Regulation Mechanism of Plant Pigments Biosynthesis: Anthocyanins, Carotenoids, and Betalains
Abstract
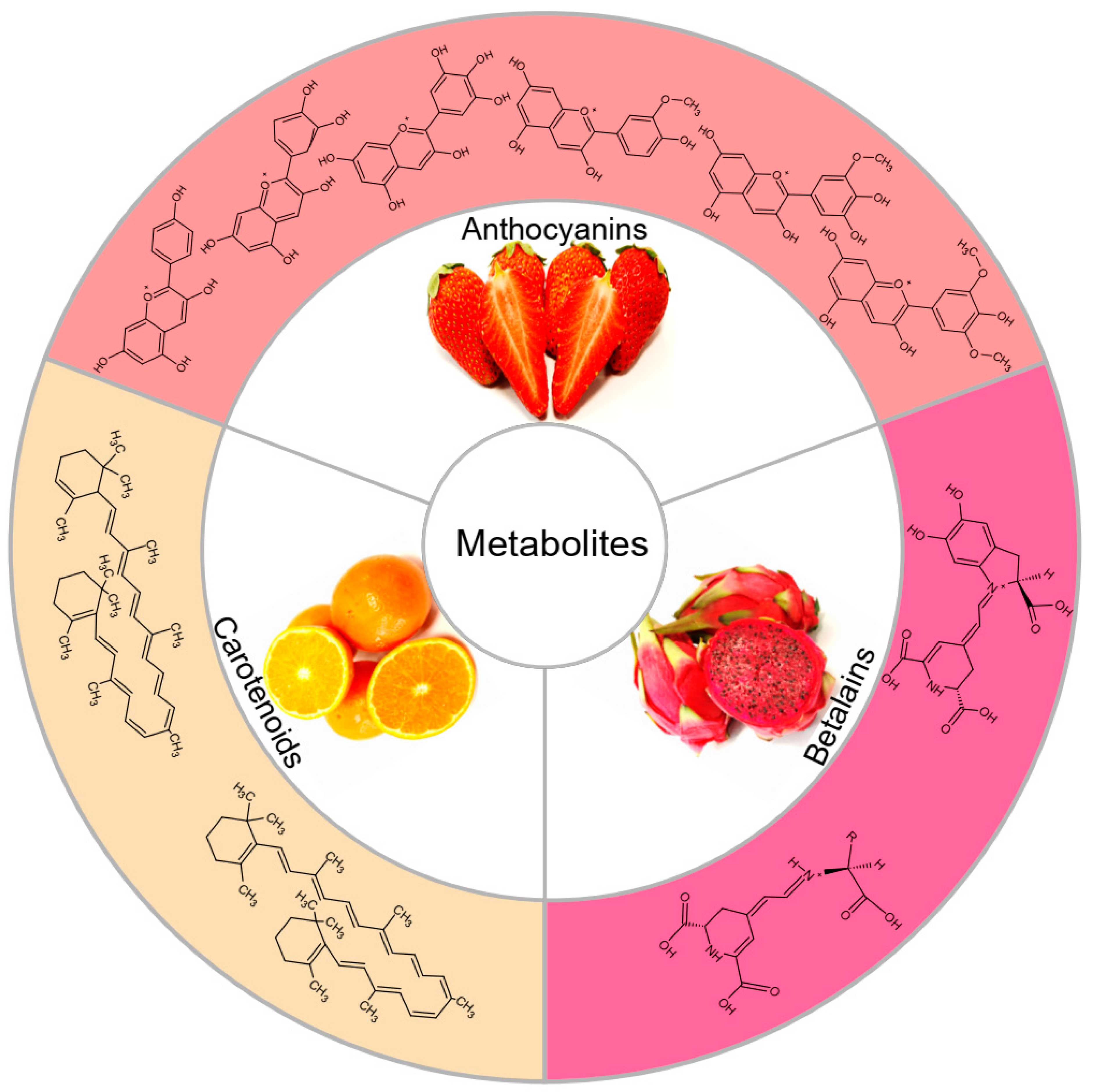
1. Anthocyanins
2. Biosynthesis of Anthocyanins
3. Transcriptional Regulation of the Anthocyanin Biosynthesis
4. Developmental Regulation of Anthocyanin Biosynthesis
5. Light and Temperature Regulation Anthocyanin Biosynthesis
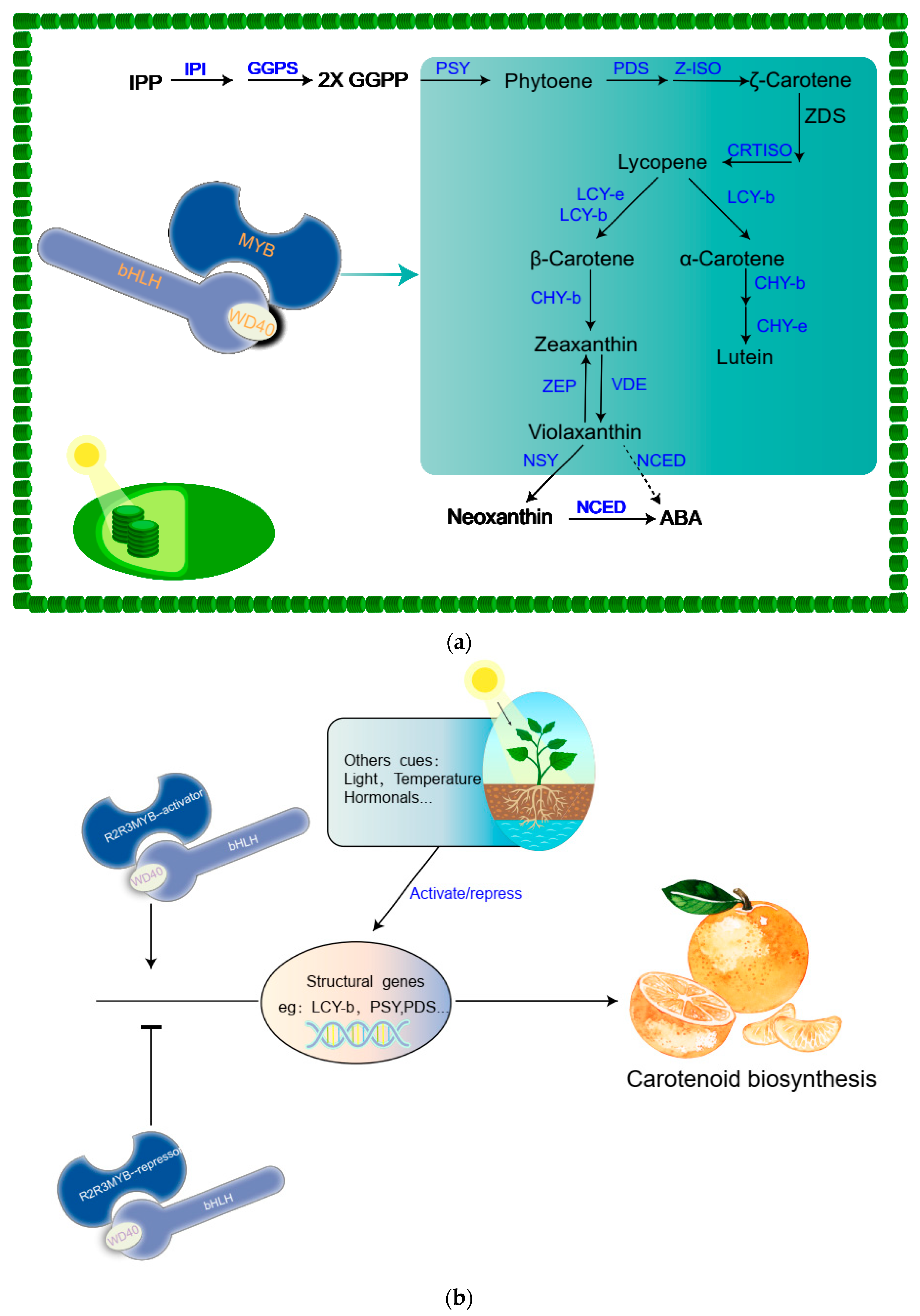
6. Carotenoids
7. Carotenoid Biosynthesis Pathway
8. Transcriptional Regulation of the Carotenoid Biosynthesis
9. Others Cues That Regulate Carotenoid Biosynthesis
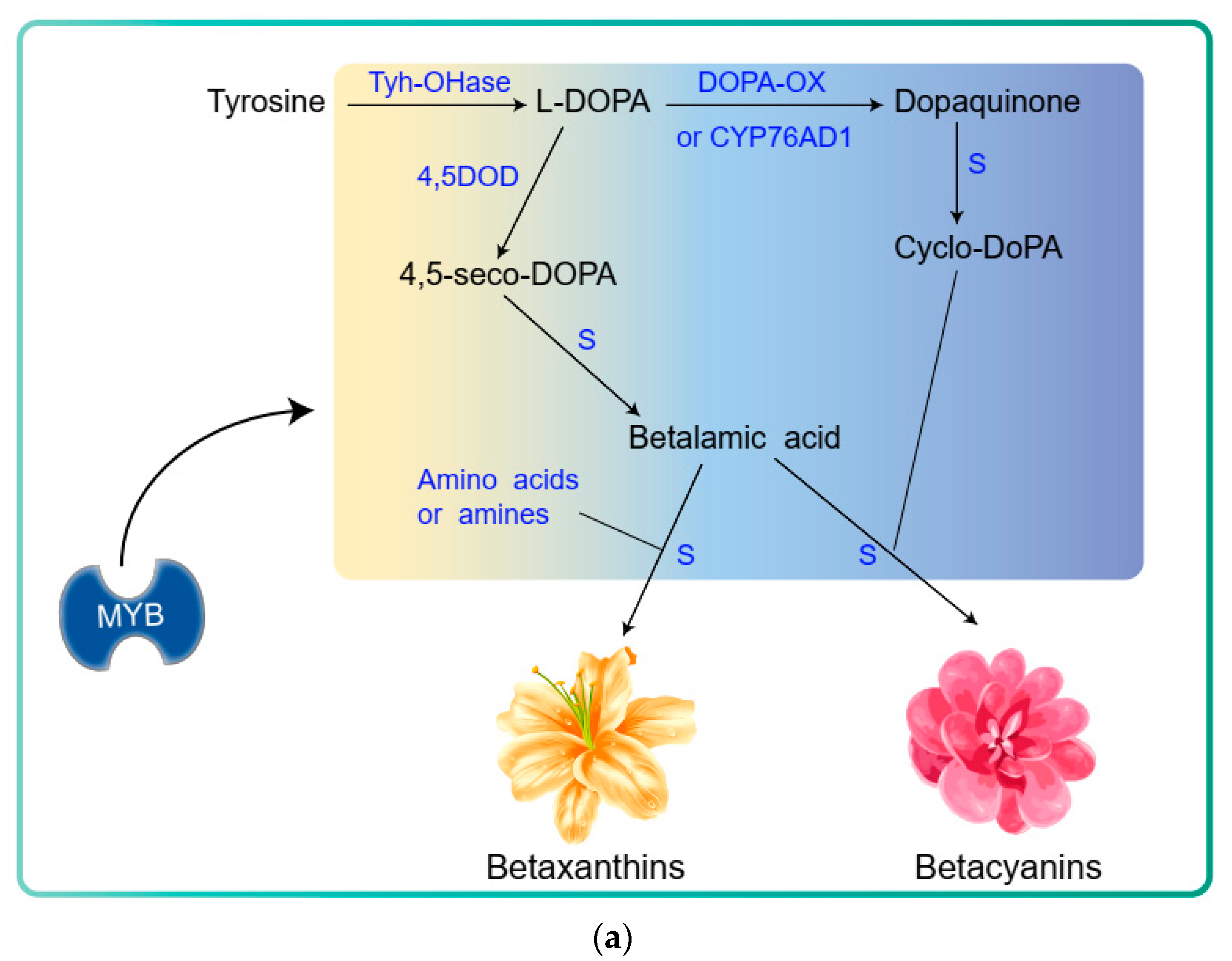
10. Betalains
11. Betalain Biosynthesis Pathway
12. Transcriptional Regulation of the Betalain Biosynthesis
13. How to Interpret Mutual Exclusion of Betalains and Anthocyanins?
14. Metabolic Engineering of Pigment Content in Plants or Microbes
15. Concluding Remarks and Future Directions
- Anthocyanins and carotenoids are regulated by MBW complexes but betalains are not regulated by MBW complexes. MYB (BvMYB1) does not interact with bHLH. Is this mechanism universal or special in the betalain pathway?
- Does BvMYB1 interact with an unknown bHLH protein?
- BvMYB1, AtPAP1, and AtMYB114 all belong to the subgroup 6 clade but BvMYB1 does not regulate anthocyanin biosynthesis. Does this result indicate a specific selection for evolution?
- WP1 associates with MtTT8, and MtWD40-1 regulates both carotenoid and anthocyanin biosynthesis. However, RCP1 positively regulates carotenoid biosynthesis and decreases anthocyanin production. How should this result be interpreted?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chaves-Silva, S.; Santos, A.L.D.; Chalfun-Júnior, A.; Zhao, J.; Peres, L.E.P.; Benedito, V.A. Understanding the genetic regulation of anthocyanin biosynthesis in plants—Tools for breeding purple varieties of fruits and vegetables. Phytochemistry 2018, 153, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Pfeiffer, A.; Sivilotti, P.; Degan, M.; Peterlunger, E.; Gaspero, G.D. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007, 30, 1381–1399. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, J.; Grotewold, E.; Ferreyra, M.L.F.; Casati, P. Flavonols protect Arabidopsis plants against UV-B deleterious effects. Mol. Plant. 2013, 6, 1376–1379. [Google Scholar] [CrossRef]
- Saslowsky, D.E.; Warek, U.; Winkel, B.S. Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 2005, 280, 23735–23740. [Google Scholar] [CrossRef]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Ruan, H.; Shi, X.; Gao, L.; Rashid, A.; Li, Y.; Lei, T.; Dai, X.; Xia, T.; Wang, Y. Functional analysis of the dihydroflavonol 4-reductase family of Camellia sinensis: Exploiting key amino acids to reconstruct reduction activity. Hortic. Res. 2022, 9, uhac098. [Google Scholar] [CrossRef]
- Sasaki, N.; Nishizaki, Y.; Ozeki, Y.; Miyahara, T. The role of acyl-glucose in anthocyanin modifications. Molecules 2014, 19, 18747–18766. [Google Scholar] [CrossRef]
- Zhao, X.; Dai, X.; Gao, L.; Guo, L.; Zhuang, J.; Liu, Y.; Ma, X.; Wang, R.; Xia, T.; Wang, Y. Functional Analysis of an Uridine Diphosphate Glycosyltransferase Involved in the Biosynthesis of Polyphenolic Glucoside in Tea Plants (Camellia sinensis). J Agric. Food Chem. 2017, 65, 10993–11001. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3′O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Dixon, R.A.; Liu, C.; Jun, J.H. Metabolic engineering of anthocyanins and condensed tannins in plants. Curr. Opin. Biotechnol. 2013, 24, 329–335. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.M.; Robinson, S.P.; Barrieu, F.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef]
- Hu, D.G.; Sun, C.H.; Ma, Q.J.; You, C.X.; Cheng, L.; Hao, Y.J. MdMYB1 Regulates Anthocyanin and Malate Accumulation by Directly Facilitating Their Transport into Vacuoles in Apples. Plant Physiol. 2016, 170, 1315–1330. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Wang, L.; Zhang, Y.; Ruan, L.; Li, H.; Wu, L.; Xu, L.; Zhang, C.; Zhou, X.; Cheng, H.; et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019, 97, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Le, G.J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Jun, J.H.; Liu, C.; Xiao, X.; Dixon, R.A. The Transcriptional Repressor MYB2 Regulates Both Spatial and Temporal Patterns of Proanthocyandin and Anthocyanin Pigmentation in Medicago truncatula. Plant Cell 2015, 27, 2860–2879. [Google Scholar]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef]
- Wang, C.K.; Han, P.L.; Zhao, Y.W.; Ji, X.L.; Yu, J.Q.; You, C.X.; Hu, D.G.; Hao, Y.J. Auxin regulates anthocyanin biosynthesis through the auxin repressor protein MdIAA26. Biochem. Biophys. Res. Commun. 2020, 533, 717–722. [Google Scholar] [CrossRef]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/bHLH/WD40 Complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shu, Q.; Lin-Wang, K.; Andrew, C.A.; Richard, V.; Espley, R.V. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1, 14. [Google Scholar] [CrossRef]
- Vale, M.; Rodrigues, J.; Badim, H.; Gerós, H.; Conde, A. Exogenous Application of Non-mature miRNA-Encoded miPEP164c Inhibits Proanthocyanidin Synthesis and Stimulates Anthocyanin Accumulation in Grape Berry Cells. Front. Plant Sci. 2021, 12, 706679. [Google Scholar] [CrossRef]
- Jia, Y.; Selva, C.; Zhang, Y.; Li, B.; McFawn, L.A.; Broughton, S.; Zhang, X.; Westcott, S.; Wang, P.; Tan, C.; et al. Uncovering the evolutionary origin of blue anthocyanins in cereal grains. Plant J. 2020, 101, 1057–1074. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, N.; Chen, M.; Zhang, R.; Sun, Q.; Xu, H.; Zhang, Z.; Wang, Y.; Sui, X.; Wang, S.; et al. Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol. J. 2020, 18, 1736–1748. [Google Scholar] [CrossRef]
- Butelli, E.; Garcia-Lor, A.; Licciardello, C.; Las Casas, G.; Hill, L.; Recupero, G.R.; Keremane, M.L.; Ramadugu, C.; Krueger, R.; Xu, Q.; et al. Changes in Anthocyanin Production during Domestication of Citrus. Plant Physiol. 2017, 173, 2225–2242. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Ramadugu, C.; Durand-Hulak, M.; Celant, A.; Recupero, G.R.; Froelicher, Y.; Martin, C. Noemi Controls Production of Flavonoid Pigments and Fruit Acidity and Illustrates the Domestication Routes of Modern Citrus Varieties. Curr. Biol. 2019, 29, 158–164.e2. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-Box Proteins Regulate Photomorphogenesis by Oppositely Modulating HY5 through their Diverse C-Terminal Domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin-Wang, K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An Apple B-Box Protein MdBBX37 Modulates Anthocyanin Biosynthesis and Hypocotyl Elongation Synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Wang, X.; Gao, J.; Guo, Y.; Huang, Z.; Du, Y. The Tomato Hoffman’s Anthocyaninless Gene Encodes a bHLH Transcription Factor Involved in Anthocyanin Biosynthesis That Is Developmentally Regulated and Induced by Low Temperatures. PLoS ONE 2016, 11, e0151067. [Google Scholar] [CrossRef] [PubMed]
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Otegui, M.S.; Grotewold, E. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal Behav. 2015, 10, e1027850. [Google Scholar] [CrossRef]
- Proveniers, M.C.; Van Zanten, M. High temperature acclimation through PIF4 signaling. Trends Plant Sci. 2013, 18, 59–64. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.; Fang, H.C.; Zhang, T.T. Nitrogen Affects Anthocyanin Biosynthesis by Regulating MdLOB52 Downstream of MdARF19 in Callus Cultures of Red-Fleshed Apple (Malus sieversii f. niedzwetzkyana). J. Plant Growth Regul. 2018, 37, 719–729. [Google Scholar] [CrossRef]
- He, Y.; Zhang, X.; Li, L.; Sun, Z.; Li, J.; Chen, X.; Hong, G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2021, 230, 205–217. [Google Scholar] [CrossRef]
- Delgado-Vargas, F.; Jiménez, A.R.; Paredes-López, O. Natural pigments: Carotenoids, anthocyanins, and betalains--characteristics, biosynthesis, processing, and stability. Crit. Rev. Food Sci. Nutr. 2000, 40, 173–289. [Google Scholar] [CrossRef]
- Sun, T.; Tadmor, Y.; Li, L. Pathways for Carotenoid Biosynthesis, Degradation, and Storage. Methods Mol. Biol. 2020, 2083, 3–23. [Google Scholar]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenase and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef]
- Rapoport, A.; Guzhova, I.; Bernetti, L.; Buzzini, P.; Kieliszek, M.; Kot, A.M. Carotenoids and Some Other Pigments from Fungi and Yeasts. Metabolites 2021, 11, 92. [Google Scholar] [CrossRef]
- Rosas-Saavedra, C.; Stange, C. Biosynthesis of Carotenoids in Plants: Enzymes and Color. Subcell. Biochem. 2016, 79, 35–69. [Google Scholar] [PubMed]
- Davies, K.M. (Ed.) Plant Pigments and Their Manipulation; Blackwell Publishing: Hoboken, NJ, USA, 2009; Volume 14, Annual Plant Reviews; p. 69. [Google Scholar]
- Ronen, G.; Carmel-Goren, L.; Zamir, D.; Hirschberg, J. An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA. 2000, 97, 11102–11107. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, G. Provitamin A biofortification of crop plants: A gold rush with many miners. Curr. Opin. Biotechnol. 2017, 44, 169–180. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef]
- Fray, R.G.; Grierson, D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 1993, 22, 589–602. [Google Scholar] [CrossRef]
- Fraser, P.D.; Romer, S.; Shipton, C.A.; Mills, P.B.; Kiano, J.W.; Misawa, N.; Drake, R.G.; Schuch, W.; Bramley, P.M. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. USA 2002, 99, 1092–1097. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Shen, Y.; Zhou, P.; Fatima, M.; Lin, J.; Yue, J.; Zhang, X.; Chen, L.Y.; Ming, R. Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res. 2019, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef]
- Wu, M.; Xu, X.; Hu, X.; Liu, Y.; Cao, H.; Chan, H.; Gong, Z.; Yuan, Y.; Luo, Y.; Feng, B.; et al. SlMYB72 Regulates the Metabolism of Chlorophylls, Carotenoids, and Flavonoids in Tomato Fruit. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Yang, H.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.; Xu, R.; et al. An R2R3-MYB transcription factor represses the transformation of alpha- and beta-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, Z.; Wang, Y.; Wang, C.; Zhu, B.; Liu, H.; Ji, W.; Wen, J.; Chu, C.; Tadege, M.; et al. The MYB Activator WHITE PETAL1 Associates with MtTT8 and MtWD40-1 to Regulate Carotenoid-Derived Flower Pigmentation in Medicago truncatula. Plant Cell 2019, 31, 2751–2767. [Google Scholar] [CrossRef]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y.W. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef]
- Lado, J.; Alós, E.; Manzi, M.; Cronje, P.J.R.; Gómez-Cadenas, A.; Rodrigo, M.J.; Zacarías, L. Light Regulation of Carotenoid Biosynthesis in the Peel of Mandarin and Sweet Orange Fruits. Front. Plant Sci. 2019, 10, 1288. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodriguez-Concepcion, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Johansson, H.; Lee, K.P.; Bou-Torrent, J.; Stewart, K.; Steel, G.; Rodriguez-Concepcion, M.; Halliday, K.J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014, 10, e1004416. [Google Scholar] [CrossRef]
- Liu, L.; Shao, Z.; Zhang, M.; Wang, Q. Regulation of carotenoid metabolism in tomato. Mol. Plant 2015, 8, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, T.; Tenga, R.; Han, M.; Zhuang, J. Low temperature effects on carotenoids biosynthesis in the leaves of green and albino tea plant (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2021, 285, 110164. [Google Scholar] [CrossRef]
- Su, L.; Diretto, G.; Purgatto, E.; Danoun, S.; Zouine, M.; Li, Z.; Roustan, J.P.; Bouzayen, M.; Giuliano, G.; Chervin, C. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol. 2015, 15, 114. [Google Scholar] [CrossRef]
- Welsch, R.; Maass, D.; Voegel, T.; Dellapenna, D.; Beyer, P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007, 145, 1073–1085. [Google Scholar] [CrossRef]
- Cruz, A.B.; Bianchetti, R.E.; Alves, F.R.R.; Purgatto, E.; Peres, L.E.P.; Rossi, M.; Freschi, L. Light, Ethylene and Auxin Signaling Interaction Regulates Carotenoid Biosynthesis During Tomato Fruit Ripening. Front. Plant Sci. 2018, 9, 1370. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, P.; Abedimanesh, S.; Mesbah-Namin, S.A.; Ostadrahimi, A. Betalains, the nature-inspired pigments, in health and diseases. Crit. Rev. Food Sci. Nutr. 2019, 59, 2949–2978. [Google Scholar] [CrossRef]
- Polturak, G.; Aharoni, A. “La Vie en Rose”: Biosynthesis, Sources, and Applications of Betalain Pigments. Mol. Plant 2018, 11, 7–22. [Google Scholar] [CrossRef]
- Jain, G.; Gould, K.S. Are betalain pigments the functional homologues of anthocyanins in plants? Environ. Exp. Bot. 2015, 119, 48–53. [Google Scholar] [CrossRef]
- Escribano, J.; Cabanes, J.; Jiménez-Atiénzar, M.; Ibañez-Tremolada, M.; Gómez-Pando, L.R.; García-Carmona, F.; Gandía-Herrero, F. Characterization of betalains, saponins and antioxidant power in differently colored quinoa (Chenopodium quinoa) varieties. Food Chem. 2017, 234, 285–294. [Google Scholar] [CrossRef]
- Gandia-Herrero, F.; Garcia-Carmona, F. Characterization of recombinant Beta vulgaris 4,5-DOPA-extradiol-dioxygenase active inthe biosynthesis of betalains. Planta 2012, 236, 91–100. [Google Scholar] [CrossRef]
- Chen, N.; Yu, Z.H.; Xiao, X.G. Cytosolic and nuclear colocalization of betalain biosynthetic enzymes in tobacco suggests that betalains are synthesized in the cytoplasm and/or nucleus of betalainic plant cells. Front. Plant Sci. 2017, 8, 831. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Timoneda, A.; Feng, T.; Sheehan, H.; Walker-Hale, N.; Pucker, B.; Lopez-Nieves, S.; Guo, R.; Brockington, S. The evolution of betalain biosynthesis in Caryophyllales. New Phytol. 2019, 224, 71–85. [Google Scholar] [CrossRef]
- Polturak, G.; Breitel, D.; Grossman, N.; Sarrion-Perdigones, A.; Weithorn, E.; Pliner, M.; Orzaez, D.; Granell, A.; Rogachev, I.; Aharoni, A. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 2016, 210, 269–283. [Google Scholar] [CrossRef]
- DeLoache, W.C.; Russ, Z.N.; Narcross, L.; Gonzales, A.M.; Martin, V.J.J.; Dueber, J.E. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015, 11, 465–471. [Google Scholar] [CrossRef]
- Sunnadeniya, R.; Bean, A.; Brown, M.; Akhavan, N.; Hatlestad, G.; Gonzalez, A.; Symonds, V.V.; Lloyd, A. Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS ONE 2016, 11, e0149417. [Google Scholar] [CrossRef]
- Brockington, S.F.; Yang, Y.; Gandía-Herrero, F.; Covshoff, S.; Hibberd, J.M.; Sage, R.F.; Wong, G.K.S.; Moore, M.J.; Smith, S.A. Lineage-specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytol. 2015, 207, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Hua, Q.; Chen, C.; Zhang, Z.; Zhang, R.; Qin, Y. Genome-Wide Characterization of R2R3-MYB Transcription Factors in Pitaya Reveals a R2R3-MYB Repressor HuMYB1 Involved in Fruit Ripening through Regulation of Betalain Biosynthesis by Repressing Betalain Biosynthesis-Related Genes. Cells 2021, 10, 1949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, C.; Xie, F.; Hua, Q.; Zhang, Z.; Qin, Y. A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. Int. J. Mol. Sci. 2021, 22, 2171. [Google Scholar] [CrossRef]
- Hatlestad, G.J.; Akhavan, N.A.; Sunnadeniya, R.M.; Elam, L.; Cargile, S.; Hembd, A.; Gonzalez, A.; McGrath, J.M.; Lloyd, A.M. The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 2015, 47, 92–96. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Timoneda, A.; Sheehan, H.; Feng, T.; Lopez-Nieves, S.; Maeda, H.A.; Brockington, S. Redirecting primary metabolism to boost production of tyrosine-derived specialised metabolites in planta. Sci. Rep. 2018, 8, 17256. [Google Scholar] [CrossRef] [PubMed]
- Shimada, S.; Otsuki, H.; Sakuta, M. Transcriptional control of anthocyanin biosynthetic genes in the Caryophyllales. J. Exp. Bot. 2007, 58, 957–967. [Google Scholar] [CrossRef]
- Shimada, S.; Takahashi, K.; Sato, Y.; Sakuta, M. Dihydroflavonol 4-reductase cDNA from non-anthocyanin-producing species in the Caryophyllales. Plant Cell Physiol. 2004, 45, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Polturak, G.; Heinig, U.; Grossman, N.; Battat, M.; Leshkowitz, D.; Malitsky, S.; Rogachev, I.; Aharoni, A. Transcriptome and metabolic profiling provides insights into betalain biosynthesis and evolution in Mirabilis jalapa. Mol. Plant 2018, 11, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Gandía-Herrero, F.; Cabanes, J.; Escribano, J.; García-Carmona, F.; Jiménez-Atiénzar, M. Encapsulation of the most potent antioxidant betalains in edible matrixes as powders of different colors. J. Agric. Food Chem. 2013, 61, 4294–4302. [Google Scholar] [CrossRef]
- Stanley, L.; Yuan, Y.W. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Rahnasto-Rilla, M.; Tyni, J.; Huovinen, M.; Jarho, E.; Kulikowicz, T.; Ravichandran, S.; Bohr, V.A.; Ferrucci, L.; Lahtela-Kakkonen, M.; Moaddel, R. Natural polyphenols as sirtuin 6 modulators. Sci. Rep. 2018, 8, 4163. [Google Scholar] [CrossRef] [PubMed]
- Tossi, V.; Tosar, L.M.; Pitta-Álvarez, S.I.; Causinb, H.F. Casting light on the pathway to betalain biosynthesis: A review. Environ. Exp. Bot. 2021, 186, 104464. [Google Scholar] [CrossRef]
- Yin, X.J.; Wang, T.T.; Zhang, M.; Zhang, Y.B.; Irfan, M.; Li, J.; Chen, L.J.; Zhang, L. Role of core structural genes for flavonoid biosynthesis and transcriptional factors in flower color of plants. Biotechnol. Biotechnol. Equip. 2021, 35, 1214–1229. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Zhang, Y.; Long, T.; Wang, S.; Yang, J. Regulation Mechanism of Plant Pigments Biosynthesis: Anthocyanins, Carotenoids, and Betalains. Metabolites 2022, 12, 871. https://doi.org/10.3390/metabo12090871
Zhao X, Zhang Y, Long T, Wang S, Yang J. Regulation Mechanism of Plant Pigments Biosynthesis: Anthocyanins, Carotenoids, and Betalains. Metabolites. 2022; 12(9):871. https://doi.org/10.3390/metabo12090871
Chicago/Turabian StyleZhao, Xuecheng, Yueran Zhang, Tuan Long, Shouchuang Wang, and Jun Yang. 2022. "Regulation Mechanism of Plant Pigments Biosynthesis: Anthocyanins, Carotenoids, and Betalains" Metabolites 12, no. 9: 871. https://doi.org/10.3390/metabo12090871
APA StyleZhao, X., Zhang, Y., Long, T., Wang, S., & Yang, J. (2022). Regulation Mechanism of Plant Pigments Biosynthesis: Anthocyanins, Carotenoids, and Betalains. Metabolites, 12(9), 871. https://doi.org/10.3390/metabo12090871





