Co-Localization of Resistance and Metabolic Quantitative Trait Loci on Carrot Genome Reveals Fungitoxic Terpenes and Related Candidate Genes Associated with the Resistance to Alternaria dauci
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Design and Crop Management
2.3. Sampling Design
2.4. Headspace Solid-Phase Microextraction Followed by Gas Chromatography–Mass Spectrometry (HS-SPME-GC-MS) for Terpene Analyses
2.5. Correlation between Metabolite Accumulation and Disease Score
2.6. Resistance and Metabolite-QTL Detection
2.7. Transcriptomic Analysis
2.8. Statistical Analysis and Highlighting of Differentially Expressed Genes within the Co-Localization Area
2.9. Fungal Growth Inhibition Assays
3. Results
3.1. Consistency of rQTL among Years
3.2. mQTL-rQTL Co-Localization Analysis Reveals Candidate Terpenes for Resistance to ALB
3.3. Differential Expression Analysis of Genes Underlying the Co-Localization Regions
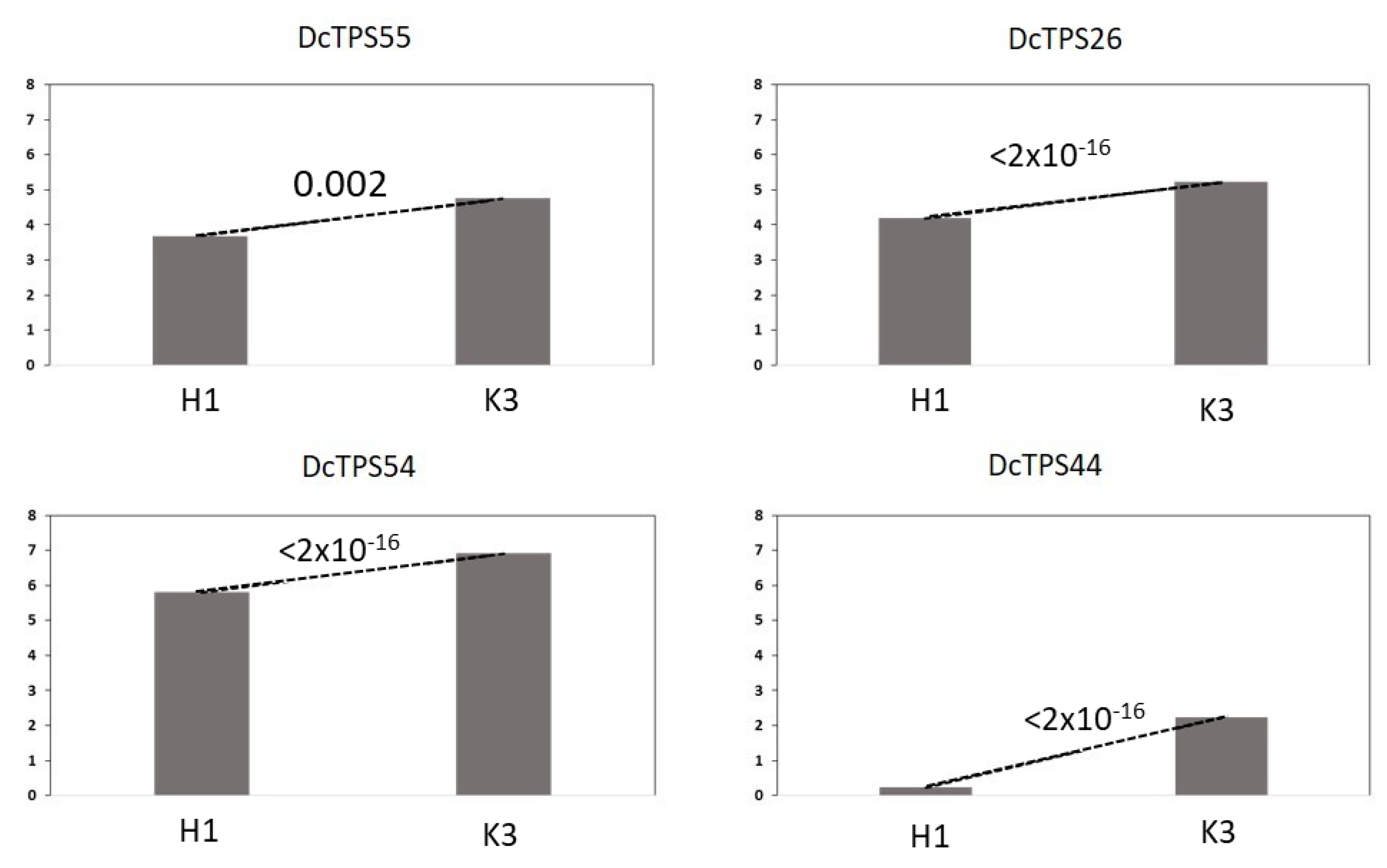
3.4. Further Analysis of TPS Genes Overexpressed in K3
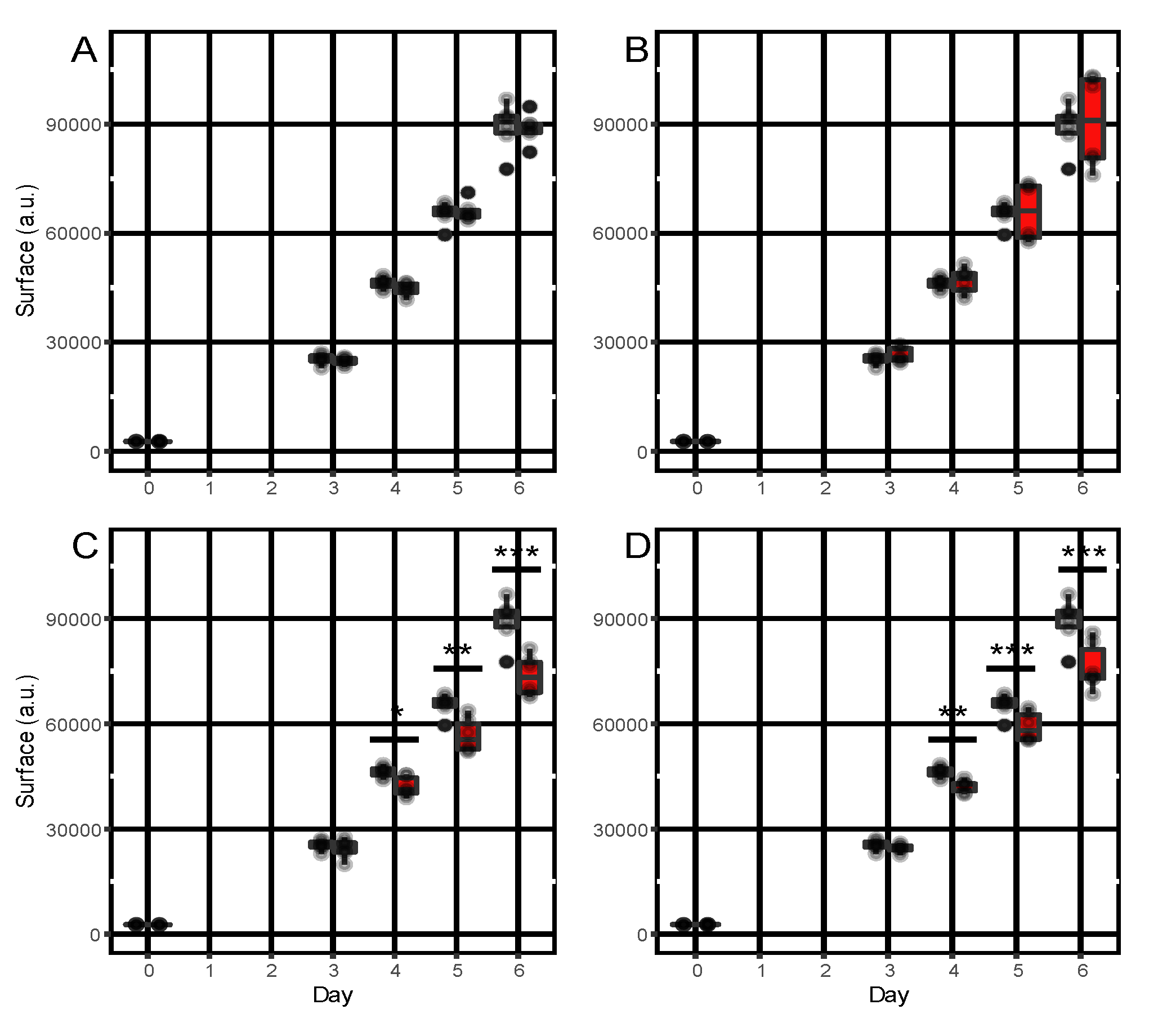
3.5. In Vitro Bioactivity of α-Pinene, Camphene, Caryophyllene and Humulene towards A. dauci
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Biotechnol. Isoprenoids 2015, 148, 63–106. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Yeh, S. Isoprene emission from plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 407–436. [Google Scholar] [CrossRef]
- Pichersky, E.; Gershenzon, J. The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 2002, 5, 237–243. [Google Scholar] [CrossRef]
- Gil, M.; Pontin, M.; Berli, F.; Bottini, R.; Piccoli, P. Metabolism of terpenes in the response of grape (Vitis vinifera L.) leaf tissues to UV-B radiation. Phytochemistry 2012, 77, 89–98. [Google Scholar] [CrossRef]
- Rodríguez, A.; Andrés, V.S.; Cervera, M.; Redondo, A.; Alquézar, B.; Shimada, T.; Gadea, J.; Rodrigo, M.; Zacarías, L.; Palou, L.; et al. The monoterpene limonene in orange peels attracts pests and microorganisms. Plant Signal. Behav. 2011, 6, 1820–1823. [Google Scholar] [CrossRef]
- Pontin, M.; Bottini, R.; Burba, J.L.; Piccoli, P. Allium sativum produces terpenes with fungistatic properties in response to infection with Sclerotium cepivorum. Phytochemistry 2015, 115, 152–160. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Yu, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 2017, 246, 803–816. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Verdonk, J.C.; Haring, M.A.; Van Tunen, A.J.; Schuurink, R.C. ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 2005, 17, 1612–1624. [Google Scholar] [CrossRef]
- Picazo-Aragonés, J.; Terrab, A.; Balao, F. Plant volatile organic compounds evolution: Transcriptional regulation, epigenetics and polyploidy. Int. J. Mol. Sci. 2020, 21, 8956. [Google Scholar] [CrossRef]
- Farrar, J.J.; Pryor, B.M.; Davis, R.M. Alternaria diseases of carrot. Plant Dis. 2004, 88, 776–784. [Google Scholar] [CrossRef]
- Le Clerc, V.; Pawelec, A.; Birolleau-Touchard, C.; Suel, A.; Briard, M. Genetic architecture of factors underlying partial resistance to Alternaria leaf blight in carrot. Theor. Appl. Genet. 2009, 118, 1251–1259. [Google Scholar] [CrossRef]
- Le Clerc, V.; Marques, S.; Suel, A.; Huet, S.; Hamama, L.; Voisine, L.; Auperpin, E.; Jourdan, M.; Barrot, L.; Prieur, R.; et al. QTL mapping of carrot resistance to leaf blight with connected populations: Stability across years and consequences for breeding. Theor. Appl. Genet. 2015, 128, 2177–2187. [Google Scholar] [CrossRef]
- Koutouan, C.E.; Le Clerc, V.; Baltenweck, R.; Claudel, P.; Halter, D.; Hugueney, P.; Hamama, L.; Suel, A.; Huet, S.; Bouvet Merlet, M.H.; et al. Link between carrot leaf secondary metabolites and resistance to Alternaria dauci. Sci. Rep. 2018, 8, 13746. [Google Scholar] [CrossRef]
- Pawelec, A.; Dubourg, C.; Briard, M. Evaluation of carrot resistance to Alternaria leaf blight in controlled environments. Plant Pathol. 2006, 55, 68–72. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 1–15. Available online: http://www.biomedcentral.com/1471-2164/7/142 (accessed on 7 December 2022). [CrossRef]
- Le Clerc, V.; Aubert, C.; Cottet, V.; Yovanopoulos, C.; Piquet, M.; Suel, A.; Huet, S.; Koutouan, C.E.; Hamama, L.; Chalot, G.; et al. Breeding for carrot resistance to Alternaria dauci without compromising taste. Mol. Breed. 2019, 39, 59. [Google Scholar] [CrossRef]
- Jourjon, M.F.; Jasson, S.; Marcel, J.; Ngom, B.; Mangin, B. MCQTL: Multi-allelic QTL mapping in multi-cross design. Bioinformatics 2005, 21, 128–130. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Lovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Gagnot, S.; Tamby, J.P.; Martin-Magniette, M.L.; Bitton, F.; Taconnat, L.; Balzergue, S.; Aubourg, S.; Renou, J.P.; Lecharny, A.; Brunaud, V. CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 2007, 36, 986–990. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Celton, J.M.; Gaillard, S.; Bruneau, M.; Pelletier, S.; Aubourg, S.; Martin-Magniette, M.L.; Navarro, L.; Laurens, F.; Renou, J.P. Widespread anti-sense transcription in apple is correlated with si RNA production and indicates a large potential for transcriptional and/or post-transcriptional control. New Phytol. 2014, 203, 287–299. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Copper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Boedo, C.; Benichou, S.; Berruyer, R.; Bersihand, S.; Dongo, A.; Simoneau, P.; Lecomte, M.; Briard, M.; Le Clerc, V.; Poupard, P. Evaluating aggressiveness and host range of Alternaria dauci in a controlled environment. Plant Pathology 2012, 61, 63–75. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Keilwagen, J.; Lehnert, H.; Berner, T.; Budahn, H.; Nothnagel, T.; Ulrich, D.; Dunemann, F. The terpene synthase gene family of carrot (Daucus carota L.): Identification of QTLs and candidate genes associated with terpenoid volatile compounds. Front. Plant Sci. 2017, 8, 1930. [Google Scholar] [CrossRef]
- Muchlinski, A.; Ibdah, M.; Ellison, S.; Yahyaa, M.; Nawade, B.; Laliberte, S.; Senalik, D.; Simon, P.; Whitehead, S.R.; Tholl, D. Diversity and function of terpene synthases in the production of carrot aroma and flavor compounds. Sci. Rep. 2020, 10, 9989. [Google Scholar] [CrossRef]
- Reichardt, S.; Budahn, H.; Lamprecht, D.; Riewe, D.; Ulrich, D.; Dunemann, F.; Kopertekh, L. The carrot monoterpene synthase gene cluster on chromosome 4 harbours genes encoding flavour-associated sabinene synthases. Hortic. Res. 2020, 7, 190. [Google Scholar] [CrossRef]
- Kearsey, M.J. The principles of QTL analysis (a minimal mathematics approach). J. Exp. Bot. 1998, 49, 1619–1623. [Google Scholar] [CrossRef]
- Keurentjes, J.J.; Fu, J.; De Vos, C.H.; Lommen, A.; Hall, R.D.; Bino, R.J.; van der Plas, L.H.W.; Jansen, R.C.; Vreugdenhil, D.; Koornneef, M. The genetics of plant metabolism. Nat. Genet. 2006, 38, 842–849. [Google Scholar] [CrossRef]
- Rowe, H.C.; Kliebenstein, D.J. Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 2008, 180, 2237–2250. [Google Scholar] [CrossRef]
- Zhang, W.; Kwon, S.T.; Chen, F.; Kliebenstein, D.J. Isolate dependency of Brassica rapa resistance QTLs to Botrytis cinerea. Front. Plant Sci. 2016, 7, 161. [Google Scholar] [CrossRef][Green Version]
- Gambliel, H.; Croteau, R. Pinene cyclases I and II. Two enzymes from sage (Salvia officinalis) which catalyze stereospecific cyclizations of geranyl pyrophosphate to monoterpene olefins of opposite configuration. J. Biol. Chem. 1984, 259, 740–748. [Google Scholar] [CrossRef]
- Huber, D.P.; Philippe, R.N.; Godard, K.A.; Sturrock, R.N.; Bohlmann, J. Characterization of four terpene synthase cDNAs from methyl jasmonate-induced Douglas-fir, Pseudotsuga menziesii. Phytochemistry 2005, 66, 1427–1439. [Google Scholar] [CrossRef]
- Matsuba, Y.; Nguyen, T.T.; Wiegert, K.; Falara, V.; Gonzales-Vigil, E.; Leong, B.; Schäfer, P.; Kudrna, D.; Wing, R.A.; Bolger, A.M.; et al. Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 2013, 25, 2022–2036. [Google Scholar] [CrossRef]
- Nützmann, H.W.; Osbourn, A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 2014, 26, 91–99. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of Specialized Metabolism by WRKY Transcription Factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef]
- Spyropoulou, E.A.; Haring, M.A.; Schuurink, R.C. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 2014, 15, 402. Available online: http://www.biomedcentral.com/1471-2164/15/402 (accessed on 7 December 2022). [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; San Andrés, V.; Cervera, M.; Redondo, A.; Alquézar, B.; Shimada, T.; Gadea, J.; Rodrigo, M.J.; Zacarias, L.; Palou, L.; et al. Terpene down-regulation in orange reveals the role of fruit aromas in mediating interactions with insect herbivores and pathogens. Plant Physiol. 2011, 156, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Shimada, T.; Cervera, M.; Alquézar, B.; Gadea, J.; Gómez-Cadenas, A.; José De Ollas, C.; Rodrigo, M.J.; Zacarias, L.; Peña, L. Terpene down-regulation triggers defense responses in transgenic orange leading to resistance against fungal pathogens. Plant Physiol. 2014, 164, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Sun, T.H.; Zhao, L.; Pan, X.W.; Lu, S. The bZIP transcription factor HY5 interacts with the promoter of the monoterpene synthase gene QH6 in modulating its rhythmic expression. Front. Plant Sci. 2015, 6, 304. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuizen, N.J.; Chen, X.; Wang, M.Y.; Matich, A.J.; Perez, R.L.; Allan, A.C.; Green, S.A.; Atkinson, R.G. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription. Plant Physiol. 2015, 167, 1243–1258. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Shen, S.; Yin, X.; Klee, H.; Zhang, B.; Chen, K.; Hancock, R. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017, 68, 4929–4938. [Google Scholar] [CrossRef]
- Yu, Z.X.; Wang, L.J.; Zhao, B.; Shan, C.M.; Zhang, Y.H.; Chen, D.F.; Chen, X.Y. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol. Plant 2015, 8, 98–110. [Google Scholar] [CrossRef]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. Antifungal activity of the components of Melaleuca alternifolia (tea tree) oil. J. Appl. Microbiol. 2003, 95, 853–860. [Google Scholar] [CrossRef]
- Sati, S.C.; Sati, N.; Ahluwalia, V.; Walia, S.; Sati, O.P. Chemical composition and antifungal activity of Artemisia nilagirica essential oil growing in northern hilly areas of India. Nat. Prod. Res. 2013, 27, 45–48. [Google Scholar] [CrossRef]
- Dahham, S.S.; Tabana, Y.M.; Iqbal, M.A.; Ahamed, M.B.; Ezzat, M.O.; Majid, A.S.; Majid, A.M. The anticancer, antioxidant and antimicrobial properties of the sesquiterpene β-caryophyllene from the essential oil of Aquilaria crassna. Molecules 2015, 20, 11808–11829. [Google Scholar] [CrossRef]
- Garcia-Rellán, D.; Verdeguer, M.; Salamone, A.; Blázquez, M.A.; Boira, H. Chemical composition, herbicidal and antifungal activity of Satureja cuneifolia essential oils from Spain. Nat. Prod. Commun. 2016, 11, 1934578X1601100636. [Google Scholar] [CrossRef]
- Feiner, A.; Pitra, N.; Matthews, P.; Pillen, K.; Wessjohann, L.A.; Riewe, D. Downy mildew resistance is genetically mediated by prophylactic production of phenylpropanoids in hop. Plant Cell Environ. 2021, 44, 323–338. [Google Scholar] [CrossRef]
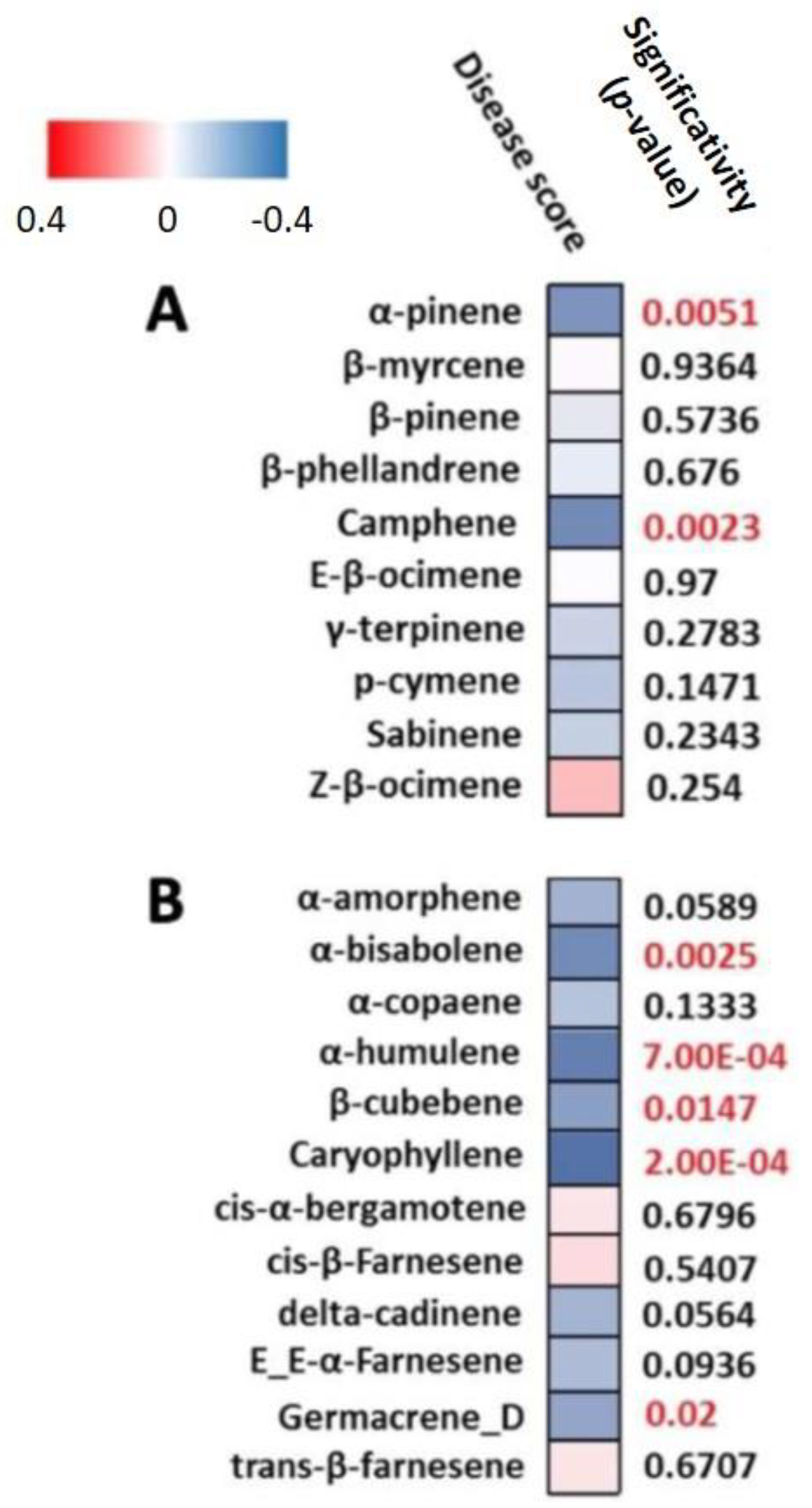
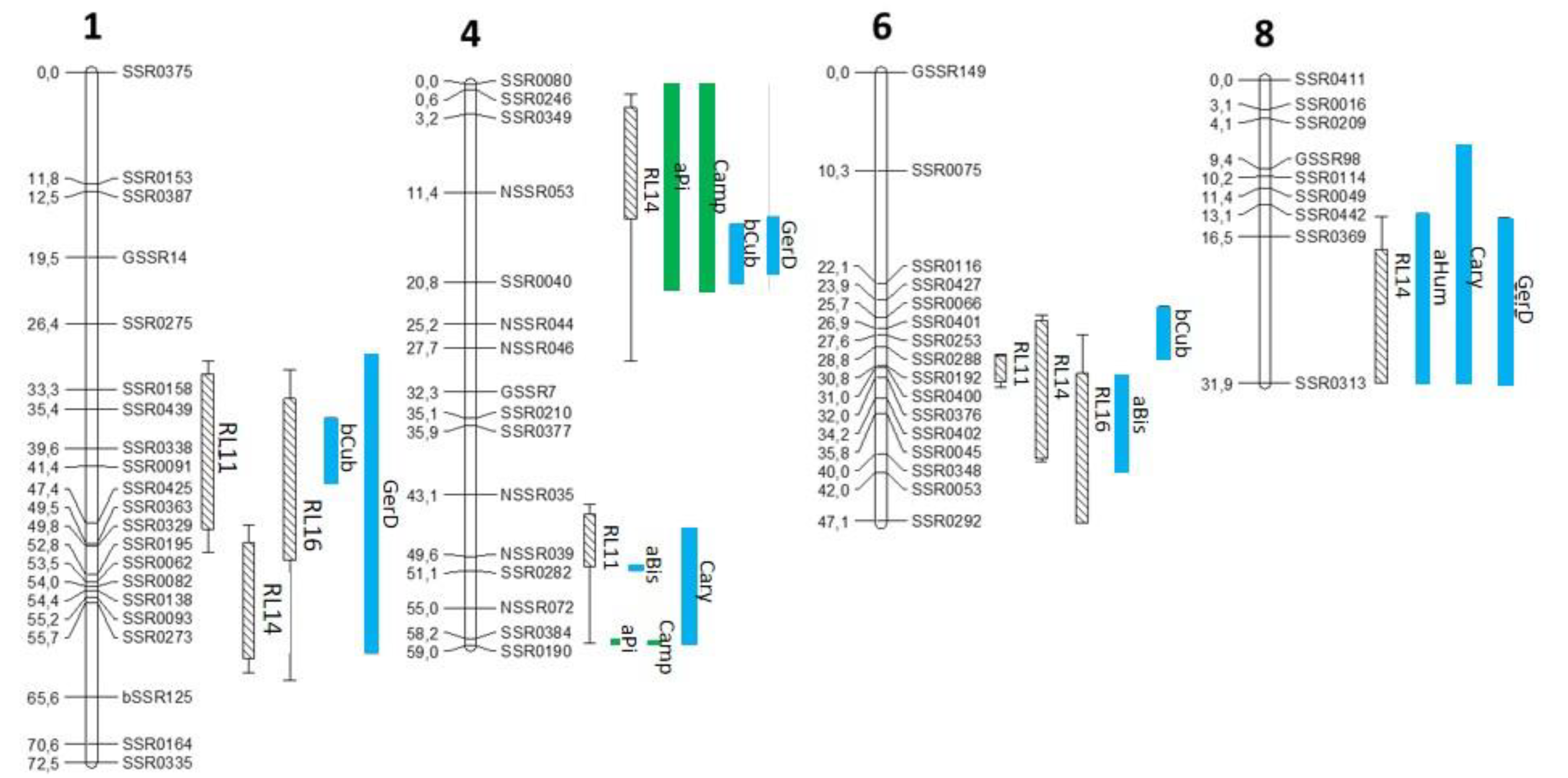
| Code of mQTL | Chr | 1-LOD SI (cM) | Max Position (cM) | R2 (%) | Global R2 (%) | Heritability H2 (%) | Additive Effect of Allele | |
|---|---|---|---|---|---|---|---|---|
| H1 | K3 | |||||||
| aPi | 4 t | 0–21.6 | 20.4 | 11.4 | 58.3 | 68 | −0.157 | 0.157 |
| 4 b | 58.3–58.7 | 58.7 | 42.9 | −0.388 | 0.388 | |||
| Camp | 4 t | 0–21.7 | 20.4 | 10.7 | 57.6 | 65 | −0.153 | 0.153 |
| 4 b | 58.4–58.7 | 58.7 | 42.5 | −0.389 | 0.389 | |||
| aBis | 4 b | 50.6–51 | 50.8 | 51.7 | 53.1 | 34 | −0.436 | 0.436 |
| 6 | 31.8–41.9 | 40.2 | 9.7 | −0.140 | 0.140 | |||
| aHum | 8 | 14.1–31.9 | 16.4 | 12.2 | 12.2 | 39 | 0.209 | −0.209 |
| bCub | 1 | 36.3–43.2 | 39.6 | 20.5 | 41.7 | 31 | −0.263 | 0.263 |
| 4 t | 14.7–20.8 | 16.4 | 26.9 | −0.273 | 0.273 | |||
| 6 | 24.6–30.1 | 28.9 | 12.6 | −0.167 | 0.167 | |||
| Cary | 4 b | 46.6–58.7 | 58.7 | 9.3 | 18.6 | 33.3 | −0.161 | 0.161 |
| 8 | 6.9–31.9 | 11.3 | 9.1 | 0.174 | −0.174 | |||
| GerD | 1 | 29.7–61 | 39.6 | 10 | 46 | 36.2 | −0.173 | 0.173 |
| 4 t | 13.9–19.8 | 16.4 | 26.4 | −0.271 | 0.271 | |||
| 8 | 14.5–31.9 | 21.4 | 18.7 | 0.235 | −0.235 | |||
| Chr | Located Genes | Genes Differentially Expressed between K3 and H1 | ||
|---|---|---|---|---|
| Underexpressed | Overexpressed | Related to Terpenes | ||
| 1 | 2181 | 207 | 206 | 5 |
| 4 t | 1229 | 76 | 92 | 2 |
| 4 b | 1718 | 154 | 196 | 10 |
| 6 | 577 | 63 | 45 | 3 |
| 8 | 1025 | 100 | 105 | 5 |
| Chr | Gene Name (Locus Number) | Function | Relative Expression | p-Value | |
|---|---|---|---|---|---|
| H1 | K3 | ||||
| 1 | bZIPHY5 (108204232) | TF | 3.94 | 4.48 | 0.004 |
| WRKY33 (108204915) | TF | 2.65 | 4.64 | <2 × 10−16 | |
| WRKY48 (108204668) | TF | 2.67 | 3.79 | <2 × 10−16 | |
| ERF71 (108206243) | TF | 0.98 | 0.15 | 0.0004 | |
| bZIP53 (108206338) | TF | 3.28 | 2.23 | <2 × 10−16 | |
| 4 t | NAC29 (108218926) | TF | 4.44 | 5.90 | 0.0004 |
| WRKY33 (108219317) | TF | 4.00 | 6.21 | <2 × 10−16 | |
| 4 b | Terpene synthase 10-like (DcTPS55;108217470) | TPS | 3.68 | 4.76 | 0.002 |
| Terpene synthase 10-like (DcTPS26;108217599) | TPS | 4.19 | 5.22 | <2 × 10−16 | |
| Terpene synthase 10-like (DcTPS54;108217617) | TPS | 5.80 | 6.92 | <2 × 10−16 | |
| β-bisabolene synthase-like (108216085) | TPS | 6.67 | 5.67 | 0.0004 | |
| WRKY7 (108215789) | TF | 5.39 | 4.74 | 0.0008 | |
| ERF054 (108216387) | TF | 3.45 | 2.1 | <2 × 10−16 | |
| NAC2 (108215781) | TF | 4.51 | 5.26 | 0.0004 | |
| ERF4 (108217832) | TF | 1.87 | 2.87 | <2 × 10−16 | |
| bZIP17 (108218833) | TF | 3.98 | 4.76 | 0.0004 | |
| bZIP27 (108217633) | TF | 3.60 | 0.51 | <2 × 10−16 | |
| 6 | bZIP61 (108225065) | TF | 4.28 | 4.89 | 0.007 |
| ERF4 (108225207) | TF | 0.82 | 3.29 | 0.002 | |
| SPL1 (108224238) | TF | 3.98 | 2.84 | <2 × 10−16 | |
| 8 | α-farnesene synthase-like(DcTPS44; 108198720) | TPS | 0.23 | 2.23 | <2 × 10−16 |
| AP2/ERF (108198780) | TF | 0.92 | 5.22 | <2 × 10−16 | |
| ERF1B-like (108198802) | TF | 1.35 | 3.21 | <2 × 10−16 | |
| MYB (108197621) | TF | 0.74 | 0.19 | 0.005 | |
| ERF_like (108197006) | TF | 4.38 | 2.66 | <2 × 10−16 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koutouan, C.E.; Le Clerc, V.; Suel, A.; Hamama, L.; Claudel, P.; Halter, D.; Baltenweck, R.; Hugueney, P.; Chich, J.-F.; Moussa, S.A.; et al. Co-Localization of Resistance and Metabolic Quantitative Trait Loci on Carrot Genome Reveals Fungitoxic Terpenes and Related Candidate Genes Associated with the Resistance to Alternaria dauci. Metabolites 2023, 13, 71. https://doi.org/10.3390/metabo13010071
Koutouan CE, Le Clerc V, Suel A, Hamama L, Claudel P, Halter D, Baltenweck R, Hugueney P, Chich J-F, Moussa SA, et al. Co-Localization of Resistance and Metabolic Quantitative Trait Loci on Carrot Genome Reveals Fungitoxic Terpenes and Related Candidate Genes Associated with the Resistance to Alternaria dauci. Metabolites. 2023; 13(1):71. https://doi.org/10.3390/metabo13010071
Chicago/Turabian StyleKoutouan, Claude Emmanuel, Valérie Le Clerc, Anita Suel, Latifa Hamama, Patricia Claudel, David Halter, Raymonde Baltenweck, Philippe Hugueney, Jean-François Chich, Sitti Anlati Moussa, and et al. 2023. "Co-Localization of Resistance and Metabolic Quantitative Trait Loci on Carrot Genome Reveals Fungitoxic Terpenes and Related Candidate Genes Associated with the Resistance to Alternaria dauci" Metabolites 13, no. 1: 71. https://doi.org/10.3390/metabo13010071
APA StyleKoutouan, C. E., Le Clerc, V., Suel, A., Hamama, L., Claudel, P., Halter, D., Baltenweck, R., Hugueney, P., Chich, J.-F., Moussa, S. A., Champlain, C., Huet, S., Voisine, L., Pelletier, S., Balzergue, S., Chevalier, W., Geoffriau, E., & Briard, M. (2023). Co-Localization of Resistance and Metabolic Quantitative Trait Loci on Carrot Genome Reveals Fungitoxic Terpenes and Related Candidate Genes Associated with the Resistance to Alternaria dauci. Metabolites, 13(1), 71. https://doi.org/10.3390/metabo13010071








