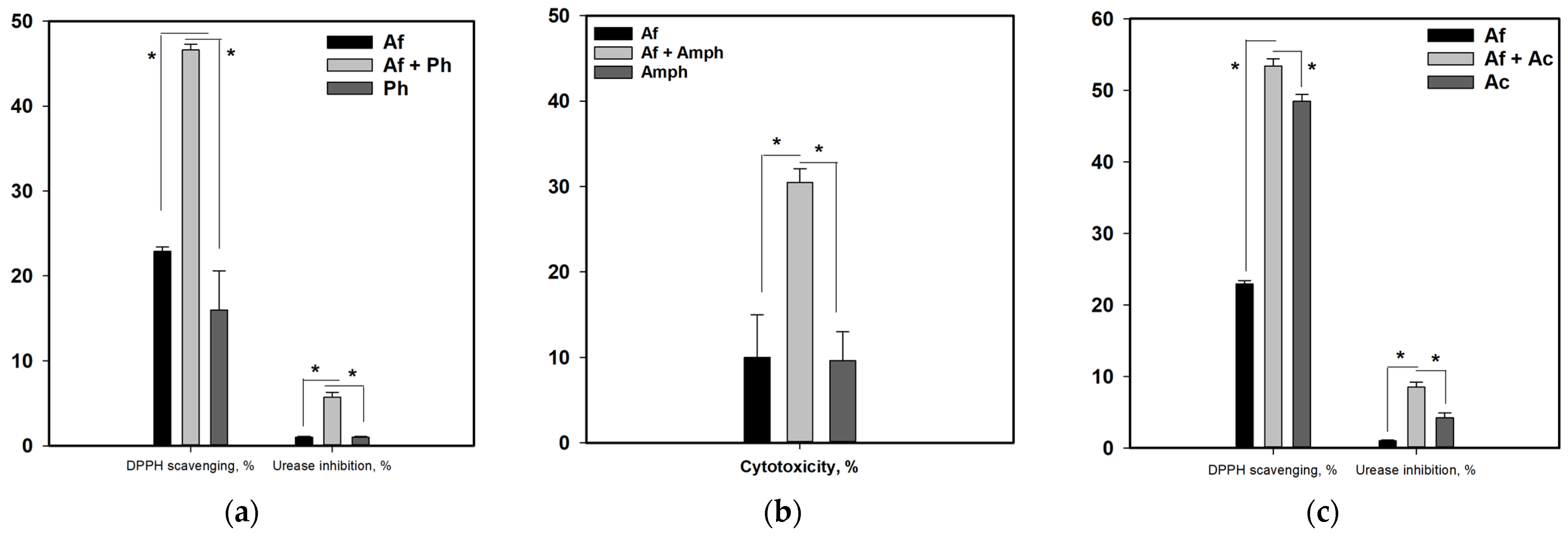
3.8. Influence of Co-Cultivation on the Biological Activity of Fungal Extracts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| N | Name | Structure | RT | Exact Mass (Measured) | Exact Mass (Calcd) | Δ, ppm | Score | |
|---|---|---|---|---|---|---|---|---|
| 1 | Af | verruculogen | 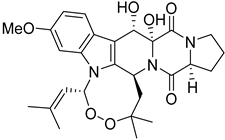 | 15.0 | 512.2414 [M+H]+ | 512.2391 | 4.5 | |
| 2 | Af AfAs AfAc | fumitremorgin C | 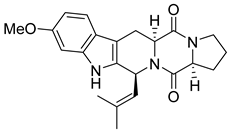 | 10.1 | 380.1971 [M+H]+ | 380.1969 | 0.5 | |
| 3 | Af AfAs | fumiquinazoline C | 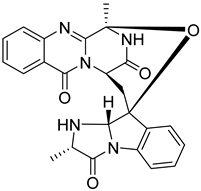 | 9.0 | 444.1671 [M+H]+ | 444.1666 | 1.1 | |
| 4 | Af AfAs | fumiquinazoline D | 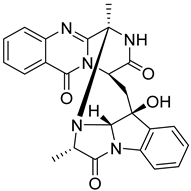 | 9.2 | 444.1671 [M+H]+ | 444.1666 | 1.1 | 0.95 a |
| 5 | Af AfAs AfPs AfAc | tryptoquivaline J | 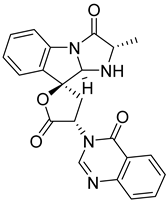 | 8.0 | 403.1396 [M+H]+ | 403.1401 | −1.2 | |
| 6 | Af AfAs AfAc | fumiquinazoline K | 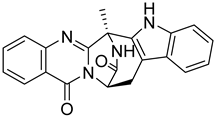 | 10.2 | 357.1338 [M+H]+ | 357.1346 | −2.2 | |
| 7 | Af | spirotriprostatin A | 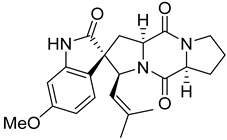 | 10.4 | 396.1920 [M+H]+ | 396.1918 | 0.5 | |
| 8 | Af AfAs AfAc | 6- methoxyspirotriprostatin B | 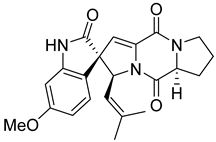 | 7.6 | 394.1754 [M+H]+ | 394.1761 | −1.8 | |
| 9 | Af AfAs AfAc | tryptoquivaline F | 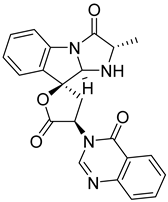 | 7.8 | 403.1396 [M+H]+ | 403.1401 | −1.2 | |
| 10 | Af AfAs | fumiquinazoline F | 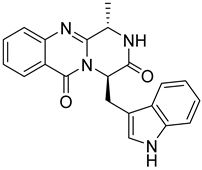 | 8.7 | 359.1494 [M+H]+ | 359.1503 | −2.5 | 0.83 a |
| 11 | Af AfAs AfAc | pseurotin A | 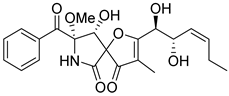 | 7.1 | 432.1652 [M+H]+ | 432.1653 | −0.2 | 0.76 a, 0.95 b |
| 12 | Af AfPh AfAs AfPs AfAc | conjugated linoleic acid (10E, 12Z) | 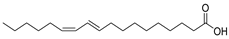 | 18.4 | 281.2487 | 281.2475 | 4.3 | 0.74 a |
| 13 | Af Ph AfPh AfPs | 2-chloroemodin | 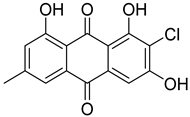 | 13.8 | 305.0200 | 305.0211 | −3.6 | 1.00 b |
| 14 | Af AfPh AfAs AfPs AfAc | scytalone | 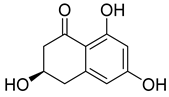 | 3.2 | 195.0649 | 195.0652 | −1.5 | 0.84 b |
| 15 | Af AfAs AfPs AfAc | pseurotin D | 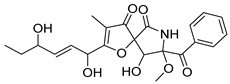 | 7.2 | 432.1650 | 432.1653 | −0.7 | 1.00 b |
| 16 | Af AfPh AfAs AfPs AfAc | agroclavine | 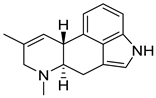 | 4.8 | 239.1519 | 239.1543 | −10.4 | 1.00 b |
| 17 | Af AfAs AfPs | fumigaclavine A | 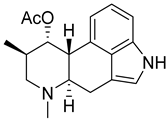 | 4.4 | 299.1749 | 299.1754 | −1.7 | 1.00 b |
| 18 | Af AfAs AfPs AfAc | pyripyropene A | 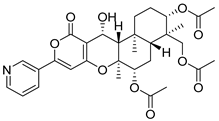 | 11.9 | 584.2507 | 584.2490 | 2.9 | 1.00 b |
| 19 | Af AfAs AfPs AfAc | fumiquinazoline G | 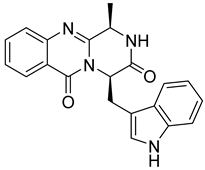 | 9.4 | 359.1480 | 359.1503 | −6.4 | 0.80 b |
| 20 | Af, Ph As, Ps Ac AfPh AfAs AfPs AfAc | ergosterol peroxide | 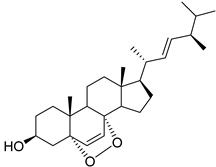 | 20.4 | 429.3348 | 429.3363 | −3.5 | 0.85 a |
| 24 | Ph AfPh | 16α-hydroxy-17β-methoxy-deoxydihydroisoaustamide | 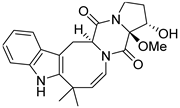 | 7.3 | 396.1921 [M+H]+ | 396.1918 | 0.8 | |
| 25 | Ph AfPh | 16β-hydroxy-17α-methoxy-deoxydihydroisoaustamide | 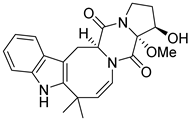 | 7.3 | 396.1921 [M+H]+ | 396.1918 | 0.8 | |
| 26 | Ph AfPh | 16α-hydroxy-17α-methoxy-deoxydihydroisoaustamide | 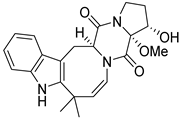 | 7.3 | 396.1921 [M+H]+ | 396.1918 | 0.8 | |
| 27 | Ph AfPh | 16,17-dihydroxy-deoxydihydroisoaustamide | 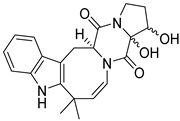 | 6.2 | 382.1748 [M+H]+ | 382.1761 | −3.4 | |
| 28 | Ph AfPh | 16β,17α-dihydroxy-deoxydihydroisoaustamide | 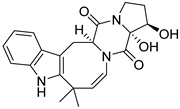 | 6.0 | 382.1748 [M+H]+ | 382.1761 | −3.4 | |
| 29 | Ph AfPh | 16α,17α-dihydroxy-deoxydihydroisoaustamide | 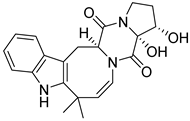 | 6.6 | 382.1748 [M+H]+ | 382.1761 | −3.4 | |
| 30 | Ph AfPh | 3β-hydroxydeoxyisoaustamide |  | 2.9 | 364.1655 [M+H]+ | 364.1656 | −0.3 | |
| 31 | Ph AfPh | deoxy-14,15-dehydroisoaustamide | 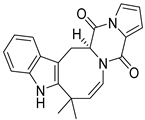 | 10.8 | 346.1544 [M+H]+ | 346.1550 | −1.7 | |
| 32 | Ph AfPh | (+)-deoxyisoaustamide | 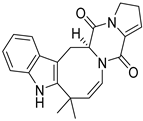 | 7.7 | 348.1713 [M+H]+ | 348.1707 | 1.7 | |
| 33 | Ph AfPh | desoxybrevianamide E |  | 9.6 | 352.2019 [M+H]+ | 352.2020 | −0.3 | |
| 34 | Ph AfPh | brevianamide F | 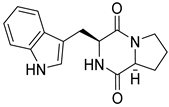 | 5.0 | 284.1388 [M+H]+ | 284.1394 | −2.1 | |
| 35 | Ph AfPh | austamide | 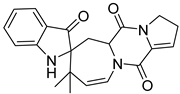 | 5.3 | 364.1635 [M+H]+ | 364.1656 | −5.8 | 0.97 a |
| 36 | Ph AfPh | citreorosein | 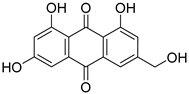 | 8.8 | 287.0539 [M+H]+ | 287.0550 | −3.8 | |
| 37 | Ph AfPh | 2-chlorocitreorosein | 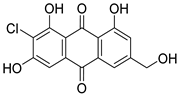 | 10.2 | 321.015 [M+H]+ | 321.0160 | −3.1 | |
| 38 | Ph AfPh | endocrocin | 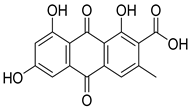 | 8.1 | 315.0488 [M+H]+ | 315.0499 | −3.5 | 0.86 a |
| 21 | Ph AfPh AfAc | emodin | 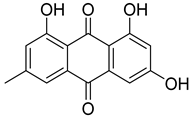 | 13.0 | 271.0605 | 271.0601 | 1.5 | 0.95 b |
| 39 | Ph AfPh | nephrolaevigatin A | 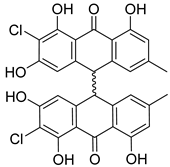 | 15.5 | 579.0665 [M+H]+ | 579.0608 | 9.8 | |
| 40 | Ph AfPh | nephrolaevigatin B | 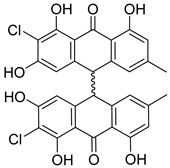 | 15.7 | 579.0626 [M+H]+ | 579.0608 | 3.1 | |
| 41 | Ph AfPh | nephrolaevigatin C | 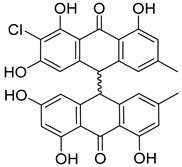 | 15.3 | 545.1005 [M+H]+ | 545.0998 | −1.3 | |
| 42 | AfPh | nephrolaevigatin D | 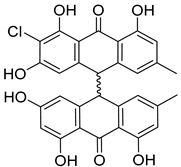 | 15.0 | 545.0992 [M+H]+ | 545.0998 | −1.1 | |
| 43 | Ph AfPh | 7-hydroxy-3-(2-hydroxypropyl)-5-methylisochromen-1-one | 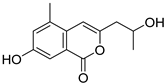 | 5.2 | 235.0949 [M+H]+ | 235.0965 | −6.8 | |
| 44 | AfPh | 3,4-dimethoxycinnamic acid | 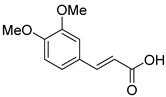 | 4.9 | 209.0805 [M+H]+ | 209.0804 | 0.5 | |
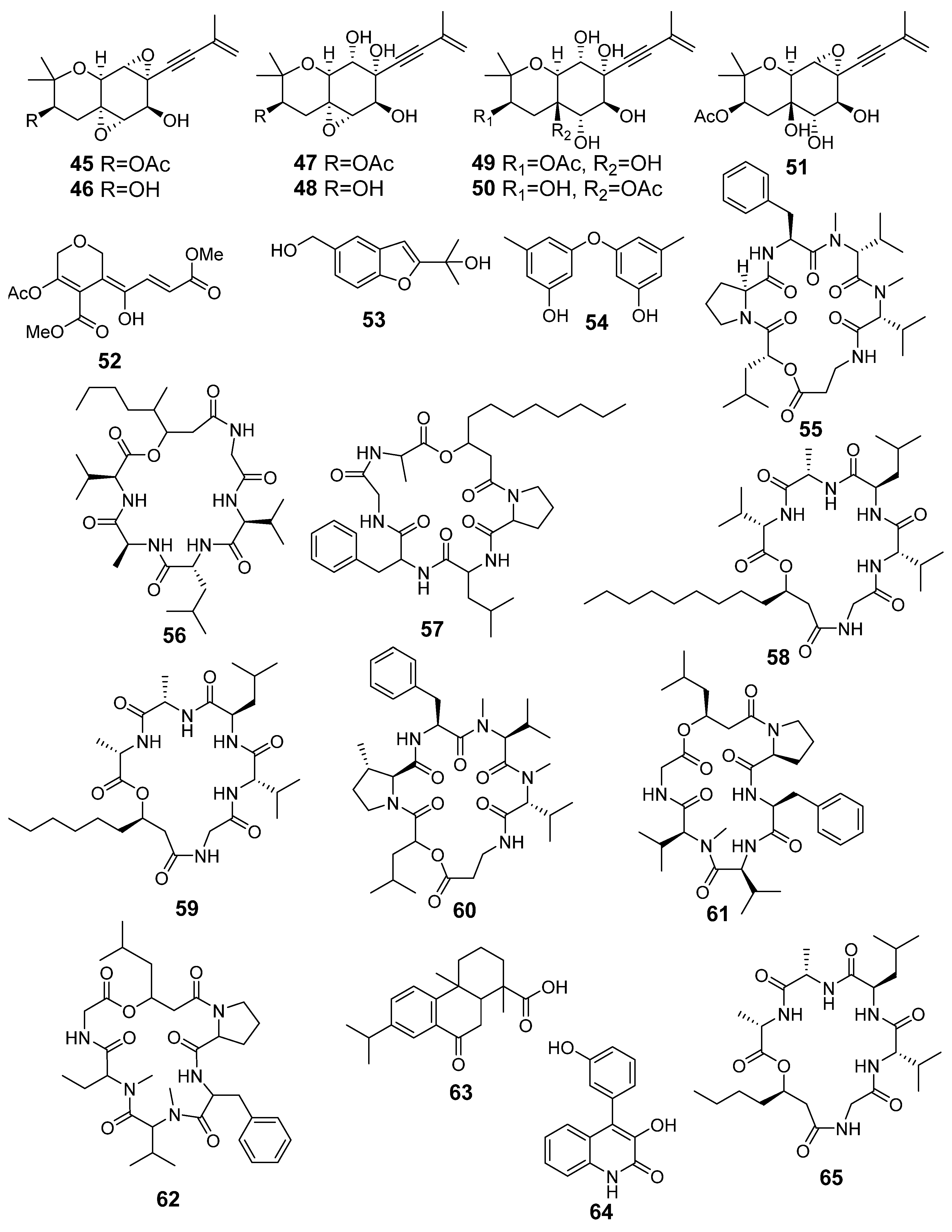
| 45 | As AfAs | oxirapentyn B | 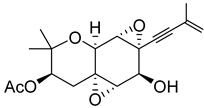 | 9.7 | 335.1487 [M+H]+ | 335.1489 | −0.6 | |
| 46 | As AfAs | oxirapentyn E | 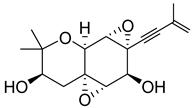 | 7.2 | 275.1279 [M-H2O+H]+ | 275.1278 | 0.4 | |
| 47 | As AfAs | oxirapentyn F | 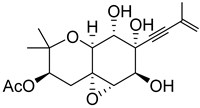 | 7.1 | 353.1587 [M+H]+ | 353.1595 | −2.3 | |
| 48 | AfAs | oxirapentyn G | 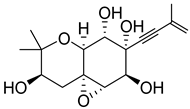 | 2.2 | 311.1504 [M+H]+ | 311.1489 | 4.8 | |
| 49 | As AfAs | oxirapentyn H | 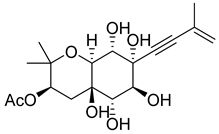 | 2.9 | 371.1704 [M+H]+ | 371.1700 | 1.1 | |
| 50 | As AfAs | oxirapentyn I | 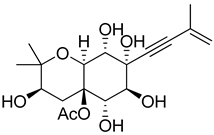 | 2.9 | 371.1704 [M+H]+ | 371.1700 | 1.1 | |
| 51 | As AfAs | oxirapentyn J | 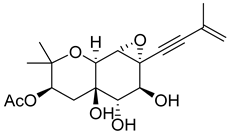 | 6.2 | 353.1587 [M+H]+ | 353.1595 | −2.3 | |
| 52 | As AfAs | isariketide A | 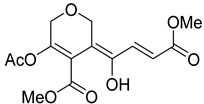 | 5.7 | 313.0910 [M+H]+ | 313.0918 | −2.6 | |
| 53 | As AfAs | acremine S | 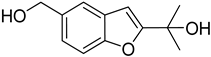 | 3.7 | 207.1014 [M+H]+ | 207.1016 | −1.0 | |
| 54 | As AfAs | diorcin |  | 10.4 | 231.1009 [M+H]+ | 231.1016 | −3.0 | |
| 55 | As AfAs | isaridin E | 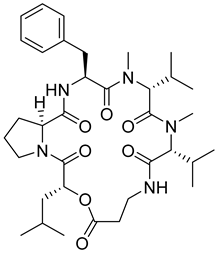 | 13.7 | 656.4010 [M+H]+ | 656.4018 | −1.2 | 0.81 a |
| 56 | As AfAs | isaridin B |  | 13.5 | 596.3997 [M+H]+ | 596.4018 | −3.5 | |
| 57 | As AfAs | isarfelin A | 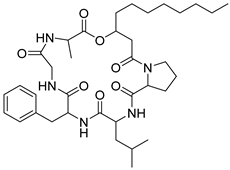 | 14.0/14.1 | 670.4187 [M+H]+ | 670.4174 | 1.9 | |
| 58 | As AfAs | isariin | 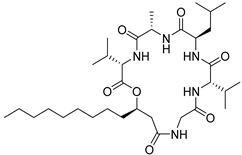 | 16.0 | 638.4495 [M+H]+ | 638.4487 | 1.3 | |
| 59 | AfAs | isariin C | 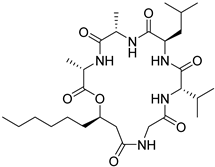 | 11.8 | 568.3692 [M+H]+ | 568.3705 | −2.3 | |
| 60 | As AfAs | psuedodestruxin C |  | 14.0/14.1 | 670.4187 [M+H]+ | 670.4174 | 1.9 | |
| 61 | As AfAs | desmethylisaridin E | 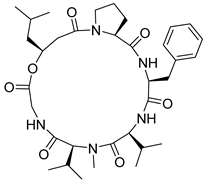 | 12.8/13.2 | 642.3870 [M+H]+ | 642.3861 | 1.4 | |
| 62 | As AfAs | isaridin F | 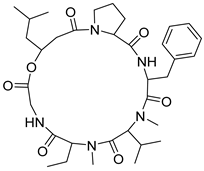 | 12.8/13.2 | 642.3870 [M+H]+ | 642.3861 | 1.4 | |
| 63 | As AfAs | 1,4a-dimethyl-9-oxo-7-propan-2-yl-3,4,10,10a-tetrahydro-2H-phenanthrene-1-carboxylic acid | 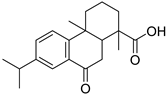 | 14.4 | 315.1947 [M+H]+ | 315.1955 | −2.5 | 0.96 a |
| 64 | As | viridicatol | 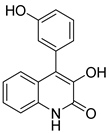 | 7.5 | 254.0795 [M+H]+ | 254.0812 | −6.7 | 0.77 a |
| 65 | AfAs | isariin D | 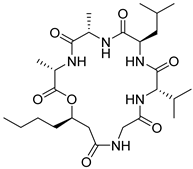 | 9.7 | 540.3395 [M+H]+ | 540.3392 | −0.6 | |
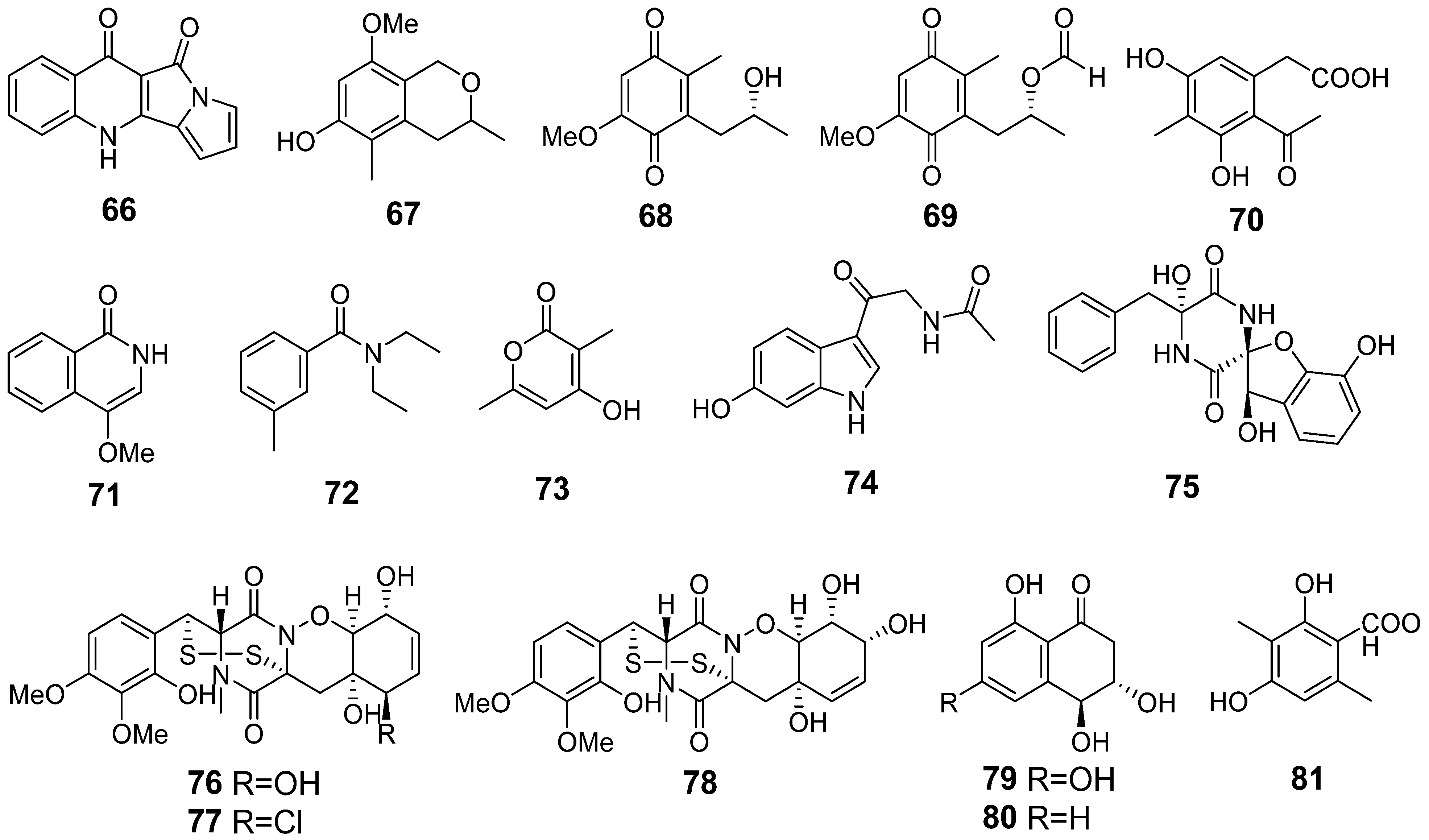
| 66 | Ps AfPs | quinolactacide | 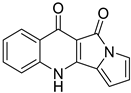 | 5.4 | 237.0670 [M+H]+ | 237.0659 | 4.6 | |
| 67 | Ps AfPs | 3,5-dimethyl-8-methoxy-3,4-dihydro-1H-isochromene-6-ol | 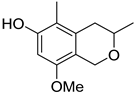 | 8.7 | 209.1175 [M+H]+ | 209.1172 | 1.4 | |
| 68 | Ps AfPs | anserinone B | 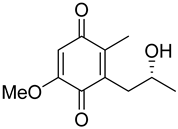 | 4.6 | 193.0856 [M−H2O+H]+ | 193.0859 | −1.6 | |
| 69 | Ps | formylanserinone B | 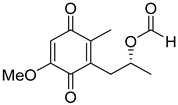 | 7.5 | 239.0913 [M+H]+ | 239.0914 | −0.4 | |
| 70 | Ps AfPs | (6-methyl curvulinic acid | 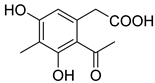 | 5.3 | 207.0651 [M−H2O+H]+ | 207.0652 | −0.5 | |
| 71 | Ps AfPs | 4-methoxyisoquinolin-1(2H)-one | 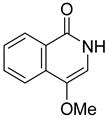 | 2.6 | 176.0702 [M+H]+ | 176.0706 | −2.3 | |
| 72 | Ps AfPs | N,N-diethyl-3-methylbenzamide | 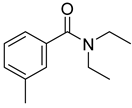 | 9.7 | 192.1390 [M+H]+ | 192.1383 | 3.6 | |
| 73 | Ps AfPs | 4-hydroxy-3,6-dimethyl-2-pyrone | 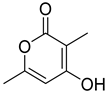 | 2.7 | 141.0548 [M+H]+ | 141.0546 | 1.4 | |
| 74 | Ps AfPs | hydroxy-N-acetyl-β-oxotriptamine | 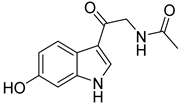 | 2.4 | 233.0925 [M+H]+ | 233.0921 | 1.7 | |
| 75 | Ps AfPs | citriperazine D | 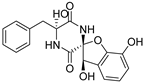 | 4.4 | 339.0979 [M−H2O+H]+ | 339.0976 | 0.9 | |
| 76 | Ps AfPs | pretrichodermamide C | 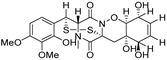 | 5.4 | 513.0988 [M+H]+ | 513.1996 | −1.6 | |
| 77 | Ps AfPs | N-methylpretrichodermamide B |  | 8.1 | 531.0662 [M+H]+ | 531.0657 | 0.9 | |
| 78 | Ps AfPs | pretrichodermamide D | 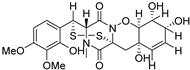 | 5.9 | 513.0988 [M+H]+ | 513.0996 | −1.6 | |
| 79 | Ps AfPs | 4-hydroxyscytalone | 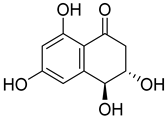 | 2.2 | 211.0603 [M+H]+ | 211.0601 | 1.0 | |
| 80 | Ps AfPs | 4-hydroxy-6-dehydroxyscytalone | 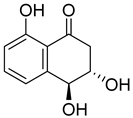 | 3.0 | 195.0654 [M+H]+ | 195.0652 | 1.0 | |
| 81 | AfPs | 3-methylorsellinic acid |  | 2.6 | 183.0659 | 183.0652 | 3.8 | |
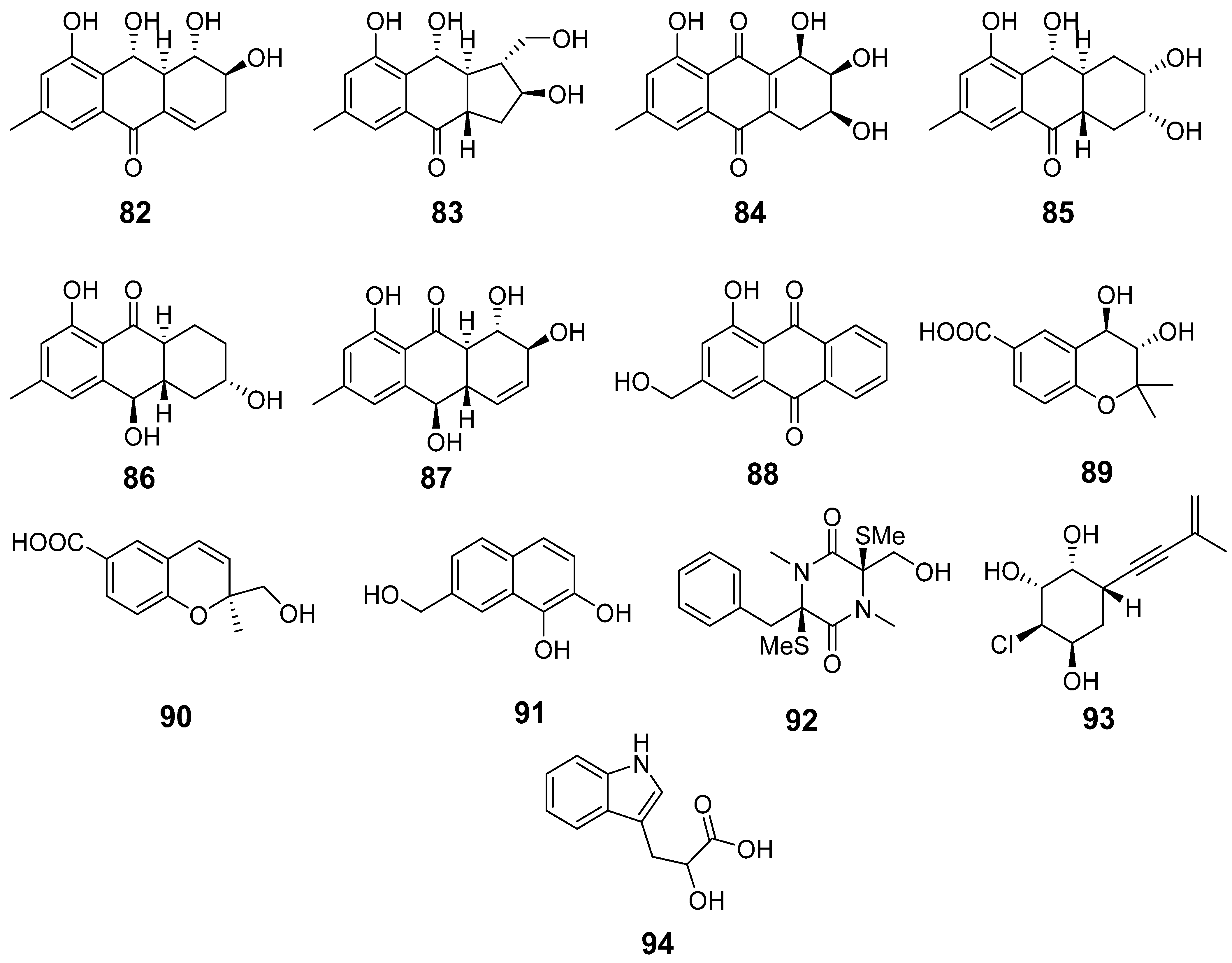
| 82 | Ac | acruciquinone A | 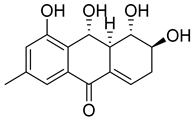 | 6.4 | 277.1063 [M+H]+ | 277.1071 | −2.9 | |
| 83 | Ac | acruciquinone C | 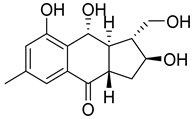 | 4.6 | 279.1226 [M+H]+ | 279.1227 | −0.4 | |
| 84 | Ac | pleosporon | 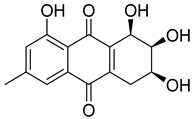 | 3.8 | 291.0853 [M+H]+ | 291.0863 | −3.4 | |
| 85 | Ac | coniothyrinone D | 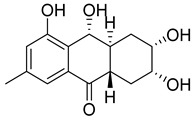 | 4.9 | 279.1226 [M+H]+ | 279.1227 | −0.4 | |
| 86 | Ac | coniothyrinone B | 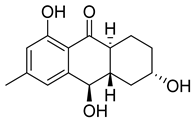 | 7.0 | 263.1272 [M+H]+ | 263.1278 | −2.3 | |
| 87 | Ac | rubrumol | 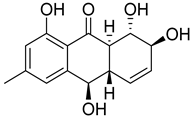 | 5.2 | 277.1062 [M+H]+ | 277.1071 | −3.2 | |
| 88 | Ac | 9,10-anthracenedione | 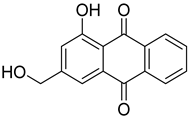 | 10.1 | 255.0652 [M+H]+ | 255.0652 | 0 | |
| 89 | Ac AfAc | trans-3,4-dihydroxy-3,4-dihydroanofinic acid | 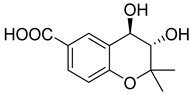 | 4.9 | 239.0895 [M+H]+ | 239.0914 | −8.0 | |
| 90 | Ac | quadricinctapyran A | 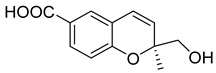 | 7.8 | 221.0819 [M+H]+ | 221.0808 | 5.0 | |
| 91 | Ac AfAc | 7-hydroxymethyl-1,2-naphthalenediol | 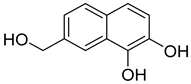 | 6.2 | 191.0707 [M+H]+ | 191.0703 | 2.1 | |
| 92 | Ac | gliovictin | 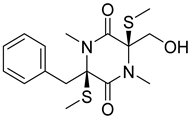 | 8.3 | 355.1160 [M+H]+ | 355.1145 | 4.2 | |
| 93 | Ac | acrucipentyn A | 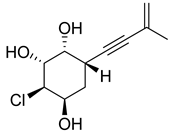 | 4.1 | 231.0777 [M+H]+ | 231.0782 | −2.2 | |
| 94 | Ac AfAc | indolelactic acid | 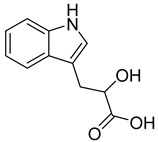 | 4.6 | 206.0810 [M+H]+ | 206.0812 | −1.0 | 0.98 a |
| 22 | AfAc | cyclotryprostatin B | 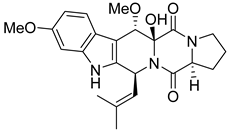 | 7.3 | 426.2035 [M+H]+ | 426.2024 | 2.6 | |
| 23 | AfAc | bisdethiobis(methylthio)gliotoxin | 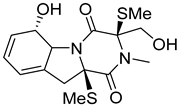 | 7.4 | 357.0936 [M+H]+ | 357.0937 | −0.3 |
References
- Polinski, J.M.; Bucci, J.P.; Gasser, M.; Bodnar, A.G. Metabarcoding assessment of prokaryotic and eukaryotic taxa in sediments from Stellwagen Bank National Marine Sanctuary. Sci. Rep. 2019, 9, 14820. [Google Scholar] [CrossRef]
- Zhou, J.; Lao, Y.-M.; Song, J.-T.; Jin, H.; Zhu, J.-M.; Cai, Z.-H. Temporal heterogeneity of microbial communities and metabolic activities during a natural algal bloom. Water Res. 2020, 183, 116020. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Sun, C.; Liu, X.; Wang, X.; Li, W.; Wei, X.; Li, Q.; Ju, J. Upregulation of a marine fungal biosynthetic gene cluster by an endobacterial symbiont. Commun. Biol. 2020, 3, 527. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Park, H.B.; Kwon, H.C.; Lee, C.-H.; Yang, H.O. Glionitrin A, an Antibiotic−Antitumor Metabolite Derived from Competitive Interaction between Abandoned Mine Microbes. J. Nat. Prod. 2009, 72, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Park, H.B.; Kim, Y.-J.; Park, J.-S.; Yang, H.O.; Lee, K.R.; Kwon, H.C. Glionitrin B, a Cancer Invasion Inhibitory Diketopiperazine Produced by Microbial Coculture. J. Nat. Prod. 2011, 74, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Caudal, F.; Tapissier-Bontemps, N.; Edrada-Ebel, R.A. Impact of Co-Culture on the Metabolism of Marine Microorganisms. Mar. Drugs 2022, 20, 153. [Google Scholar] [CrossRef]
- Tekaia, F.; Latgé, J.-P. Aspergillus fumigatus: Saprophyte or pathogen? Curr. Opin. Microbiol. 2005, 8, 385–392. [Google Scholar] [CrossRef]
- Boysen, J.M.; Saeed, N.; Hillmann, F. Natural products in the predatory defence of the filamentous fungal pathogen Aspergillus fumigatus. Beilstein J. Org. Chem. 2021, 17, 1814–1827. [Google Scholar] [CrossRef]
- Tamiya, H.; Ochiai, E.; Kikuchi, K.; Yahiro, M.; Toyotome, T.; Watanabe, A.; Yaguchi, T.; Kamei, K. Secondary metabolite profiles and antifungal drug susceptibility of Aspergillus fumigatus and closely related species, Aspergillus lentulus, Aspergillus udagawae, and Aspergillus viridinutans. J. Infect. Chemother. 2015, 21, 385–391. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Kalinovskii, A.I.; Pivkin, M.V.; Dmitrenok, P.S.; Kuznetsova, T.A. Fumitremorgins from the marine isolate of the fungus Aspergillus fumigatus. Chem. Nat. Compd. 2004, 40, 615–617. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Kalinovskii, A.I.; Pivkin, M.V.; Dmitrenok, P.S.; Kuznetsova, T.A. Alkaloids from the marine isolate of the fungus Aspergillus fumigatus. Chem. Nat. Compd. 2005, 41, 236–238. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Zhuravleva, O.I.; Antonov, A.S.; Kalinovsky, A.I.; Pivkin, M.V.; Menchinskaya, E.S.; Aminin, D.L. New metabolites from the marine-derived fungus Aspergillus fumigatus. Nat. Prod. Commun. 2012, 7, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Belousova, E.B.; Zhuravleva, O.I.; Yurchenko, E.A.; Oleynikova, G.K.; Antonov, A.S.; Kirichuk, N.N.; Chausova, V.E.; Khudyakova, Y.V.; Menshov, A.S.; Popov, R.S.; et al. New Anti-Hypoxic Metabolites from Co-Culture of Marine-Derived Fungi Aspergillus carneus KMM 4638 and Amphichorda sp. KMM 4639. Biomolecules 2023, 13, 741. [Google Scholar] [CrossRef]
- Oyedeji, A.B.; Green, E.; Adebiyi, J.A.; Ogundele, O.M.; Gbashi, S.; Adefisoye, M.A.; Oyeyinka, S.A.; Adebo, O.A. Metabolomic approaches for the determination of metabolites from pathogenic microorganisms: A review. Food Res. Int. 2021, 140, 110042. [Google Scholar] [CrossRef]
- Yurchenko, A.; Smetanina, O.; Ivanets, E.; Kalinovsky, A.; Khudyakova, Y.; Kirichuk, N.; Popov, R.; Bokemeyer, C.; von Amsberg, G.; Chingizova, E.; et al. Pretrichodermamides D–F from a Marine Algicolous Fungus Penicillium sp. KMM 4672. Mar. Drugs 2016, 14, 122. [Google Scholar] [CrossRef]
- Nesterenko, L.E.; Popov, R.S.; Zhuravleva, O.I.; Kirichuk, N.N.; Chausova, V.E.; Krasnov, K.S.; Pivkin, M.V.; Yurchenko, E.A.; Isaeva, M.P.; Yurchenko, A.N. A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum. Fermentation 2023, 9, 337. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Oleinikova, G.K.; Antonov, A.S.; Kirichuk, N.N.; Pelageev, D.N.; Rasin, A.B.; Menshov, A.S.; Popov, R.S.; Kim, N.Y.; Chingizova, E.A.; et al. New Antibacterial Chloro-Containing Polyketides from the Alga-Derived Fungus Asteromyces cruciatus KMM 4696. J. Fungi 2022, 8, 454. [Google Scholar] [CrossRef]
- Hong, S.-B.; Cho, H.-S.; Shin, H.-D.; Frisvad, J.C.; Samson, R.A. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 477–486. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Rank, C.; Nielsen, K.F.; Larsen, T.O. Metabolomics of Aspergillus fumigatus. Med. Mycol. 2009, 47, S53–S71. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Smetanina, O.F.; Kalinovsky, A.I.; Pushilin, M.A.; Glazunov, V.P.; Khudyakova, Y.V.; Kirichuk, N.N.; Ermakova, S.P.; Dyshlovoy, S.A.; Yurchenko, E.A.; et al. Oxirapentyns F-K from the Marine-Sediment-Derived Fungus Isaria felina KMM 4639. J. Nat. Prod. 2014, 77, 1321–1328. [Google Scholar] [CrossRef]
- Abe, M.; Imai, T.; Ishii, N.; Usui, M.; Okuda, T.; Oki, T. Quinolactacide, a new quinolone insecticide from Penicillium citrinum Thom F 1539. Biosci. Biotechnol. Biochem. 2005, 69, 1202–1205. [Google Scholar] [CrossRef]
- Girich, E.V.; Yurchenko, A.N.; Smetanina, O.F.; Trinh, P.T.; Ngoc, N.T.; Pivkin, M.V.; Popov, R.S.; Pislyagin, E.A.; Menchinskaya, E.S.; Chingizova, E.A.; et al. Neuroprotective Metabolites from Vietnamese Marine Derived Fungi of Aspergillus and Penicillium Genera. Mar. Drugs 2020, 18, 608. [Google Scholar] [CrossRef]
- Yurchenko, E.A.; Menchinskaya, E.S.; Pislyagin, E.A.; Trinh, P.T.H.; Ivanets, E.V.; Smetanina, O.F.; Yurchenko, A.N. Neuroprotective Activity of Some Marine Fungal Metabolites in the 6-Hydroxydopamin- and Paraquat-Induced Parkinson’s Disease Models. Mar. Drugs 2018, 16, 457. [Google Scholar] [CrossRef]
- Hirota, A.; Nemoto, A.; Tsuchiya, Y.; Hojo, H.; Abe, N. Isolation of a 2-pyrone compound as an antioxidant from a fungus and its new reaction product with 1,1-diphenyl-2-picrylhydrazyl radical. Biosci. Biotechnol. Biochem. 1999, 63, 418–420. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Berdyshev, D.V.; Smetanina, O.F.; Ivanets, E.V.; Zhuravleva, O.I.; Rasin, A.B.; Khudyakova, Y.V.; Popov, R.S.; Dyshlovoy, S.A.; von Amsberg, G.; et al. Citriperazines A-D produced by a marine algae-derived fungus Penicillium sp. KMM 4672. Nat. Prod. Res. 2020, 34, 1118–1123. [Google Scholar] [CrossRef]
- Gautschi, J.T.; Amagata, T.; Amagata, A.; Valeriote, F.A.; Mooberry, S.L.; Crews, P. Expanding the Strategies in Natural Product Studies of Marine-Derived Fungi: A Chemical Investigation of Penicillium Obtained from Deep Water Sediment. J. Nat. Prod. 2004, 67, 362–367. [Google Scholar] [CrossRef] [PubMed]
- El-Neketi, M.; Ebrahim, W.; Lin, W.; Gedara, S.; Badria, F.; Saad, H.E.A.; Lai, D.; Proksch, P. Alkaloids and polyketides from Penicillium citrinum, an endophyte isolated from the Moroccan plant Ceratonia siliqua. J. Nat. Prod. 2013, 76, 1099–1104. [Google Scholar] [CrossRef]
- Masuma, R.; Tabata, N.; Tomoda, H.; Haneda, K.; Iwai, Y.; Omura, S. Arohynapenes A and B, new anticoccidial agents produced by Penicillium sp.: Taxonomy, fermentation, and structure elucidation. J. Antibiot. 1994, 47, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Smetanina, O.F.; Yurchenko, A.N.; Ivanets, E.V.; Gerasimenko, A.V.; Trinh, P.T.H.; Ly, B.M.; Nhut, N.D.; Van, T.T.T.; Yurchenko, E.A.; Afiyatullov, S.S. Aromatic Metabolites of Marine Fungus Penicillium sp. KMM 4672 Associated with a Brown Alga Padina sp. Chem. Nat. Compd. 2017, 53, 600–602. [Google Scholar] [CrossRef]
- Orfali, R.S.; Aly, A.H.; Ebrahim, W.; Abdel-Aziz, M.S.; Müller, W.E.G.; Lin, W.; Daletos, G.; Proksch, P. Pretrichodermamide C and N-methylpretrichodermamide B, two new cytotoxic epidithiodiketopiperazines from hyper saline lake derived Penicillium sp. Phytochem. Lett. 2015, 11, 168–172. [Google Scholar] [CrossRef]
- Bai, H.-Y.; Zheng, W.-H.; Han, S.; Bao, F.; Sun, L.-L.; Zhang, K.-X.; Wang, L.-Y.; Du, H.; Li, Y.-M.; Feng, S.-L. Metabolomic Determination of Specialized Metabolites Using Liquid Chromatography-Tandem Mass Spectrometry in the Traditional Chinese Medicines Astragali Radix and Hedysari Radix. Nat. Prod. Commun. 2020, 15, 1934578X19901192. [Google Scholar] [CrossRef]
- Abraham, W.R.; Arfmann, H.A. Hydroxy-(methylbutenynyl)-benzoic acid and derivatives from Curvularia fallax. Phytochemistry 1990, 29, 2641. [Google Scholar] [CrossRef]
- Golyshin, P.N.; Golyshina, O.V.; Timmis, K.N.; Chernikova, T.; Waliczek, A.; Ferrer, M.; Beloqui, A.; Guazzaroni, M.E.; Vieites, J.M.; Pazos, F.; et al. Preparation of Fluorescent Probe-Based Reactome Array for Detection, Immobilization and Isolation of Enzymes. WO2010105851, 22 September 2010. [Google Scholar]
- Zhuravleva, O.I.; Chingizova, E.A.; Oleinikova, G.K.; Starnovskaya, S.S.; Antonov, A.S.; Kirichuk, N.N.; Menshov, A.S.; Popov, R.S.; Kim, N.Y.; Berdyshev, D.V.; et al. Anthraquinone Derivatives and Other Aromatic Compounds from Marine Fungus Asteromyces cruciatus KMM 4696 and Their Effects against Staphylococcus aureus. Mar. Drugs 2023, 21, 431. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, A.; Chen, H.; Wang, M.; Ding, G.; Sun, L.; Li, L.; Dai, M. Anthraquinones from the saline-alkali plant endophytic fungus Eurotium rubrum. J. Antibiot. 2017, 70, 1138–1141. [Google Scholar] [CrossRef]
- Sun, P.; Huo, J.; Kurtan, T.; Mandi, A.; Antus, S.; Tang, H.; Draeger, S.; Schulz, B.; Hussain, H.; Krohn, K.; et al. Structural and stereochemical studies of hydroxyanthraquinone derivatives from the endophytic fungus Coniothyrium sp. Chirality 2013, 25, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ondeyka, J.G.; Zink, D.L.; Basilio, A.; Vicente, F.; Collado, J.; Platas, G.; Huber, J.; Dorso, K.; Motyl, M.; et al. Isolation, structure and antibacterial activity of pleosporone from a pleosporalean ascomycete discovered by using antisense strategy. Bioorg. Med. Chem. 2009, 17, 2162–2166. [Google Scholar] [CrossRef] [PubMed]
- Prompanya, C.; Dethoup, T.; Gales, L.; Lee, M.; Pereira, J.A.C.; Silva, A.M.S.; Pinto, M.M.M.; Kijjoa, A. New Polyketides and New Benzoic Acid Derivatives from the Marine Sponge-Associated Fungus Neosartorya quadricincta KUFA 0081. Mar. Drugs 2016, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Fenical, W. Isolation of gliovictin from the marine deuteromycete Asteromyces cruciatus. Phytochemistry 1987, 26, 3347. [Google Scholar] [CrossRef]
- Rutherford, J.C. The Emerging Role of Urease as a General Microbial Virulence Factor. PLoS Pathog. 2014, 10, e1004062. [Google Scholar] [CrossRef]
- Cox Gary, M.; Mukherjee, J.; Cole Garry, T.; Casadevall, A.; Perfect John, R. Urease as a Virulence Factor in Experimental Cryptococcosis. Infect. Immun. 2000, 68, 443–448. [Google Scholar] [CrossRef]
- Bao, J.; Wang, J.; Zhang, X.Y.; Nong, X.H.; Qi, S.H. New Furanone Derivatives and Alkaloids from the Co-Culture of Marine-Derived Fungi Aspergillus sclerotiorum and Penicillium citrinum. Chem. Biodivers. 2017, 14, e1600327. [Google Scholar] [CrossRef]
- Knowles, S.L.; Raja, H.A.; Roberts, C.D.; Oberlies, N.H. Fungal-fungal co-culture: A primer for generating chemical diversity. Nat. Prod. Rep. 2022, 39, 1557–1573. [Google Scholar] [CrossRef]
- Raffa, N.; Keller, N.P. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog. 2019, 15, e1007606. [Google Scholar] [CrossRef]
- Nikolaou, E.; Agrafioti, I.; Stumpf, M.; Quinn, J.; Stansfield, I.; Brown, A.J.P. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 2009, 9, 44. [Google Scholar] [CrossRef]
- Lv, J.-M.; Hu, D.; Gao, H.; Kushiro, T.; Awakawa, T.; Chen, G.-D.; Wang, C.-X.; Abe, I.; Yao, X.-S. Biosynthesis of helvolic acid and identification of an unusual C-4-demethylation process distinct from sterol biosynthesis. Nat. Commun. 2017, 8, 1644. [Google Scholar] [CrossRef]
- Thambi, A.; Chakraborty, K. Anti-inflammatory decurrencyclics A-B, two undescribed nor-dammarane triterpenes from triangular sea bell Turbinaria decurrens. Nat. Prod. Res. 2023, 37, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef] [PubMed]
- Langenfeld, A.; Blond, A.; Gueye, S.; Herson, P.; Nay, B.; Dupont, J.; Prado, S. Insecticidal cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2011, 74, 825–830. [Google Scholar] [CrossRef]
- Baute, R.; Deffieux, G.; Merlet, D.; Baute, M.-A.; Neveu, A. New insecticidal cyclodepsipeptides from the fungus Isaria felina I. production, isolation and insecticidal properties of Isariins B, C and D. J. Antibiot. 1981, 34, 1261–1265. [Google Scholar] [CrossRef]
- Smith, E.B.; Dolan, S.K.; Fitzpatrick, D.A.; Doyle, S.; Jones, G.W. Towards understanding the gliotoxin detoxification mechanism: In vivo thiomethylation protects yeast from gliotoxin cytotoxicity. Microb. Cell 2016, 3, 120. [Google Scholar] [CrossRef]
- Owens, R.A.; O’Keeffe, G.; Smith, E.B.; Dolan, S.K.; Hammel, S.; Sheridan, K.J.; Fitzpatrick, D.A.; Keane, T.M.; Jones, G.W.; Doyle, S. Interplay between gliotoxin resistance, secretion, and the methyl/methionine cycle in Aspergillus fumigatus. Eukaryot. Cell 2015, 14, 941–957. [Google Scholar]
- Li, X.; Kim, S.-K.; Nam, K.W.; Kang, J.S.; Choi, H.D.; Son, B.W. A New Antibacterial Dioxopiperazine Alkaloid Related to Gliotoxin from a Marine Isolate of the Fungus Pseudallescheria. J. Antibiot. 2006, 59, 248–250. [Google Scholar] [CrossRef]
- Jain, R.; Valiante, V.; Remme, N.; Docimo, T.; Heinekamp, T.; Hertweck, C.; Gershenzon, J.; Haas, H.; Brakhage, A.A. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol. Microbiol. 2011, 82, 39–53. [Google Scholar] [CrossRef]
- Carberry, S.; Molloy, E.; Hammel, S.; O’Keeffe, G.; Jones, G.W.; Kavanagh, K.; Doyle, S. Gliotoxin effects on fungal growth: Mechanisms and exploitation. Fungal Genet. Biol. 2012, 49, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Dolan, S.K.; O’Keeffe, G.; Jones, G.W.; Doyle, S. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015, 23, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Yaakoub, H.; Mina, S.; Calenda, A.; Bouchara, J.P.; Papon, N. Oxidative stress response pathways in fungi. Cell. Mol. Life Sci. 2022, 79, 333. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Al-Rashida, M.; Uroos, M.; Qazi, S.U.; Naz, S.; Ishtiaq, M.; Khan, K.M. A patent update on therapeutic applications of urease inhibitors (2012–2018). Expert Opin. Ther. Pat. 2019, 29, 181–189. [Google Scholar] [CrossRef]
- Zhou, C.; Bhinderwala, F.; Lehman, M.K.; Thomas, V.C.; Chaudhari, S.S.; Yamada, K.J.; Foster, K.W.; Powers, R.; Kielian, T.; Fey, P.D. Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog. 2019, 15, e1007538. [Google Scholar] [CrossRef]
- Gogineni, V.; Chen, X.; Hanna, G.; Mayasari, D.; Hamann, M.T. Role of symbiosis in the discovery of novel antibiotics. J. Antibiot. 2020, 73, 490–503. [Google Scholar] [CrossRef]
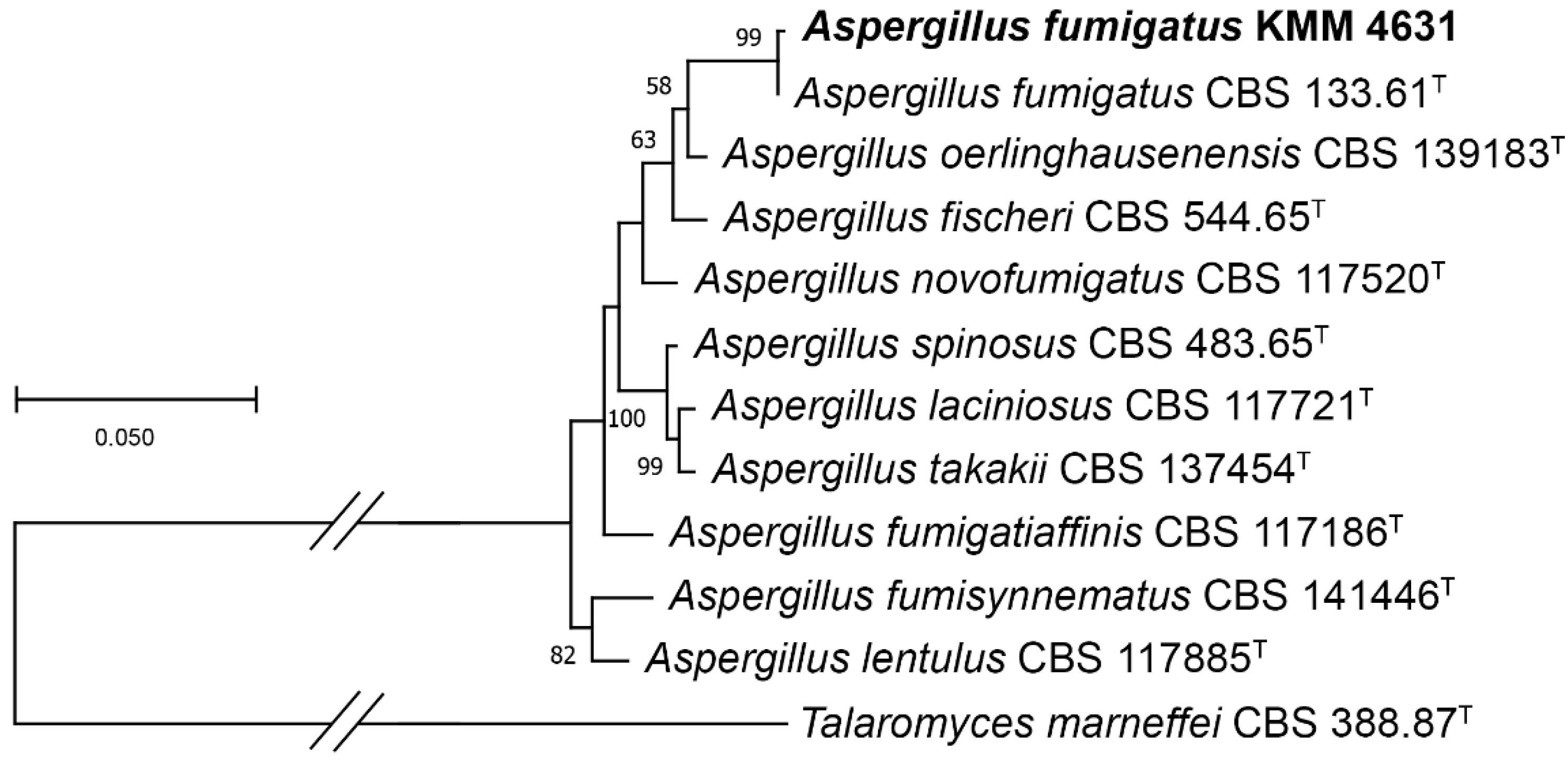
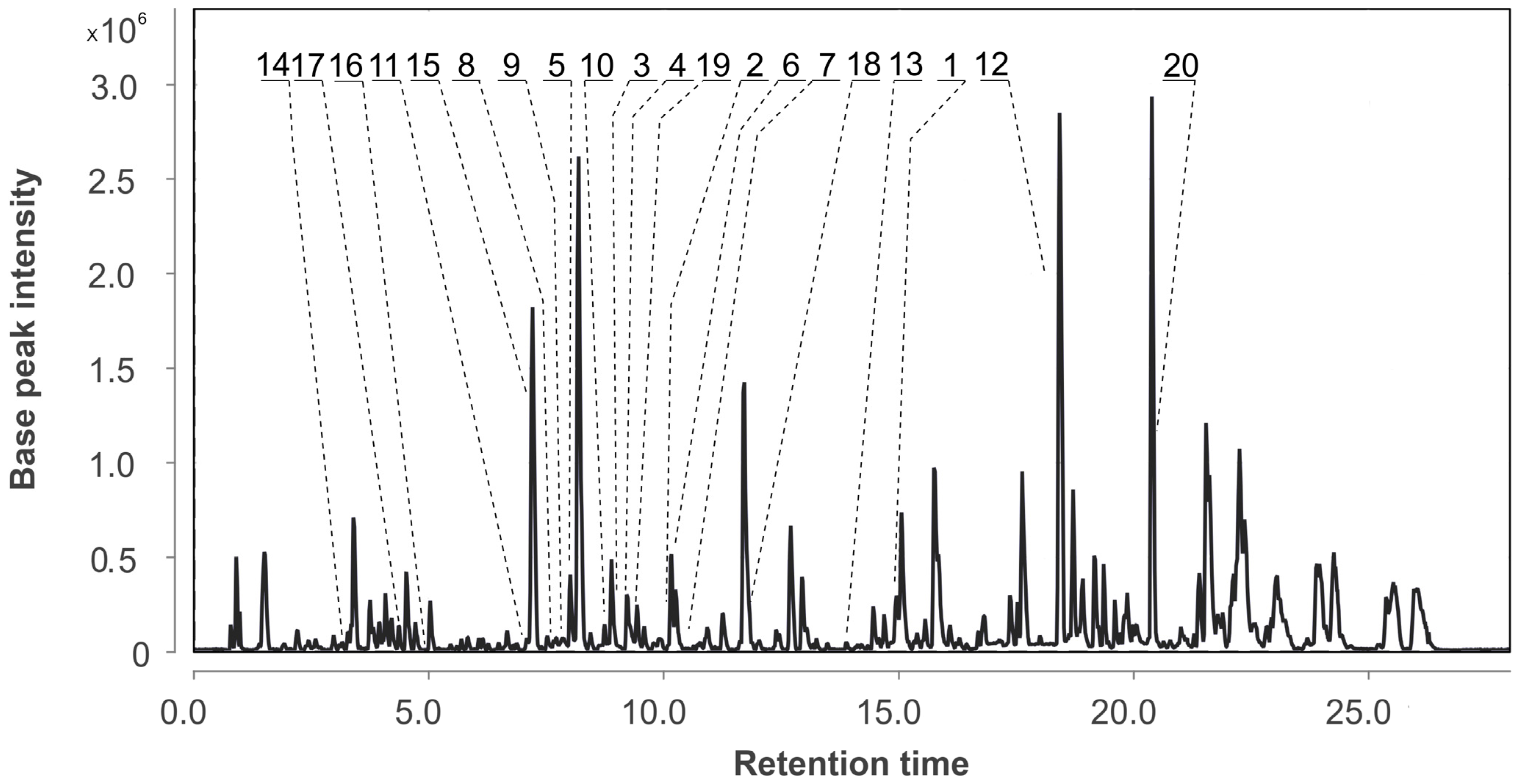
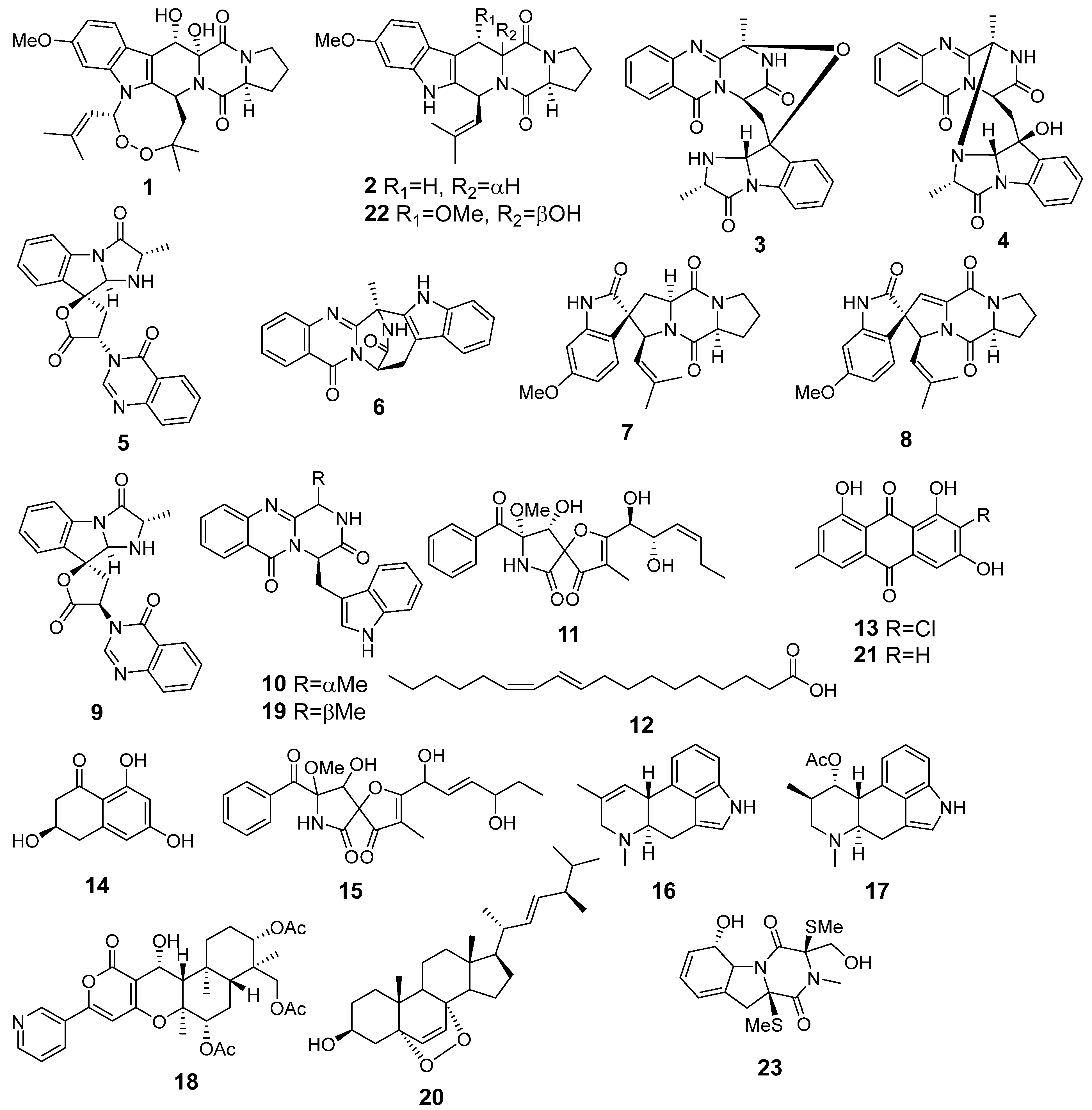

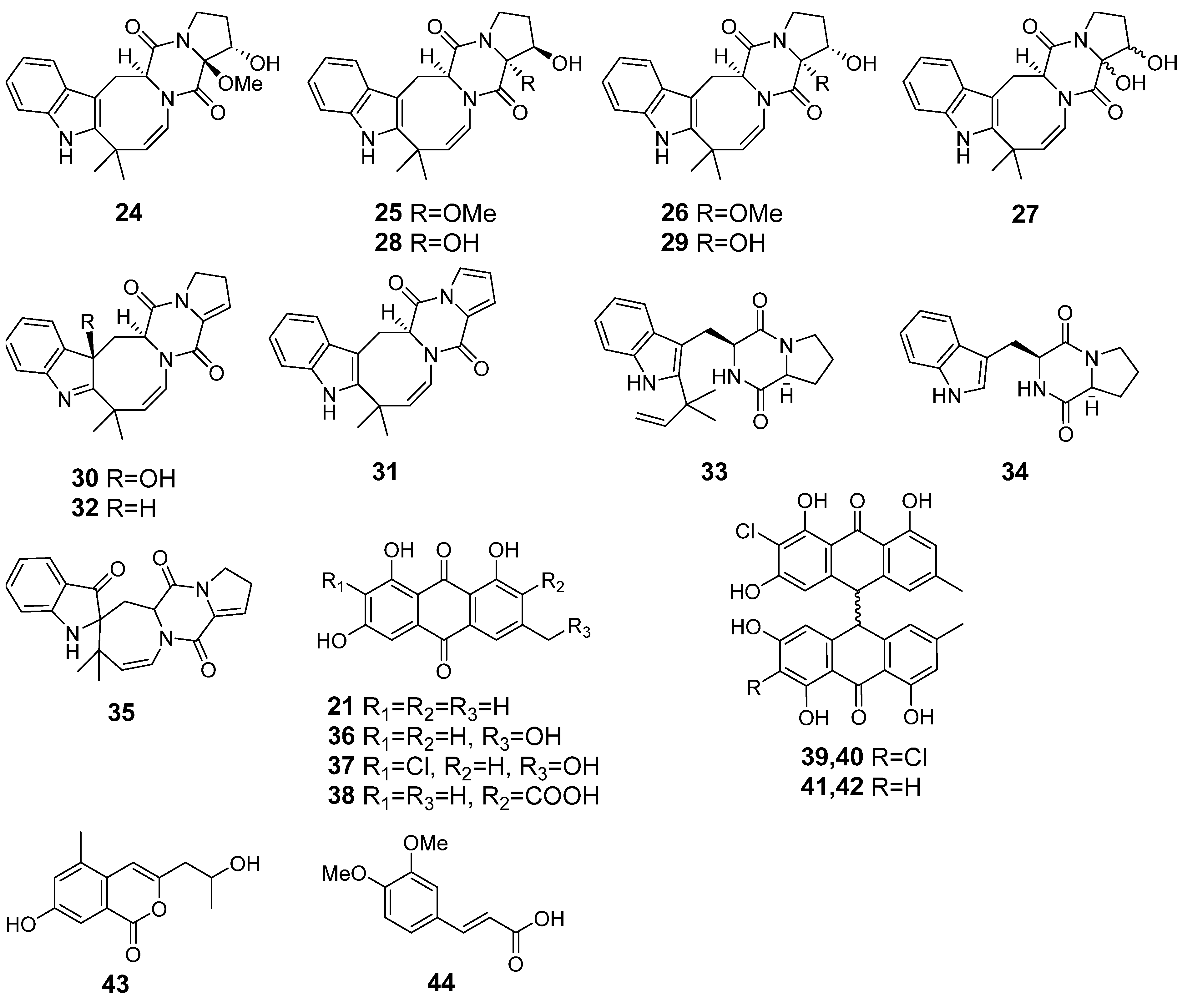
| Species | Strain Number | GenBank Accession Number | |||
|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | ||
| Aspergillus takakii | CBS 137454T | MN431378 | AB787221 | AB787566 | MN969097 |
| Aspergillus laciniosus | CBS 117721T | AB299413 | AY870756 | AY870716 | MN969080 |
| Aspergillus spinosus | CBS 483.65T | EF669988 | EF669844 | EF669914 | EF669775 |
| Aspergillus fumisynnematus | CBS 141446T | AB250779 | AB248076 | AB259968 | MN969073 |
| Aspergillus lentulus | CBS 117885T | EF669969 | EF669825 | EF669895 | EF669756 |
| Aspergillus fumigatiaffinis | CBS 117186T | MN431367 | DQ094885 | DQ094891 | MN969072 |
| Aspergillus oerlinghausenensis | CBS 139183T | KT359601 | KT359603 | KT359605 | MN969162 |
| Aspergillus fumigatus | CBS 133.61T | EF669931 | EF669791 | EF669860 | EF669719 |
| Aspergillus fumigatus | KMM 4631 | OR578448 | OQ466614 | OR600992 | OR600993 |
| Aspergillus fischeri | CBS 544.65T | EF669936 | EF669796 | EF669865 | EF669724 |
| Aspergillus novofumigatus | CBS 117520T | MN431372 | DQ094886 | DQ094893 | MN969083 |
| Talaromyces marneffei | CBS 388.87T | JN899344 | JX091389 | KF741958 | KM023283 |
| Fungal Culture | Sample Code | Mass of Crude Extract, mg |
|---|---|---|
| Aspergillus fumigatus KMM 4631 | Af | 21.1 |
| Aspergillus fumigatus KMM 4631+ Amphichorda sp. KMM 4639 | AfAs | 14.5 |
| Amphichorda sp. KMM 4639 | As | 12.7 |
| Aspergillus fumigatus KMM 4631+ Penicillium sp. KMM 4672 | AfPs | 87.0 |
| Penicillium sp. KMM 4672 | Ps | 102.1 |
| Aspergillus fumigatus KMM 4631+ Penicillium hispanicum KMM 4689 | AfPh | 365.0 |
| Penicillium hispanicum KMM 4689 | Ph | 233.1 |
| Aspergillus fumigatus KMM 4631+ Asteromyces cruciatus KMM 4696 | AfAc | 39.5 |
| Asteromyces cruciatus KMM 4696 | Ac | 56.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yurchenko, A.N.; Nesterenko, L.E.; Popov, R.S.; Kirichuk, N.N.; Chausova, V.E.; Chingizova, E.A.; Isaeva, M.P.; Yurchenko, E.A. The Metabolite Profiling of Aspergillus fumigatus KMM4631 and Its Co-Cultures with Other Marine Fungi. Metabolites 2023, 13, 1138. https://doi.org/10.3390/metabo13111138
Yurchenko AN, Nesterenko LE, Popov RS, Kirichuk NN, Chausova VE, Chingizova EA, Isaeva MP, Yurchenko EA. The Metabolite Profiling of Aspergillus fumigatus KMM4631 and Its Co-Cultures with Other Marine Fungi. Metabolites. 2023; 13(11):1138. https://doi.org/10.3390/metabo13111138
Chicago/Turabian StyleYurchenko, Anton N., Liliana E. Nesterenko, Roman S. Popov, Natalya N. Kirichuk, Viktoria E. Chausova, Ekaterina A. Chingizova, Marina P. Isaeva, and Ekaterina A. Yurchenko. 2023. "The Metabolite Profiling of Aspergillus fumigatus KMM4631 and Its Co-Cultures with Other Marine Fungi" Metabolites 13, no. 11: 1138. https://doi.org/10.3390/metabo13111138
APA StyleYurchenko, A. N., Nesterenko, L. E., Popov, R. S., Kirichuk, N. N., Chausova, V. E., Chingizova, E. A., Isaeva, M. P., & Yurchenko, E. A. (2023). The Metabolite Profiling of Aspergillus fumigatus KMM4631 and Its Co-Cultures with Other Marine Fungi. Metabolites, 13(11), 1138. https://doi.org/10.3390/metabo13111138