Elucidating the Differentiation Synthesis Mechanisms of Differently Colored Resistance Quinoa Seedings Using Metabolite Profiling and Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sampling
2.2. Morphological Data Collection
2.3. Differences in Physiological Indicators of Quinoa Cultivars during Seedling Stage
2.4. Metabolite Extraction Detection and Qualitative and Quantitative Analysis
2.5. Transcriptome Sequencing and Data Analysis
2.5.1. RNA Extraction, Library Construction, and Sequencing
2.5.2. Analysis of RNA-Seq Data
2.6. Real-Time Fluorescence Quantitative PCR Analysis
2.7. Association Analysis of Metabolite Profiling and Transcriptome
2.8. Statistical Analysis
3. Results
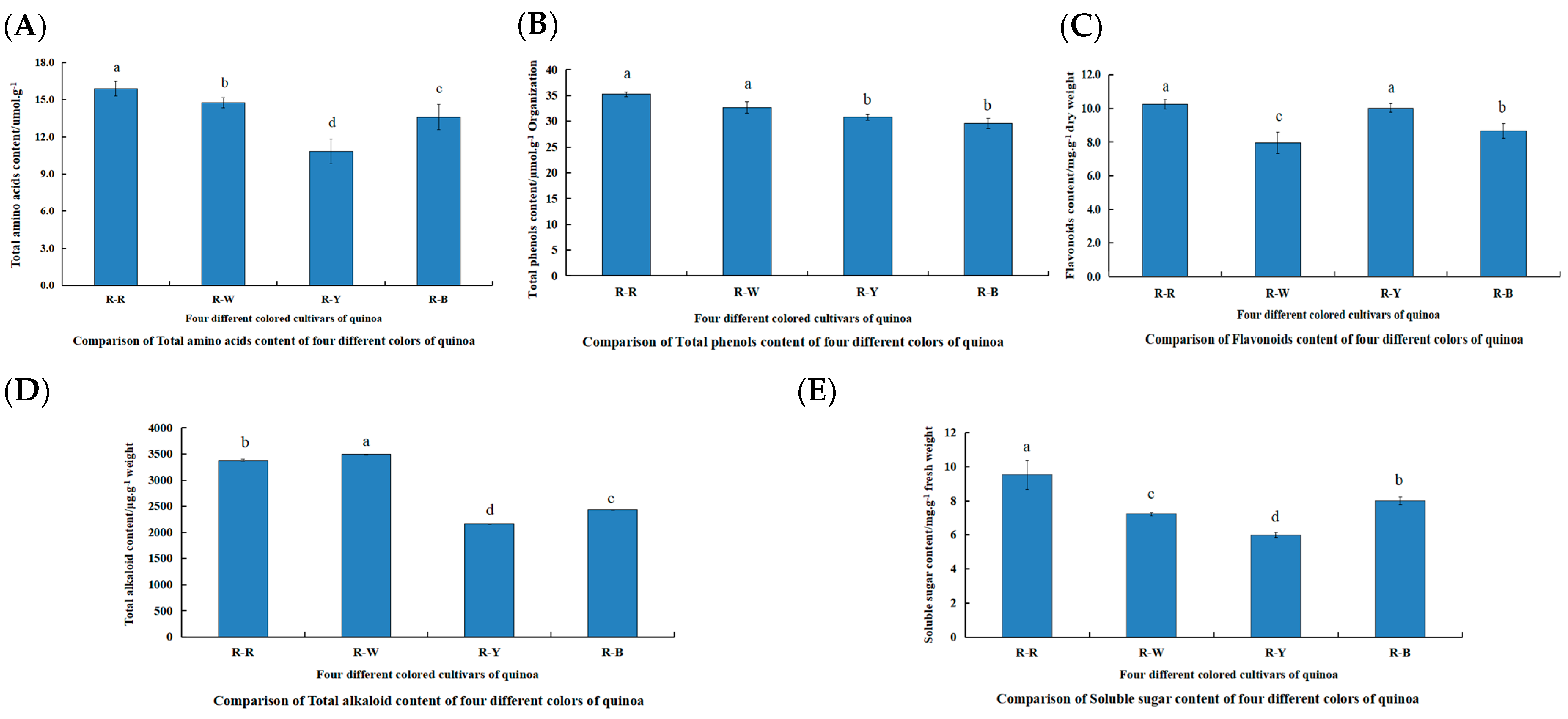
3.1. Differences in Metabolite Content during the Seedling Stage of Different Quinoa Cultivars
3.2. Qualitative and Quantitative Analysis of Metabolites in Different Quinoa Seedling Cultivars
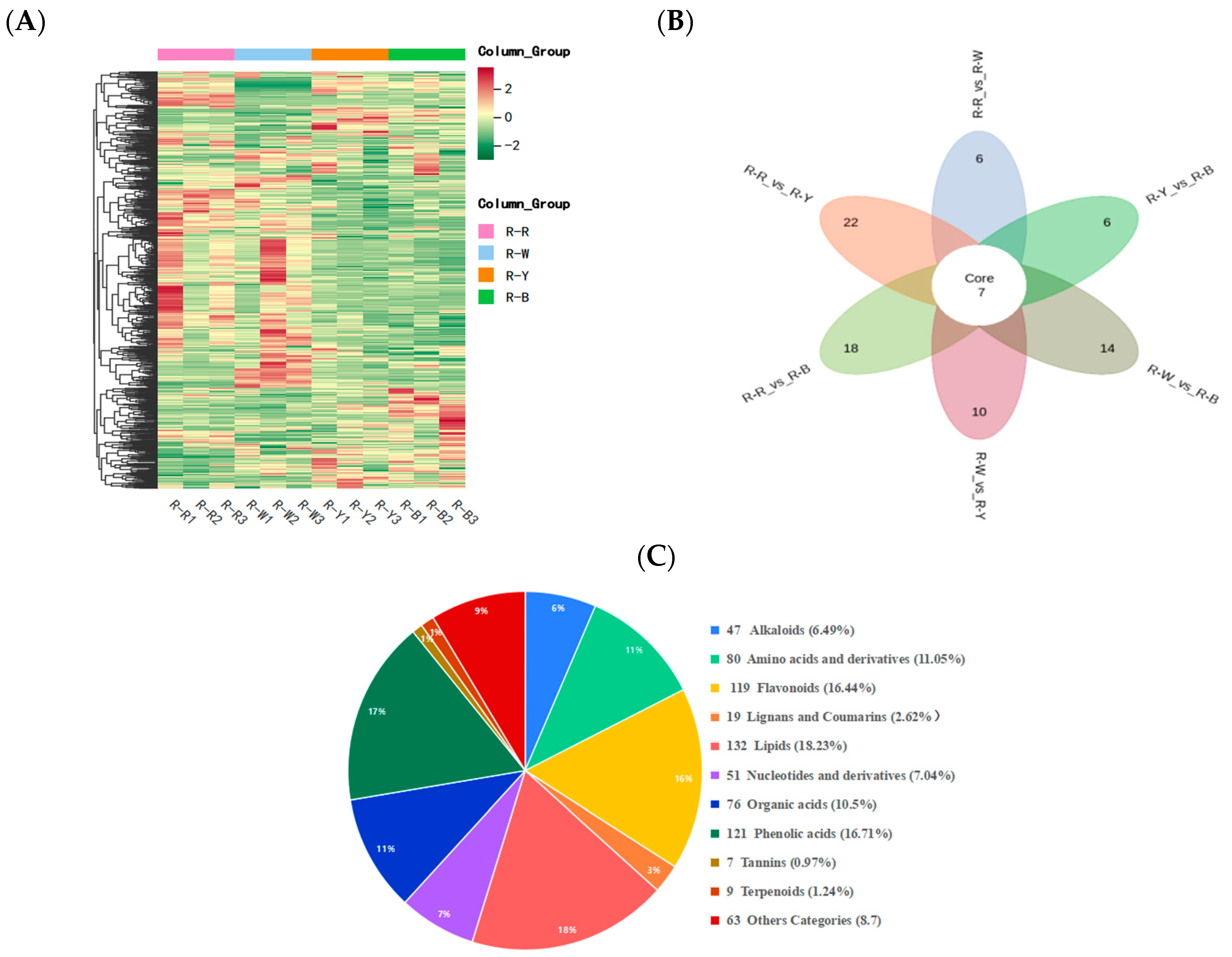
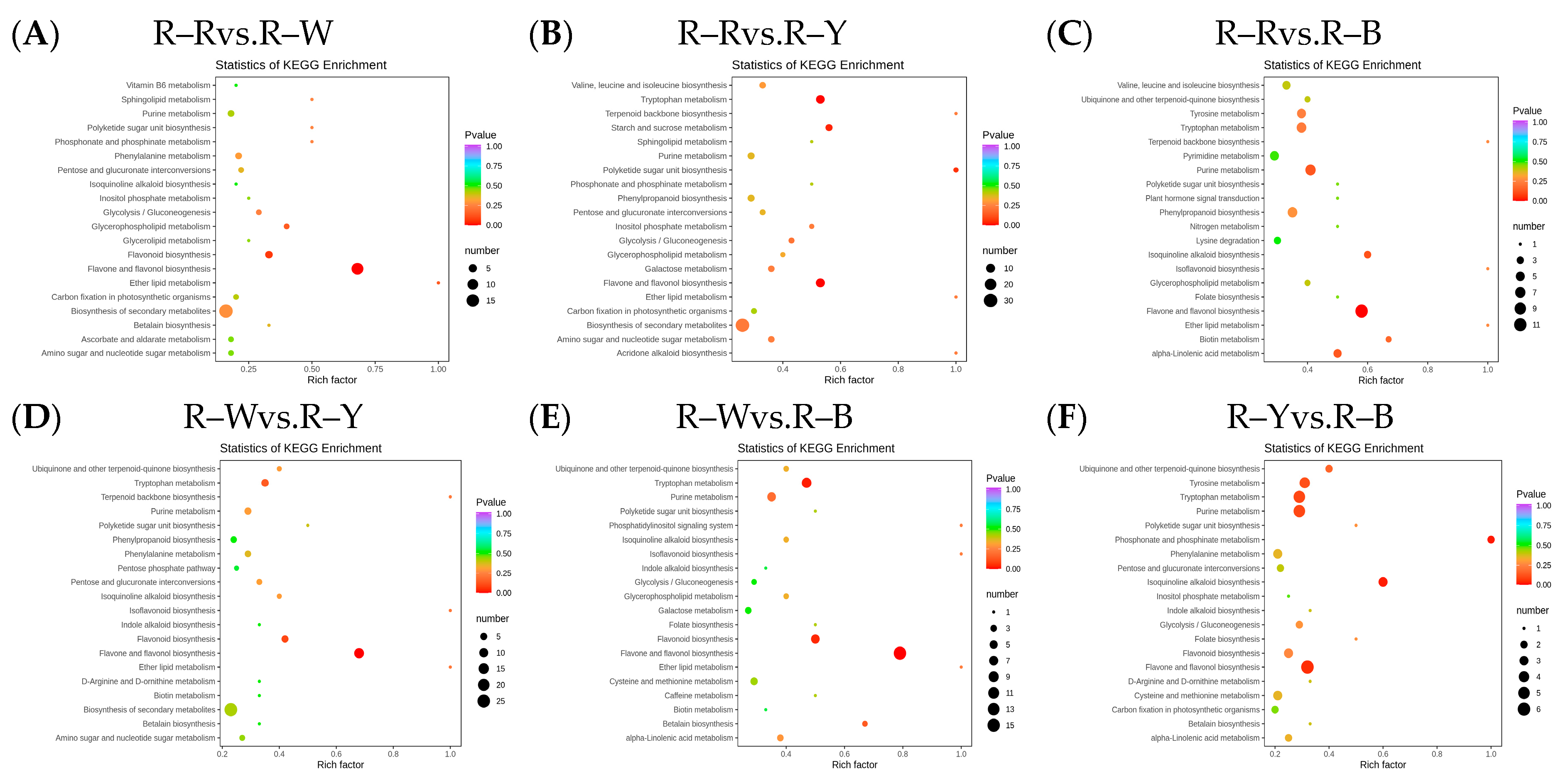
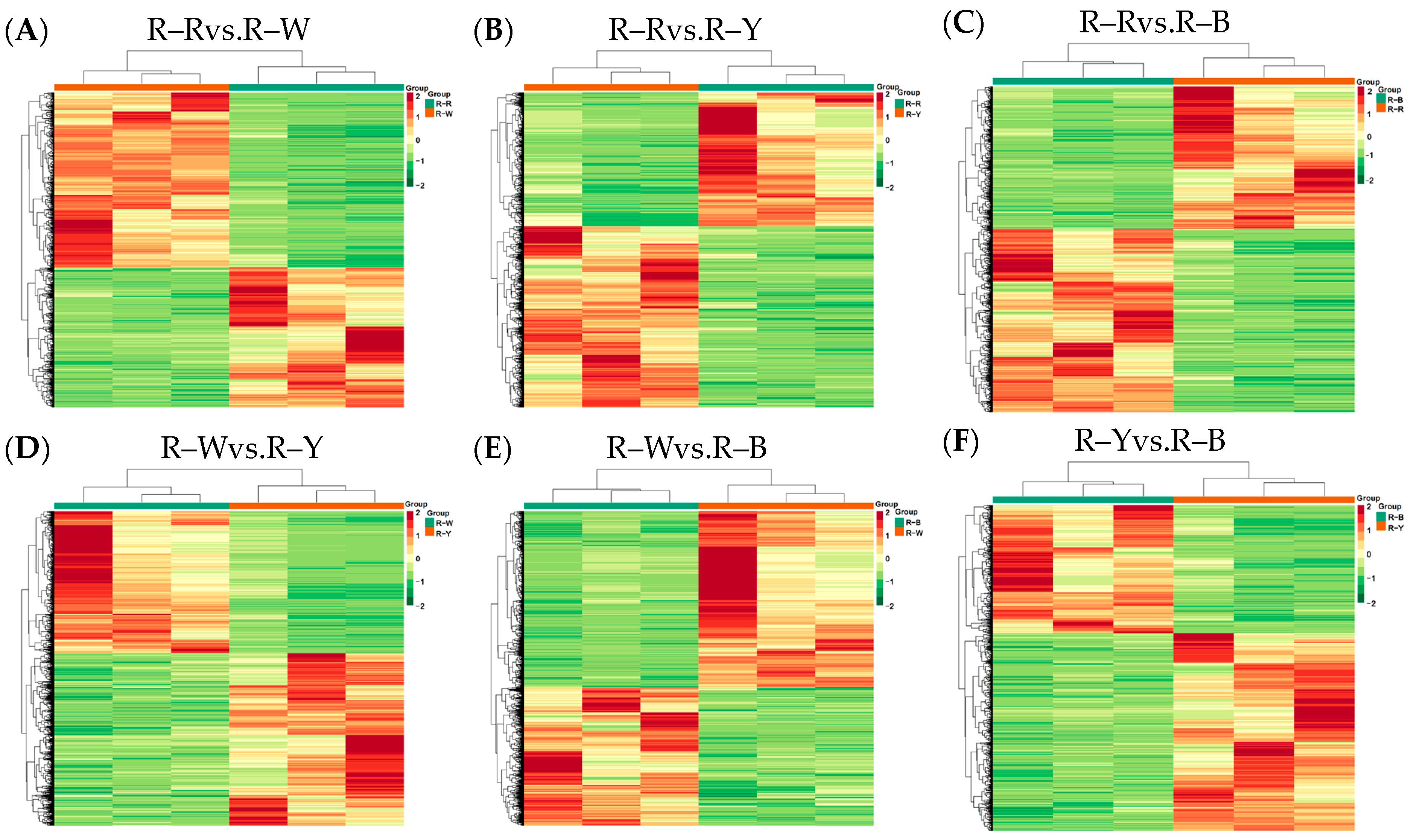
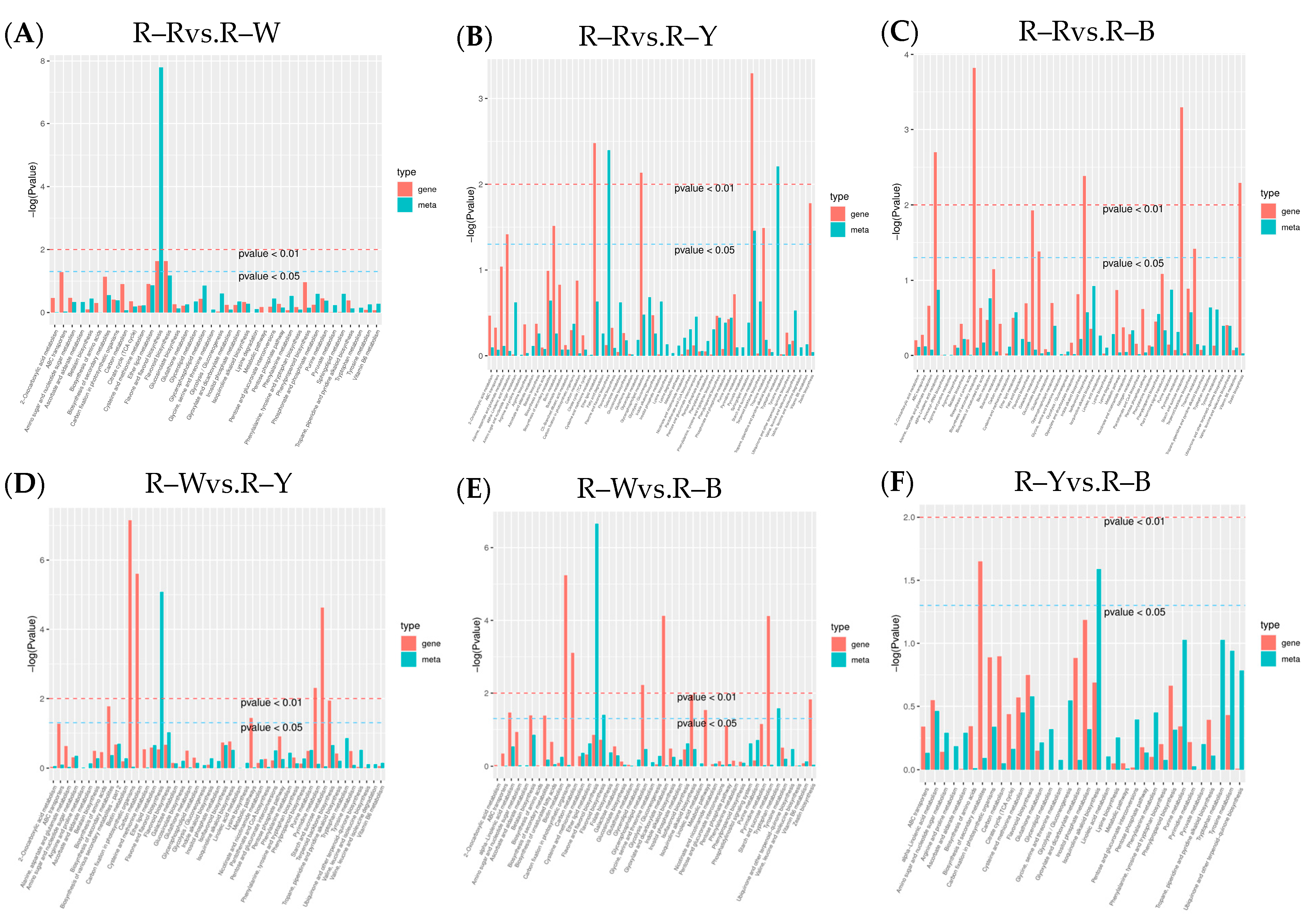
3.3. Differential Analysis and Enrichment of Metabolites in Different Quinoa Seedling Cultivars
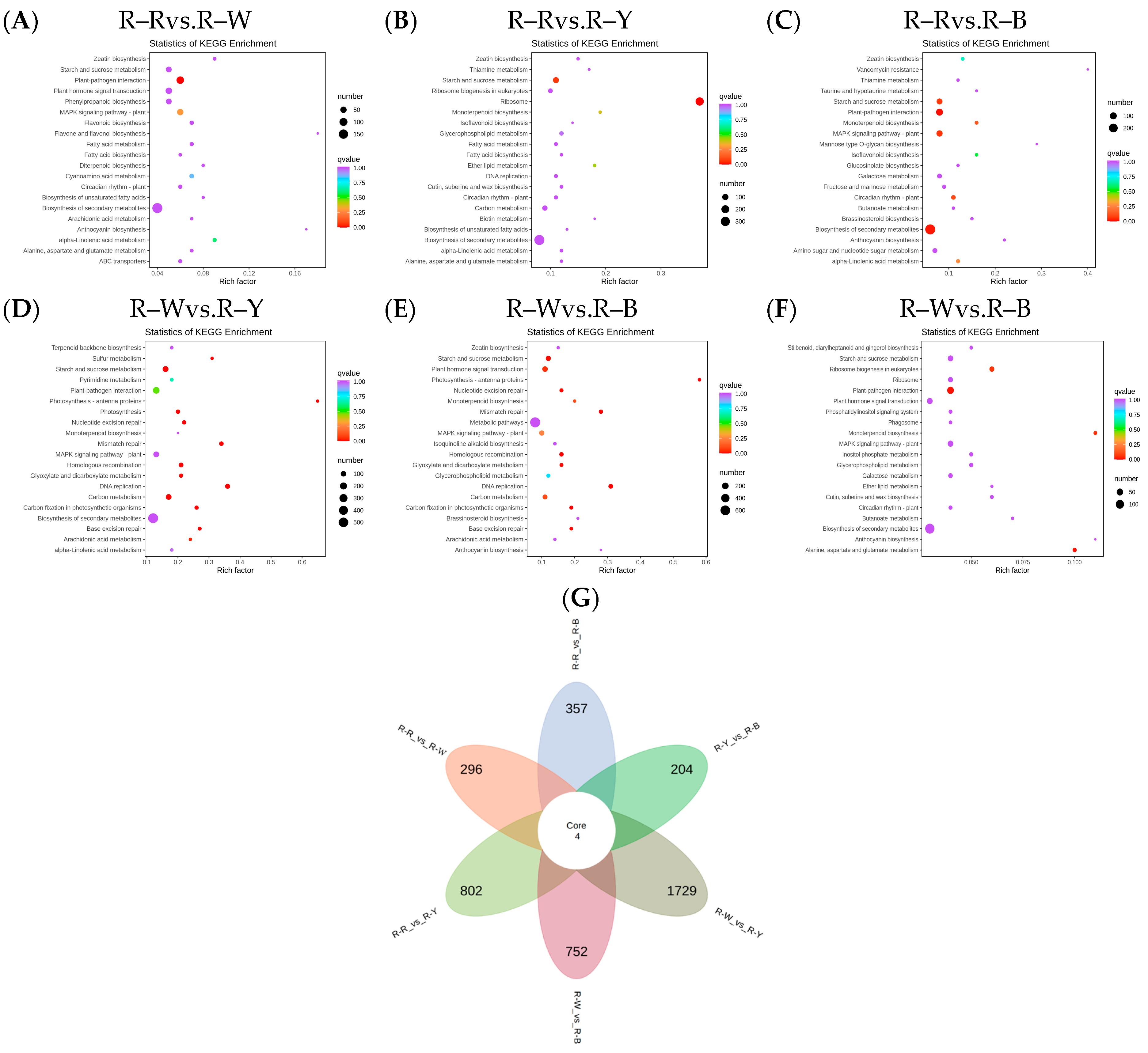
3.4. Transcriptome Analysis of Quinoa Seedlings among Different Cultivars
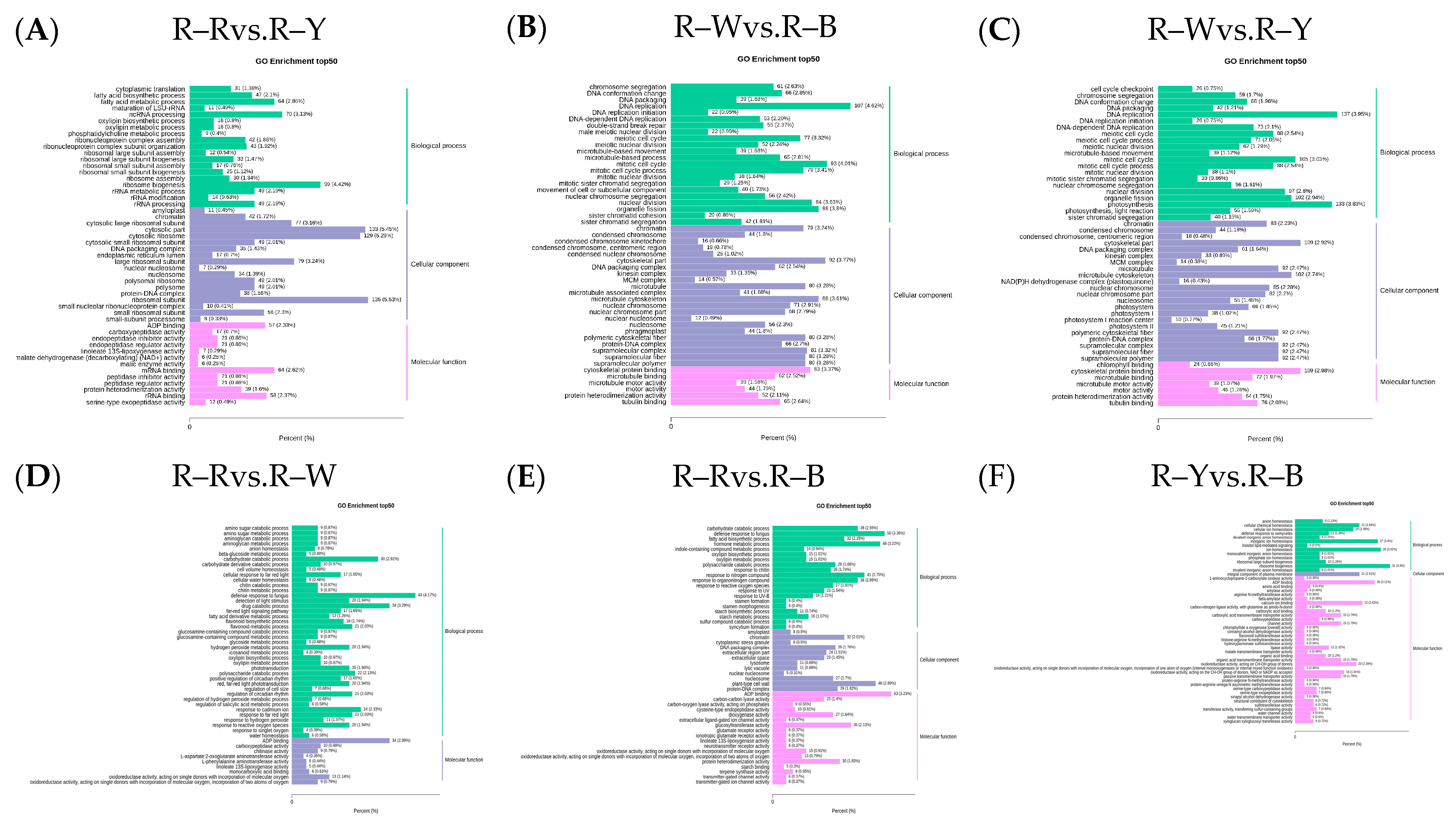
3.5. Differential Gene Expression Analysis of the Transcriptomes from Different Quinoa Seedling Cultivars
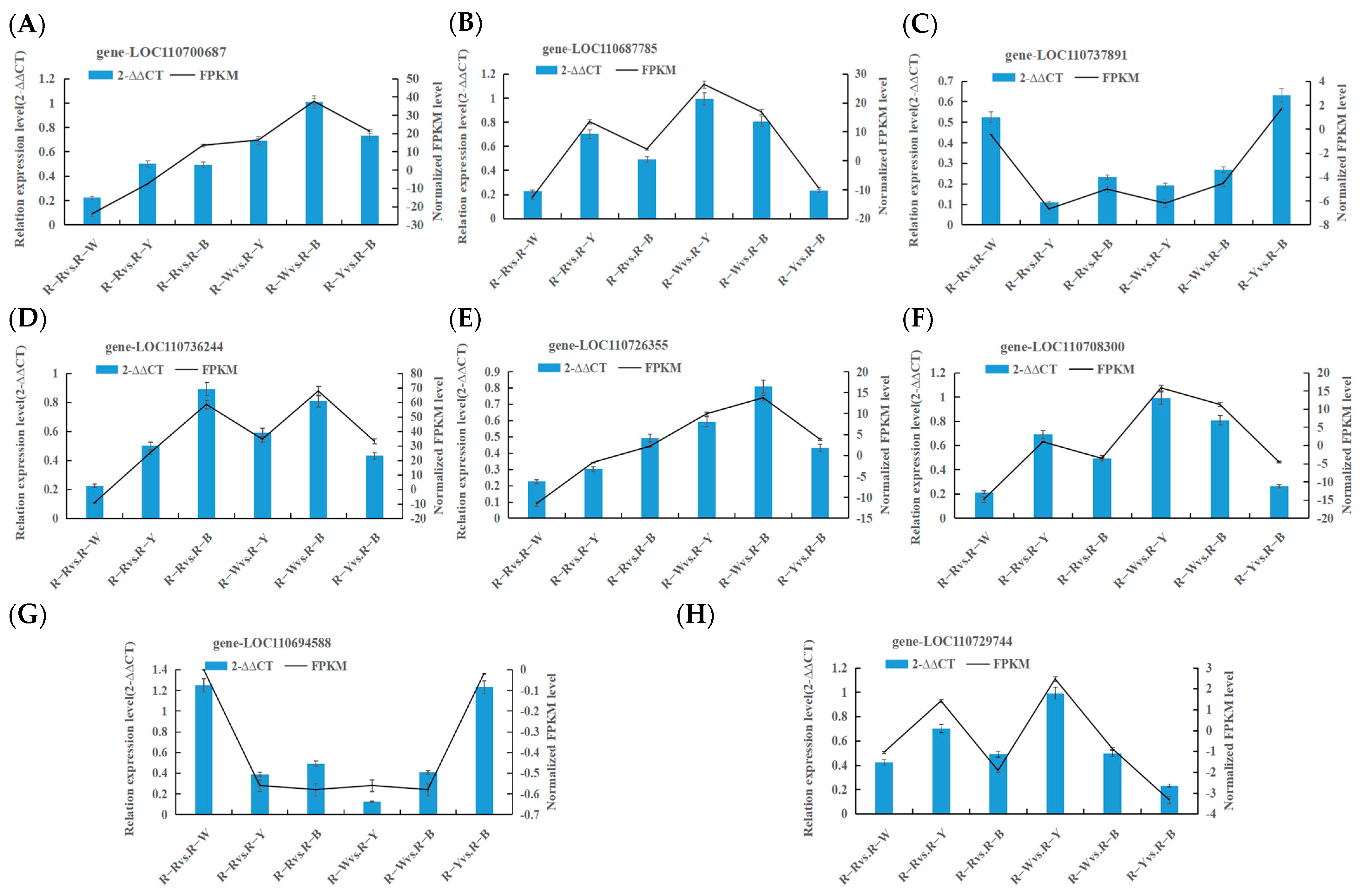
3.6. Real-Time Fluorescence Quantitative PCR Validation
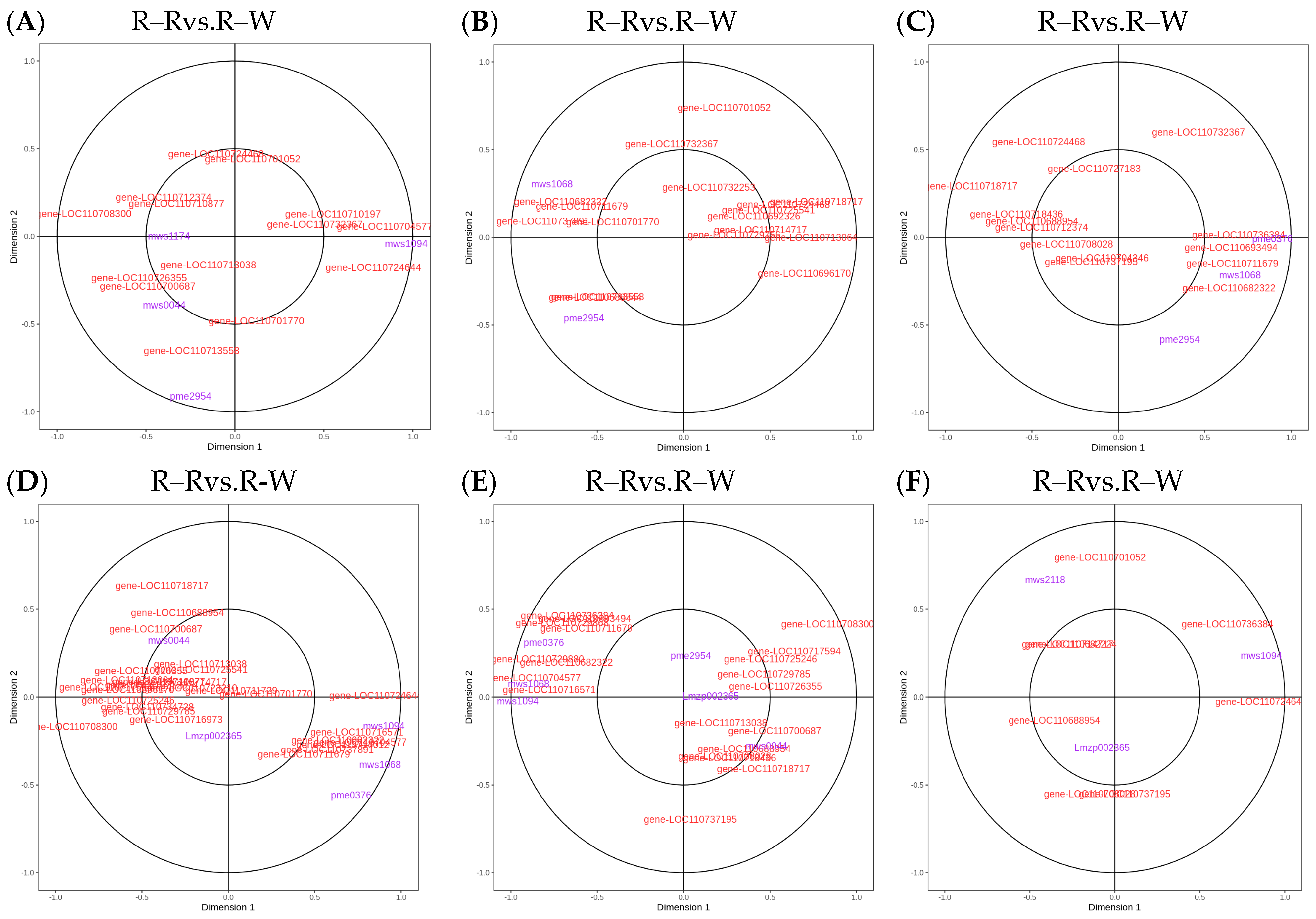
3.7. Correlation Analysis of the Metabolome and Transcriptomes of Different Quinoa Seedling Cultivars
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atul, B.; Sudhir, S.; Deepak, O. Chenopodium quinoa–An Indian perspective. Ind. Crops Prod. 2006, 23, 73–87. [Google Scholar] [CrossRef]
- Roman, V.J.; den Toom, L.A.; Gamiz, C.C.; van der Pijl, N.; Visser, R.G.; van Loo, E.N.; van der Linden, C.G. Differential responses to salt stress in ion dynamics, growth and seed yield of European quinoa varieties. Environ. Exp. Bot. 2020, 177, 104146. [Google Scholar] [CrossRef]
- Adolf, V.I.; Shabala, S.; Andersen, M.N.; Razzaghi, F.; Jacobsen, S.E. Varietal differences of quinoa’s tolerance to saline conditions. Plant Soil 2012, 357, 117–129. [Google Scholar] [CrossRef]
- Hariadi, Y.; Marandon, K.; Tian, Y.; Jacobsen, S.E.; Shabala, S. Ionic and osmotic relations in quinoa (Chenopodium quinoa Willd.) plants grown at various salinity levels. J. Exp. Bot. 2011, 62, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Vega-Gálvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martínez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef]
- Alandia, G.; Rodriguez, J.P.; Jacobsen, S.E.; Bazile, D.; Condori, B. Global expansion of quinoa and challenges for the Andean region. Glob. Food Secur. 2020, 26, 100429. [Google Scholar] [CrossRef]
- Pirozi, M.R.; Borges, J.T.; HM, P.S.A.; Chaves, J.B.; Coimbra, J.S. Quinoa: Nutritional, functional, and antinutritional aspects. Crit. Rev. Food Sci. Nutr. 2017, 57, 1618–1630. [Google Scholar] [CrossRef]
- James, L.E.A. Quinoa (Chenopodium quinoa Willd.): Composition, chemistry, nutritional, and functional properties. Adv. Food Nutr. Res. 2009, 58, 1–31. [Google Scholar] [CrossRef]
- Escuredo, O.; Martín, M.I.G.; Moncada, G.W.; Fischer, S.; Hierro, J.M.H. Amino acid profile of the quinoa (Chenopodium quinoa Willd.) using near infrared spectroscopy and chemometric techniques. J. Cereal Sci. 2014, 60, 67–74. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United States/World Health Organization/United Nations University, Energy and Protein Requirements; Report of a Joint FAO/WHO/UNU Meeting; World Health Organization: Geneva, Switzerland, 1985. [Google Scholar]
- Repo-Carrasco, R.; Espinoza, C.; Jacobsen, S.E. Nutritional value and use of the Andean crops quinoa (Chenopodium quinoa) and kañiwa (Chenopodium pallidicaule). Food Rev. Int. 2003, 19, 179–189. [Google Scholar] [CrossRef]
- Guo, H.; Hao, Y.; Yang, X.; Ren, G.; Richel, A. Exploration on bioactive properties of quinoa protein hydrolysate and peptides: A review. Crit. Rev. Food Sci. Nutr. 2023, 63, 2896–2909. [Google Scholar] [CrossRef] [PubMed]
- Relatives, W.C. Genomic and Breeding Resources by Ch. Kole.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 35–61. [Google Scholar]
- González, J.A.; Bruno, M.; Valoy, M.; Prado, F.E. Genotypic variation of gas exchange parameters and leaf stable carbon and nitrogen isotopes in ten quinoa cultivars grown under drought. J. Agron. Crop Sci. 2011, 197, 81–93. [Google Scholar] [CrossRef]
- Abd El-Samad, E.H.; Hussin, S.A.; El-Naggar, A.M.; El-Bordeny, N.E.; Eisa, S.S. The potential use of quinoa as a new non-traditional leafy vegetable crop. Biosci. Res. 2018, 15, 3387–3403, ISSN: 2218-3973. [Google Scholar]
- Adamczewska-Sowińska, K.; Sowiński, J.; Jama-Rodzeńska, A. The effect of sowing date and harvest time on leafy greens of quinoa (Chenopodium quinoa willd.) yield and selected nutritional parameters. Agriculture 2021, 11, 405. [Google Scholar] [CrossRef]
- Pathan, S.; Eivazi, F.; Valliyodan, B.; Paul, K.; Ndunguru, G.; Clark, K. Nutritional composition of the green leaves of quinoa (Chenopodium quinoa Willd.). J. Food Res. 2019, 8, 55–65. [Google Scholar] [CrossRef]
- Pandey, S.; Gupta, R.K. Screening of nutritional, phytochemical, antioxidant and antibacterial activity of Chenopodium album (Bathua). J. Pharmacogn. Phytochem. 2014, 3, 1–9. [Google Scholar]
- Poonia, A.; Upadhayay, A. Chenopodium album Linn: Review of nutritive value and biological properties. J. Food Sci. Technol. 2015, 52, 3977–3985. [Google Scholar] [CrossRef]
- Saini, S.; Saini, K. Chenopodium album Linn: An outlook on weed cum nutritional vegetable along with medicinal properties. Emergent Life Sci. Res. 2020, 6, 28–33. [Google Scholar] [CrossRef]
- Yadav, R.K.; Tomar, B.S.; Pachauri, N.; Jain, V. Studies of nutritional properties and antioxidant potential in green leafy vegetables. J. Sci. Food Agric. 2018, 2, 7–13. [Google Scholar] [CrossRef]
- Pathan, S.; Siddiqui, R.A. Nutritional composition and bioactive components in quinoa (Chenopodium quinoa Willd.) greens: A Review. Nutrients 2022, 14, 558. [Google Scholar] [CrossRef]
- Tang, Y.; Li, X.; Zhang, B.; Chen, P.X.; Liu, R.; Tsao, R. Characterisation of phenolics, betanins and antioxidant activities in seeds of three Chenopodium quinoa Willd. genotypes. Food Chem. 2015, 166, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Li, X.; Zhang, H.; Zhang, H.; Zhou, J.; Ma, X.; Li, L. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem. X 2023, 17, 100594. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Han, P.; Li, Y.; Wang, W.; Lai, D.; Zhou, L. Quinoa secondary metabolites and their biological activities or functions. Molecules 2019, 24, 2512. [Google Scholar] [PubMed]
- Al-Qabba, M.M.; El-Mowafy, M.A.; Althwab, S.A.; Alfheeaid, H.A.; Aljutaily, T.; Barakat, H. Phenolic profile, antioxidant activity, and ameliorating efficacy of chenopodium quinoa sprouts against CCl4-induced oxidative stress in rats. Nutrients 2020, 12, 2904. [Google Scholar] [CrossRef]
- Chandrasekara, A.; Shahidi, F. Bioactivities and antiradical properties of millet grains and hulls. J. Agric. Food Chem. 2011, 59, 9563–9571. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, B.; Li, X.; Chen, P.X.; Zhang, H.; Liu, R.; Tsao, R. Bound phenolics of quinoa seeds released by acid, alkaline, and enzymatic treatments and their antioxidant and α-glucosidase and pancreatic lipase inhibitory effects. J. Agric. Food Chem. 2016, 64, 1712–1719. [Google Scholar] [CrossRef]
- Morales, D.; Miguel, M.; Garcés-Rimón, M. Pseudocereals: A novel source of biologically active peptides. Crit. Rev. Food Sci. Nutr. 2021, 61, 1537–1544. [Google Scholar] [CrossRef]
- Lin, T.A.; Ke, B.J.; Cheng, C.S.; Wang, J.J.; Wei, B.L.; Lee, C.L. Red quinoa bran extracts protects against carbon tetrachloride-induced liver injury and fibrosis in mice via activation of antioxidative enzyme systems and blocking TGF-β1 pathway. Nutrients 2019, 11, 395. [Google Scholar] [CrossRef]
- Han, Y.; Chi, J.; Zhang, M.; Zhang, R.; Fan, S.; Huang, F.; Liu, L. Characterization of saponins and phenolic compounds: Antioxidant activity and inhibitory effects on α-glucosidase in different varieties of colored quinoa (Chenopodium quinoa Willd). Biosci. Biotechnol. Biochem. 2019, 83, 2128–2139. [Google Scholar] [CrossRef]
- Rocchetti, G.; Miragoli, F.; Zacconi, C.; Lucini, L.; Rebecchi, A. Impact of cooking and fermentation by lactic acid bacteria on phenolic profile of quinoa and buckwheat seeds. Food Res. Int. 2019, 119, 886–894. [Google Scholar] [CrossRef]
- Preczenhak, A.P.; Orsi, B.; Lima, G.P.P.; Tezotto-Uliana, J.V.; Minatel, I.O.; Kluge, R.A. Cysteine enhances the content of betalains and polyphenols in fresh-cut red beet. Food Chem. 2019, 286, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I. The reported colour formation mechanism in pitaya fruit through co-accumulation of anthocyanins and betalains is inconsistent and fails to establish the co-accumulation. BMC Genom. 2022, 23, 740. [Google Scholar] [CrossRef] [PubMed]
- Kanner, J.; Harel, S.; Granit, R. Betalains-A new of dietary cationized antioxidants. J. Agric. Food Chem. 2001, 49, 5178–5185. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Gonzalez, N.; Jaime-Fonseca, M.R.; San Martin-Martinez, E.; Zepeda, L.G. Extraction, stability, and separation of betalains from Opuntia joconostle cv. using response surface method-ology. J. Agric. Food Chem. 2013, 61, 11995–12004. [Google Scholar] [CrossRef]
- Bao, X.W.; Han, X.X.; Du, G.M.; Wei, C.Y.; Zhu, X.; Ren, W.; Zeng, L.J.; Zhang, Y.T. Antioxidant activities and immunomodulatory effects in mice of betalain in vivo. Food Sci.Chin. 2019, 40, 196–201. [Google Scholar]
- Rahimi, P.; Abedimanesh, S.; Mesbah-Namin, S.A.; Ostadrahimi, A. Betalains, the nature-inspired pigments, in health and disease. Crit. Rev. Food Sci. Nutr. 2019, 59, 2949–2978. [Google Scholar] [CrossRef]
- Winkler, C.; Wirleitner, B.; Schroecksnadel, K.; Schennach, H.; Fuchs, D. In vitro effects of beetroot juice on stimulated and unstimulated peripheral blood mononuclear cells. Appl. Biochem. Biotechnol. 2005, 1, 180–185. [Google Scholar]
- Esatbeyoglu, T.; Wagner, A.E.; Motafakkerazad, R.; Nakajima, Y.; Matsugo, S.; Rimbach, G. Free radical scavenging and antioxidant activity of betalain: Electon spin resonance spectroscopy studies and studies in cultured cells. Food Chem. Toxicol. 2014, 73, 119–126. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Rai, A.; Saito, K.; Yamazaki, M. Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 2017, 90, 764–787. [Google Scholar] [CrossRef]
- Hirasawa, T.; Saito, M.; Yoshikawa, K.; Furusawa, C.; Shmizu, H. Integrated analysis of the transcriptome and metabolome of Corynebacterium glutamicum during penicillin-induced glutamic acid production. Biotechnol. J. 2018, 13, 1700612. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kong, Z.; Liu, J.; Zhang, P.; Wang, Q.; Huan, X.; Li, L.; Qin, P. Non-targeted metabolomics of quinoa seed filling period based on liquid chromatog-raphy-mass spectrometry. Food Res Int. 2020, 137, 109743. [Google Scholar] [CrossRef]
- Wu, Q.; Bai, X.; Zhao, W.; Shi, X.; Xiang, D.; Wan, Y.; Wu, X.; Sun, Y.; Zhao, J.; Peng, L. Investigation into the underlying regulatory mechanisms shaping inflorescence architecture in Chenopodium quinoa. BMC Genom. 2019, 20, 658. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, L.; Chen, H.; Chen, S.; Liu, Y. Analysis of flavonoid metabolites in citrus peels (Citrus reticulata “Dahongpao”) using UPLC-ESI-MS/MS. Molecules 2019, 24, 2680. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- González, I.; Déjean, S.; Martin, P.G.; Baccini, A. CCA: An R package to extend canonical correlation analysis. J. Stat. Softw. 2008, 23, 1–14. [Google Scholar] [CrossRef]
- Bouhaddani, S.E.; Houwing-Duistermaat, J.; Salo, P.; Perola, M.; Jongbloed, G.; Uh, H.W. Evaluation of O2PLS in Omics data integration[C]//BMC bioinformatics. BioMed Cent. 2016, 17, 117–132. [Google Scholar] [CrossRef]
- Yan, C.; Guo, J.; Zhang, M.L. Research progress of flavonoids in buckwheat. Food Nutr. China 2015, 21, 65–69. [Google Scholar]
- Ren, Q.; Li, Y.; Wu, C.; Wang, C.; Zhang, J. Metabolism of secondary metabolites isolated from Tartary buckwheat and its extract. Food Chem. 2014, 154, 134–144. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Shirooie, S. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar]
- You, Q.; Wang, B.; Chen, F.; Huang, Z.; Wang, X.; Luo, P.G. Comparison of anthocyanins and phenolics in organically and conventionally grown blueberries in selected cultivars. Food Chem. 2011, 125, 201–208. [Google Scholar] [CrossRef]
- Cai, Y.; Sun, M.; Corke, H. Identification and distribution of simple and acylated betacyanins in the Amaranthaceae. J. Agric. Food Chem. 2001, 49, 1971–1978. [Google Scholar] [CrossRef]
- Repo-Carrasco-Valencia, R.; Hellström, J.K.; Pihlava, J.M.; Mattila, P.H. Flavonoids and other phenolic compounds in Andean indigenous grains: Quinoa (Chenopodium quinoa), kañiwa (Chenopodium pallidicaule) and kiwicha (Amaranthus caudatus). Food Chem. 2010, 120, 128–133. [Google Scholar] [CrossRef]
- Benavente-Garcia, O.; Castillo, J. Update on uses and properties of citrus flavonoids: New findings in anticancer, cardiovascular, and anti-inflammatory activity. J. Agric. Food Chem. 2008, 56, 6185–6205. [Google Scholar]
- Renard, C.M.; Wende, G.; Booth, E.J. Cell wall phenolics and polysaccharides in different tissues of quinoa (Chenopodium quinoa Willd). J. Sci. Food Agric. 1999, 79, 2029–2034. [Google Scholar] [CrossRef]
- Gómez-Caravaca, A.M.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Caboni, M.F. Simultaneous determination of phenolic compounds and saponins in quinoa (Chenopodium quinoa Willd.) by a liquid chromatography–diode array detection–electrospray ionization–time-of-flight mass spectrometry methodology. J. Agric. Food Chem. 2011, 59, 10815–10825. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Hou, H.; Ma, X.; Sun, S.; Wang, H.; Kong, L. Metabolomics and gene expression analysis reveal the accumulation patterns of phenylpropanoids and flavonoids in different colored-grain wheats (Triticum aestivum L. ). Food Res. Int. 2020, 138, 109711. [Google Scholar] [CrossRef] [PubMed]
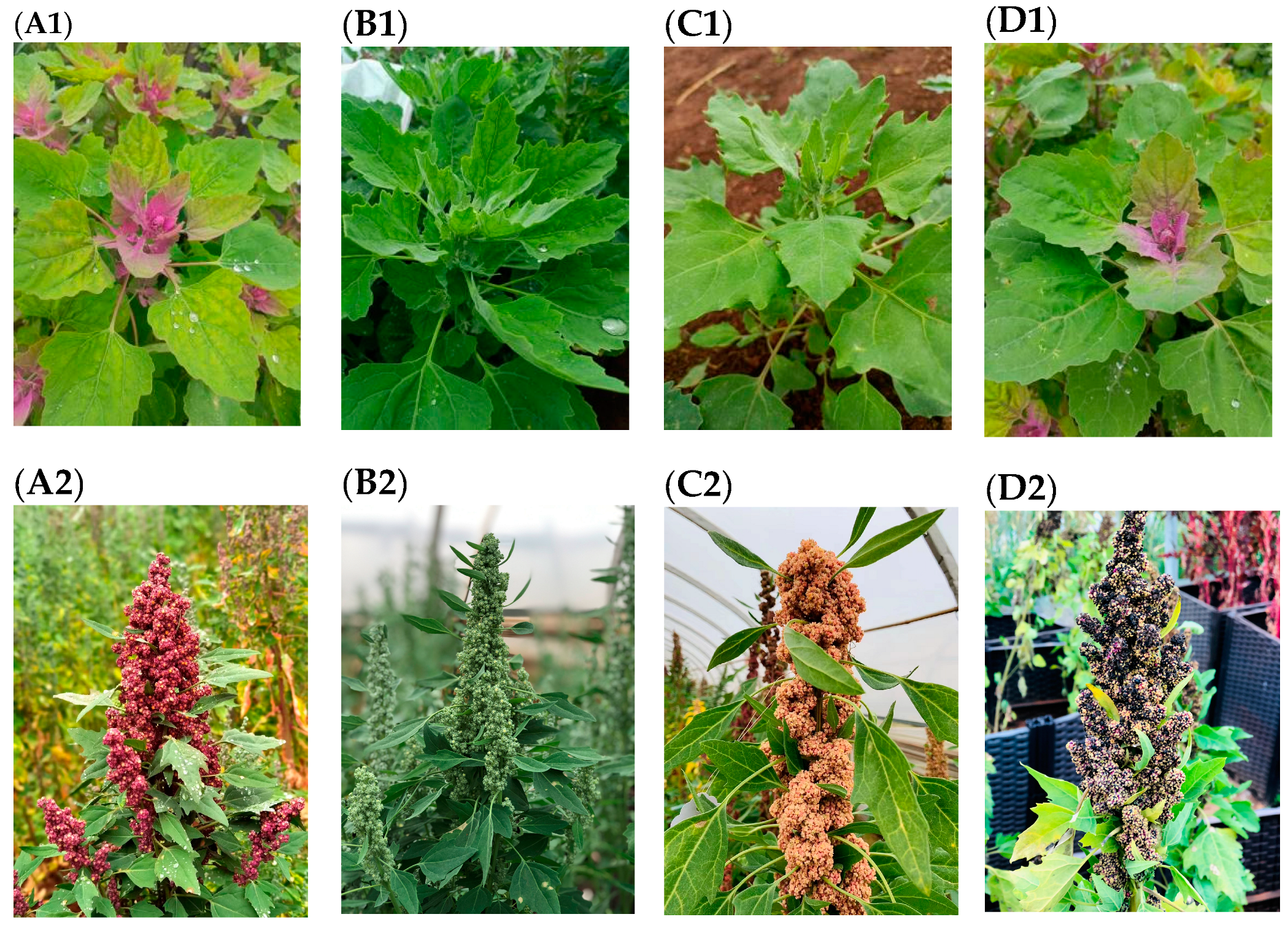

| Quinoa Color Classification | Plant Height (cm) M ± SD | Stem Thickness (cm) M ± SD | Leaf Area (LAI) (mm2) M ± SD | Relative Chlorophyll Content (SPAD) |
|---|---|---|---|---|
| Red quinoa (R–R) | 54.93 ± 1.36 c | 1.27 ± 1.01 a | 3137.10 ± 22.19 a | 49.1 |
| White quinoa (R–W) | 69.67 ± 0.59 a | 1.10 ± 1.16 bc | 2809.21 ± 19.11 ab | 44.43 |
| Yellow quinoa (R–Y) | 56.55 ± 1.12 b | 1.05 ± 1.29 c | 2782.80 ± 29.36 c | 44.87 |
| Black quinoa (R–B) | 65.90 ± 1.57 ab | 1.22 ± 2.13 ab | 2811.27 ± 16.56 ab | 46.67 |
| Group | R–Rvs.R–W | R–Rvs.R–Y | R–Rvs.R–B | R–Wvs.R–Y | R–Wvs.R–B | R–Yvs.R–B |
|---|---|---|---|---|---|---|
| Number of DAMs | 169 | 249 | 272 | 250 | 273 | 112 |
| Upregulated DAMs | 61 | 76 | 78 | 97 | 111 | 50 |
| Downregulated DAMs | 108 | 173 | 194 | 153 | 162 | 62 |
| Number of DAMs annotated by kegg | 43 | 73 | 83 | 68 | 74 | 47 |
| Amino acid and its derivatives | 5 | 20 | 19 | 12 | 15 | 10 |
| Phenolic acids | 34 | 48 | 55 | 42 | 47 | 22 |
| Flavonoid | 94 | 54 | 56 | 93 | 92 | 30 |
| Alkaloids | 10 | 17 | 17 | 12 | 13 | 14 |
| Organic acid | 6 | 24 | 21 | 14 | 9 | 2 |
| Lipids | 4 | 45 | 66 | 43 | 64 | 17 |
| Group | Index | Compounds | Content Comparison | Log2FC |
|---|---|---|---|---|
| R–Rvs.R–W | mws1174 | 3-O-Acetylpinobanksin | R > B > Y > W | −3.09 |
| R–Rvs.R–Y | mws1078 | Anthranilic Acid | R > W > B > Y | −1.13 |
| mws0051 | Acacetin | R > W > B > Y | −2.04 | |
| R–Rvs.R–B | mws0047 | Apigenin-7-O-neohesperidoside | R > W > Y > B | −1.01 |
| pme1651 | Indole 3-acetic acid (IAA) | Y > W > R > B | −1.24 | |
| mws0853 | Sinapyl alcohol | R > W > Y > B | 3.02 | |
| R–Wvs.R–Y | pme1439 | p-Coumaric acid | W > R > B > Y | −1.38 |
| R–Wvs.R–B | NK10253223 | 2-Amino-3-methoxybenzoic acid | W > R > Y > B | −1.18 |
| R–Yvs.R–B | mws2118 | Phloretin-2′-O-glucoside | Y > W > R > B | −1.16 |
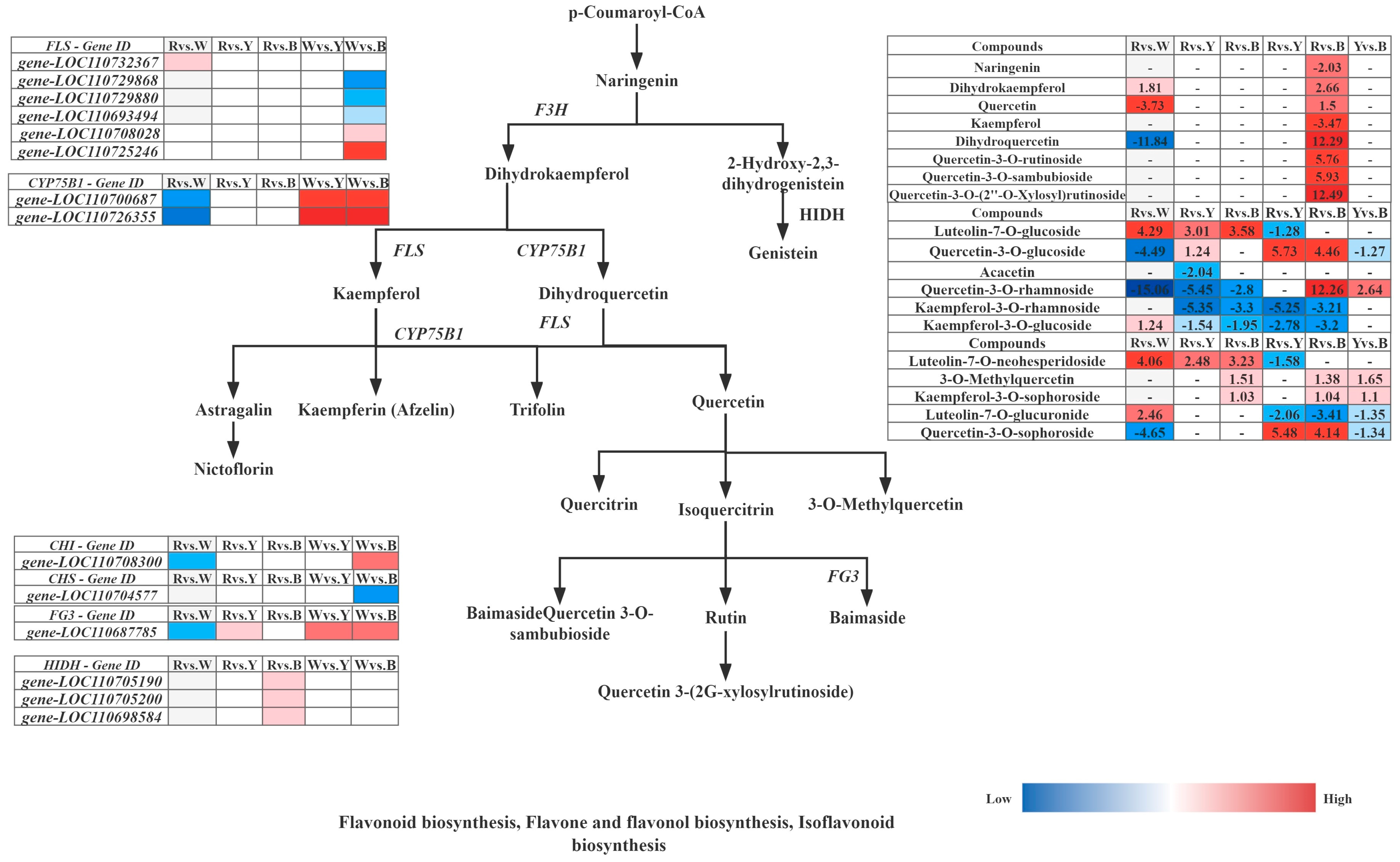
| Index | Compounds | Log2FC of Relative Metabolites | |||||
|---|---|---|---|---|---|---|---|
| Rvs.W | Rvs.Y | Rvs.B | Wvs.Y | Wvs.B | Yvs.B | ||
| Flavonoid biosynthesis (ko00941) | |||||||
| mws1094 | Aromadendrin (Dihydrokaempferol) | 1.81 | - | - | −1.66 | −2.66 | - |
| pme2954 | Quercetin | −3.73 | - | - | - | 1.50 | - |
| mws1174 | 3-O-Acetylpinobanksin | −3.09 | - | - | - | - | - |
| mws0044 | Dihydroquercetin(Taxifolin) | −1.18 | - | - | 1.13 | 1.23 | - |
| Lmzp002365 | Hesperetin-7-O-glucoside | - | - | - | 3.84 | 2.82 | - |
| pme0376 | Naringenin (5,7,4′-Trihydroxyflavanone) | - | - | - | −1.07 | −2.03 | - |
| mws1068 | Kaempferol (3,5,7,4′-Tetrahydroxyflavone) | - | - | - | −4.03 | −3.47 | - |
| Flavone and flavonol biosynthesis (ko00944) | |||||||
| pme2954 | Quercetin | - | −3.21 | −2.24 | - | - | - |
| pme2459 | Luteolin-7-O-glucoside (Cynaroside) | - | 3.01 | 3.58 | - | - | - |
| mws0091 | Quercetin-3-O-glucoside (Isoquercitrin) | - | 1.24 | - | - | - | −1.27 |
| mws0051 | Acacetin | - | −2.04 | - | - | - | - |
| mws0045 | Quercetin-3-O-rhamnoside(Quercitrin) | - | −5.45 | - | - | - | - |
| mws0919 | Kaempferol-3-O-rhamnoside (Afzelin) | - | −5.35 | −3.30 | - | - | - |
| Lmsn002815 | Kaempferol-3-O-rutinoside(Nicotiflorin) | - | 1.61 | 1.43 | - | - | - |
| mws2209 | Kaempferol-3-O-glucoside (Astragalin) | - | −1.54 | −1.95 | - | - | - |
| mws1068 | Kaempferol (3,5,7,4′-Tetrahydroxyflavone) | - | −3.05 | −2.50 | - | - | - |
| pmp001079 | Luteolin-7-O-neohesperidoside (Lonicerin) | - | 2.48 | 3.23 | - | - | - |
| Lmmn004912 | 3-O-Methylquercetin | - | - | 1.51 | - | - | 1.65 |
| mws0045 | Quercetin-3-O-rhamnoside(Quercitrin) | - | - | −2.80 | - | - | 2.64 |
| mws0047 | Apigenin-7-O-neohesperidoside (Rhoifolin) | - | - | −1.01 | - | - | - |
| Lmyn001269 | Kaempferol-3-O-sophoroside | - | - | 1.03 | - | - | 1.10 |
| mws4167 | Luteolin-7-O-glucuronide | - | - | - | - | - | −1.35 |
| Lmtp003677 | Quercetin-3-O-sophoroside (Baimaside) | - | - | - | - | - | −1.34 |
| Tryptophan metabolism (ko00380) | |||||||
| pmb0774 | N-Hydroxytryptamine | - | −2.28 | - | - | −3.53 | - |
| pme2024 | Serotonin | - | −2.24 | - | - | −3.59 | - |
| Zmtn001624 | N-Acetylisatin | - | −2.16 | - | - | - | - |
| pme3083 | 2-(Formylamino)benzoic acid | - | −1.36 | - | - | −1.84 | - |
| mws0005 | Tryptamine | - | −3.27 | - | - | −4.12 | - |
| Zmtn001464 | 4,8-Dihydroxyquinoline-2-carboxylic acid | - | 3.27 | - | - | −1.51 | - |
| mws0677 | N-Acetyl-5-hydroxytryptamine | - | −1.18 | - | - | - | - |
| mws1078 | Anthranilic Acid | - | −1.13 | - | - | - | - |
| pme1228 | 5-Hydroxy-L-tryptophan | - | 1.07 | - | - | −1.01 | - |
| NK10253223 | 2-Amino-3-methoxybenzoic acid | - | - | - | - | −1.18 | - |
| mws0596 | 3-Hydroxyanthranilic acid | - | - | - | - | 1.15 | - |
| Isoquinoline alkaloid biosynthesis (ko00950) | |||||||
| pme3827 | 3,4-Dihydroxy-L-phenylalanine (L-Dopa) | - | - | - | - | - | 2.84 |
| pme1002 | L-Tyramine | - | - | - | - | - | 1.28 |
| Hmgn001653 | Protocatechualdehyde | - | - | - | - | - | −1.96 |
| Phosphonate and phosphinate metabolism (ko00440) | |||||||
| mws2125 | Phosphoenolpyruvate | - | - | - | - | - | 1.95 |
| pmb0302 | 2-Aminoethylphosphonate | - | - | - | - | - | 1.04 |
| Group | R–Rvs.R–W | R–Rvs.R–Y | R–Rvs.R–B | R–Wvs.R–Y | R–Wvs.R–B | R–Yvs.R–B |
|---|---|---|---|---|---|---|
| Number of DEGs | 2370 | 4507 | 3125 | 6473 | 4436 | 1492 |
| Upregulated DEGs | 1318 | 2592 | 1760 | 3547 | 1949 | 587 |
| Downregulated DEGs | 1052 | 1915 | 1365 | 2926 | 2487 | 905 |
| Quantity | Gene-ID | Gene Description | Primer | 5′ to 3′ |
|---|---|---|---|---|
| 1 | LOC110700687 | flavonoid 3′-monooxygenase-like | Forward Primer | TTGACTGACACTGAGATTA |
| Reverse Primer | GATTGCGGATTAGTTCTG | |||
| 2 | LOC110726355 | flavonoid 3′-monooxygenase-like | Forward Primer | GGAAGAACACAAGGCTAACT |
| Reverse Primer | CCTCACCATCACAATTATCTCT | |||
| 3 | LOC110708300 | fatty-acid-binding protein 1-like | Forward Primer | CTGATGTCACTGAACCTAA |
| Reverse Primer | CCTCCTCAATCCAATACC | |||
| 4 | LOC110687785 | anthocyanidin 3-O-glucosyltransferase-like | Forward Primer | TGCTATCTTAATCACTCA |
| Reverse Primer | CCATCTTCATCTCTTCTA | |||
| 5 | LOC110737891 | fatty-acid-binding protein 3, chloroplastic-like | Forward Primer | CGACTCCTGTTGATGAAT |
| Reverse Primer | CAAGCCAAGTTAGAAGAATC | |||
| 6 | LOC110736244 | phospholipase A1-Igamma3, chloroplastic-like | Forward Primer | GTCTAATATCCTCTCCTAA |
| Reverse Primer | CTTCTTACCGTTCTACTA | |||
| 7 | LOC110729744 | carboxylesterase 1-like | Forward Primer | GATTGTTGTGTCTGTTGAG |
| Reverse Primer | AGCATCCATAGCATCATC | |||
| 8 | LOC110694588 | uncharacterized LOC110694588 | Forward Primer | TCCACAAGTTCTGTTCAC |
| Reverse Primer | GCAGTAACCGCATCTATA | |||
| Internal reference gene | TUB-6 | beta-6 tubulin | Forward Primer | TGAGAACGCAGATGAGTGTATG |
| Reverse Primer | GAAACGAAGACAGCAAGTGACA |
| Meta ID | Compounds | Accumulation Comparison | Log2FC | |||||
|---|---|---|---|---|---|---|---|---|
| Rvs.W | Rvs.Y | Rvs.B | Wvs.Y | Wvs.B | Yvs.B | |||
| pme0376 | Naringenin (5,7,4′-Trihydroxyflavanone) | W > R > Y > B | - | - | - | - | −2.03 | - |
| mws1094 | Dihydrokaempferol | W > Y > R > B | 1.81 | - | - | - | 2.66 | - |
| pme2954 | Quercetin | Y > W > B > R | −3.73 | - | - | - | 1.50 | - |
| mws1068 | Kaempferol | W > Y > B > R | - | - | - | - | −3.47 | - |
| mws0044 | Dihydroquercetin(Taxifolin) | B > R > Y > W | −11.84 | - | - | - | 12.29 | - |
| mws0059 | Quercetin-3-O-rutinoside (Rutin) | Y > R > B > W | - | - | - | - | 5.76 | - |
| Lmjp002596 | Quercetin-3-O-sambubioside | Y > B > R > W | - | - | - | - | 5.93 | - |
| Hmcp001618 | Quercetin-3-O-(2″-O-Xylosyl)rutinoside | B > Y > R > W | - | - | - | - | 12.49 | - |
| pme2459 | Luteolin-7-O-glucoside (Cynaroside) | W > B > Y > R | 4.29 | 3.01 | 3.58 | −1.28 | - | - |
| mws0091 | Quercetin-3-O-glucoside (Isoquercitrin) | Y > R > B > W | −4.49 | 1.24 | - | 5.73 | 4.46 | −1.27 |
| mws0051 | Acacetin | B > R > Y > W | - | −2.04 | - | - | - | - |
| mws0045 | Quercetin-3-O-rhamnoside(Quercitrin) | R > B > Y > W | −15.06 | −5.45 | −2.80 | - | 12.26 | 2.64 |
| mws0919 | Kaempferol-3-O-rhamnoside (Afzelin) | R > W > B > Y | - | −5.35 | −3.30 | −5.25 | −3.21 | - |
| mws2209 | Kaempferol-3-O-glucoside (Astragalin) | W > R > Y > B | 1.24 | −1.54 | −1.95 | −2.78 | −3.20 | - |
| pmp001079 | Luteolin-7-O-neohesperidoside | W > B > Y > R | 4.06 | 2.48 | 3.23 | −1.58 | - | - |
| Lmmn004912 | 3-O-Methylquercetin | B > W > R > Y | - | - | 1.51 | - | 1.38 | 1.65 |
| Lmyn001269 | Kaempferol-3-O-sophoroside | B > R > W > Y | - | 1.03 | - | 1.04 | 1.10 | |
| mws4167 | Luteolin-7-O-glucuronide | W > B > Y > R | 2.46 | - | - | −2.06 | −3.41 | −1.35 |
| Lmtp003677 | Quercetin-3-O-sophoroside (Baimaside) | W > Y > R > B | −4.65 | - | 5.48 | 4.14 | −1.34 | |
| mws1138 | Betanin (Betanidin-5-O-glucoside) | R > B > Y > W | −4.06 | −1.36 | - | 2.70 | 3.92 | - |
| mws0014 | Ferulic acid | R > B > W > Y | −2.62 | −3.31 | −2.13 | - | - | 1.18 |
| mws2212 | Caffeic acid | R > W > Y > B | −1.29 | −1.43 | −2.28 | - | - | - |
| mws0027 | Syringic acid | R > Y > W > B | - | - | −1.48 | - | - | - |
| Group | Gene Name | KEGG | Meta Name | Compounds | PCC |
|---|---|---|---|---|---|
| R–Rvs.R–W R–Wvs.R–Y R–Wvs.R-B | gene-LOC110700687 | K05280 flavonoid 3′-monooxygenase [EC:1.14.14.82]|(RefSeq) flavonoid 3′-monooxygenase-like (A) | mws0059 | Quercetin-3-O-rutinoside | 0.835 |
| mws0044 | Dihydroquercetin | 0.9 | |||
| Lmjp002596 | Quercetin-3-O-sambubioside | 0.8 | |||
| Hmcp001618 | Quercetin-3-O-(2″-O-Xylosyl)rutinoside | 0.83 | |||
| gene-LOC110687785 | K22794 flavonol-3-O-glucoside/galactoside glucosyltransferase [EC:2.4.1.239 2.4.1.-]|(RefSeq) anthocyanidin 3-O-glucoside 2″-O-glucosyltransferase-like (A) | Hmcp001618 | Quercetin-3-O-(2″-O-Xylosyl)rutinoside | 0.822 | |
| Lmjp002596 | Quercetin-3-O-sambubioside | 0.81 | |||
| R–Rvs.R–Y | gene-LOC110737891 | K01859 chalcone isomerase [EC:5.5.1.6]|(RefSeq) probable chalcone--flavonone isomerase 3 (A) | mws1068 | Kaempferol (3,5,7,4′-Tetrahydroxyflavone) | 0.889 |
| R–Rvs.R-B, R–Wvs.R–Y, R–Wvs.R-B | gene-LOC110736244 | K16818 phospholipase A1 [EC:3.1.1.32]|(RefSeq) phospholipase A(1) DAD1, chloroplastic (A) | mws0120 | Choline Alfoscerate | −0.8 |
| R–Rvs.R-B, R–Yvs.R-B | gene-LOC110729744 | K22097 3-O-acetylpapaveroxine carboxylesterase [EC:3.1.1.105]|(RefSeq) carboxylesterase 1-like (A) | pme3827 | 3,4-Dihydroxy-L-phenylalanine (L-Dopa) | −0.884 |
| R–Rvs.R–Y, R–Rvs.R-B, R–Wvs.R–Y, R–Wvs.R–B | gene-LOC110694588 | K14085 aldehyde dehydrogenase family 7 member A1 [EC:1.2.1.31 1.2.1.8 1.2.1.3]|(RefSeq) aldehyde dehydrogenase family 2 member C4-like (A) | pme2024 | Serotonin | 0.827 |
| pmb0774 | N-Hydroxytryptamine | 0.834 | |||
| R–Rvs.R–B, R–Wvs.R–B | gene-LOC110682171 | K06123 1-acylglycerone phosphate reductase [EC:1.1.1.101]|(RefSeq) hypothetical protein (A) | pmb0484 | Choline | −0.822 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Liu, J.; Zhang, P.; Wang, Q.; Li, L.; Xie, H.; Li, H.; Wang, H.; Cheng, S.; Qin, P. Elucidating the Differentiation Synthesis Mechanisms of Differently Colored Resistance Quinoa Seedings Using Metabolite Profiling and Transcriptome Analysis. Metabolites 2023, 13, 1065. https://doi.org/10.3390/metabo13101065
Liu J, Liu J, Zhang P, Wang Q, Li L, Xie H, Li H, Wang H, Cheng S, Qin P. Elucidating the Differentiation Synthesis Mechanisms of Differently Colored Resistance Quinoa Seedings Using Metabolite Profiling and Transcriptome Analysis. Metabolites. 2023; 13(10):1065. https://doi.org/10.3390/metabo13101065
Chicago/Turabian StyleLiu, Junna, Jian Liu, Ping Zhang, Qianchao Wang, Li Li, Heng Xie, Hanxue Li, Hongxin Wang, Shunhe Cheng, and Peng Qin. 2023. "Elucidating the Differentiation Synthesis Mechanisms of Differently Colored Resistance Quinoa Seedings Using Metabolite Profiling and Transcriptome Analysis" Metabolites 13, no. 10: 1065. https://doi.org/10.3390/metabo13101065
APA StyleLiu, J., Liu, J., Zhang, P., Wang, Q., Li, L., Xie, H., Li, H., Wang, H., Cheng, S., & Qin, P. (2023). Elucidating the Differentiation Synthesis Mechanisms of Differently Colored Resistance Quinoa Seedings Using Metabolite Profiling and Transcriptome Analysis. Metabolites, 13(10), 1065. https://doi.org/10.3390/metabo13101065





