Genomic Comparative Analysis of Cordyceps pseudotenuipes with Other Species from Cordyceps
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. Strain Culture
2.3. Genome Sequencing and Assembly
2.4. Gene Prediction and Annotation
2.5. Secondary Metabolite Biosynthesis Gene Cluster Analysis
2.6. Cluster Analysis
2.7. Synteny Analysis
3. Results
3.1. Basic Features of the C. pseudotenuipes Genome
3.1.1. Genome Sequencing and Assembly
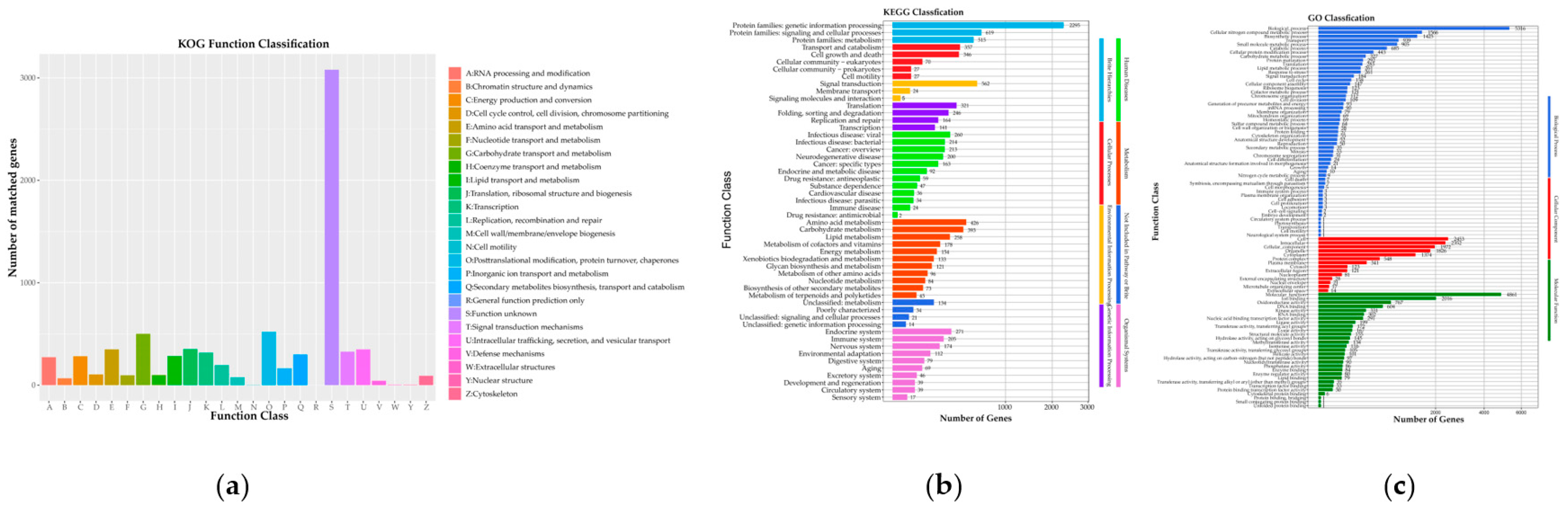
3.1.2. C. pseudotenuipes Genome Annotation
3.1.3. C. pseudotenuipes Additional Annotation
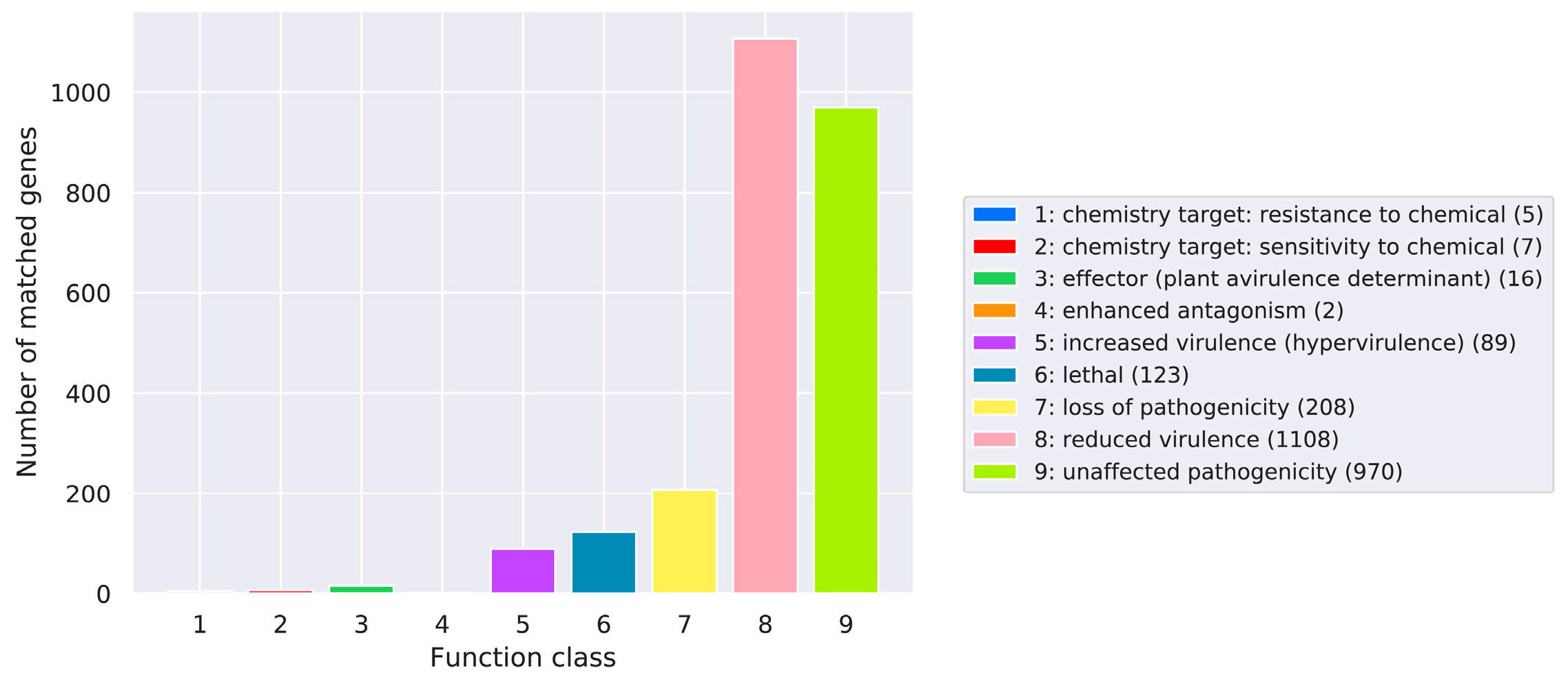
Pathogen Host Interactions (PHI)
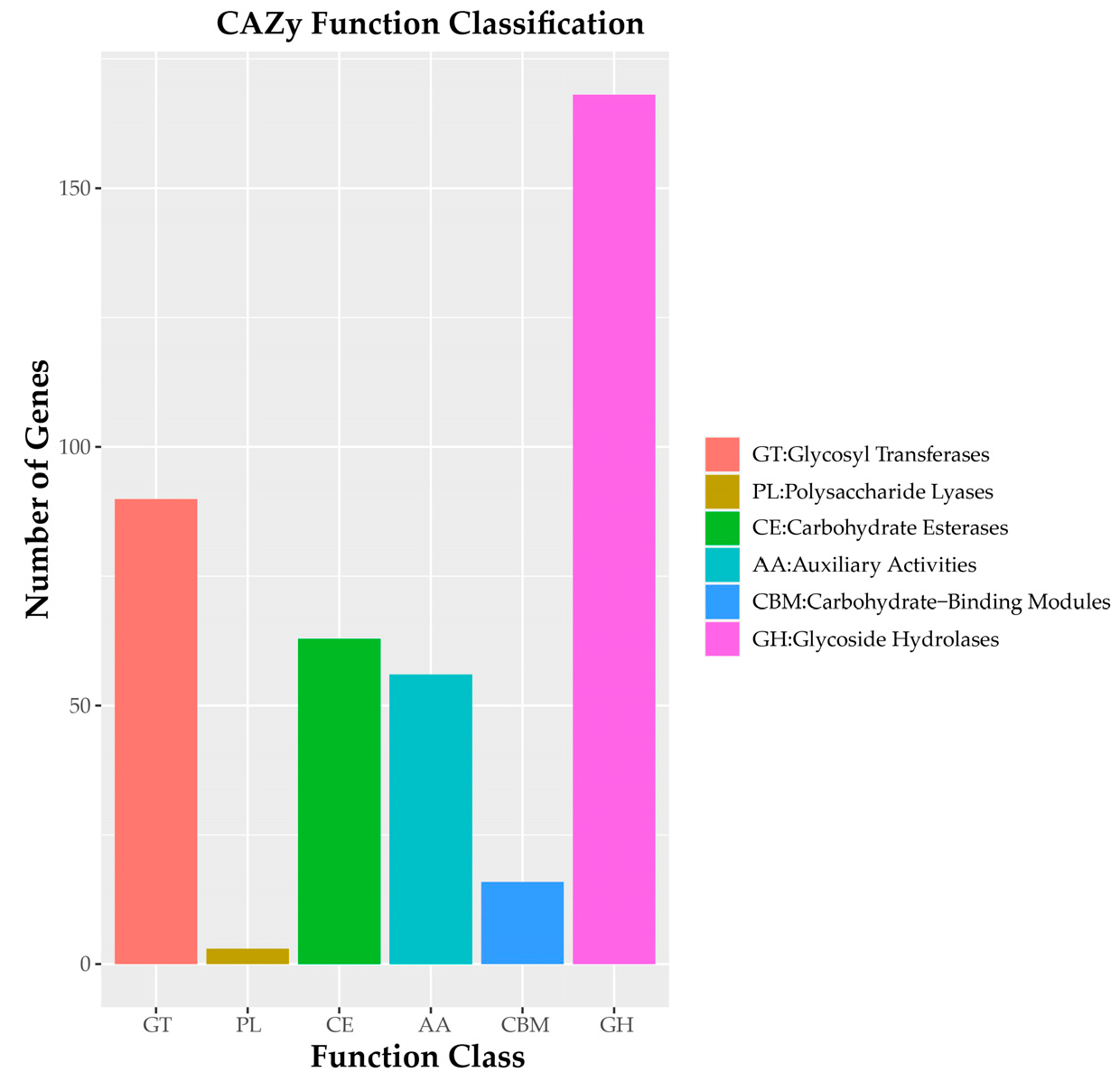
Carbohydrate Genes
3.2. Characteristics of C. pseudotenuipes, C. tenuipes, C. cicadae, and C. militaris Genomes
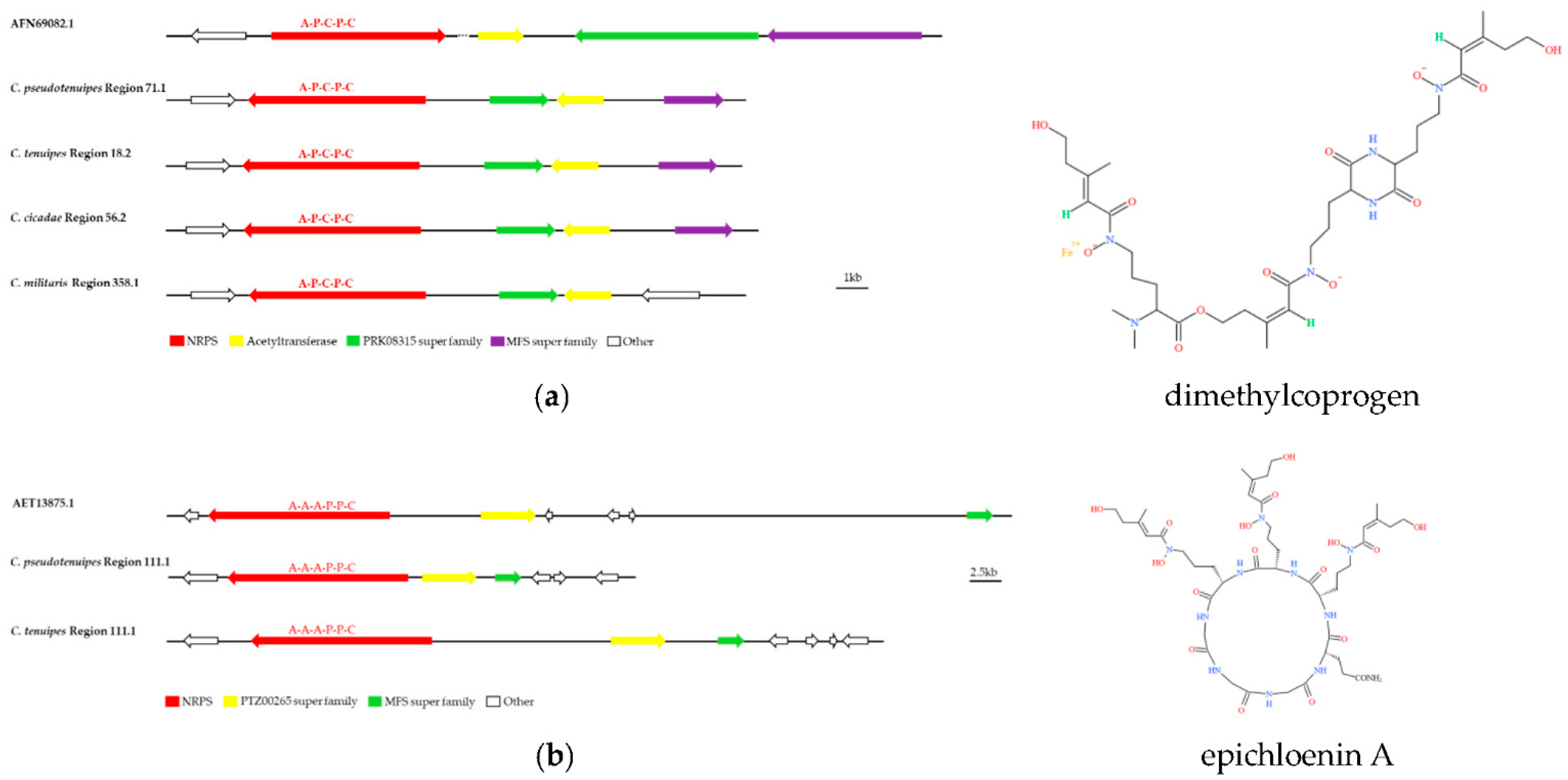
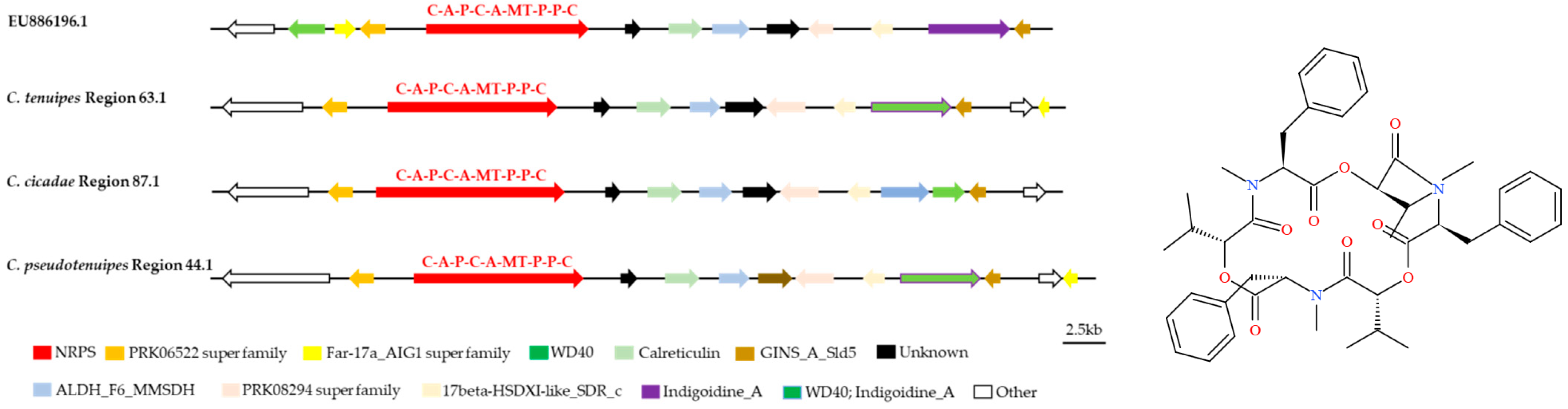
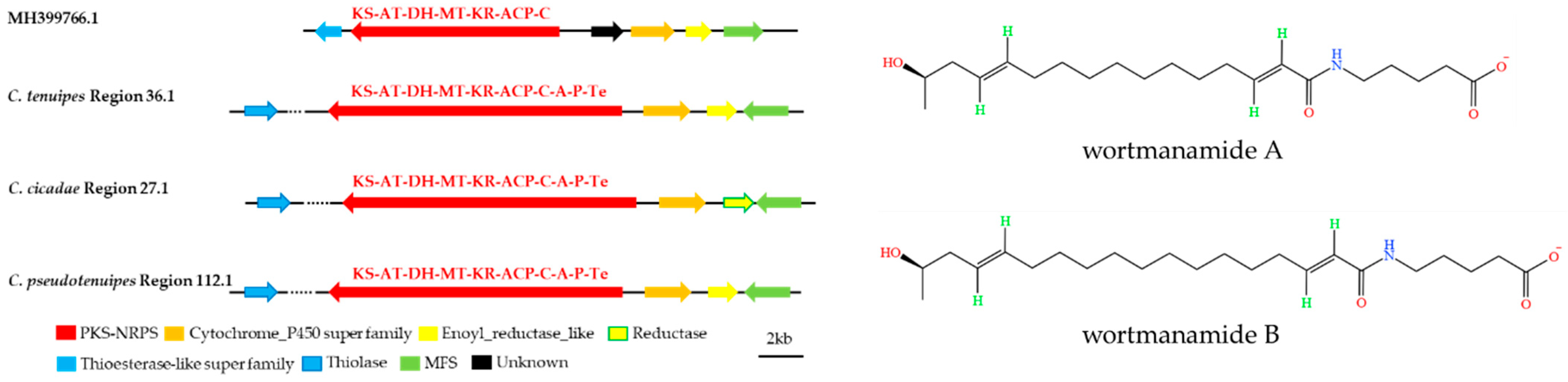
3.3. Analysis of Secondary Metabolite Biosynthesis Gene Cluster
3.4. Cluster Analysis
3.5. Synteny Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, Q.-Y.; Wang, Y.; Wang, Z.-Q.; Tang, D.-X.; Zhao, Z.-Y.; Wu, H.-J.; Yu, H. Morphology and Phylogeny Reveal Five Novel Species in the Genus Cordyceps (Cordycipitaceae, Hypocreales) From Yunnan, China. Front. Microbiol. 2022, 13, 846909. [Google Scholar] [CrossRef]
- Wang, Y.B.; Wang, Y.; Fan, Q.; Duan, D.E.; Zhang, G.D.; Dai, R.Q.; Dai, Y.D.; Zeng, W.B.; Chen, Z.H.; Li, D.D.; et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic po-sition of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- Kepler, R.M.; Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.-H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef]
- Lu, M.Y.; Chen, C.C.; Lee, L.Y.; Lin, T.W.; Kuo, C.F. N (6)-(2-Hydroxyethyl) adenosine in the Medicinal Mushroom Cordyceps cicadae Attenuates Lipopolysaccharide-Stimulated Pro-inflammatory Responses by Suppressing TLR4-Mediated NF-κB Sig-naling Pathways. J. Nat. Prod. 2015, 78, 2452–2460. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.; Yu, Q.; Zheng, B.; Wang, H.; Wang, J.; Chen, S.; Nie, S.; Xie, M. Cultured Cordyceps sinensis polysaccharides modulate intestinal mucosal immunity and gut microbiota in cyclophosphamide-treated mice. Carbohydr. Polym. 2020, 235, 115957. [Google Scholar] [CrossRef]
- Zhang, J.; Jian, T.; Zhang, Y.; Zhang, G.; Ling, J. Dynamic content changes of cordycepin and adenosine and transcriptome in Cordyceps kyushuensis Kob at different fermentation stages. Bioprocess Biosyst. Eng. 2021, 44, 1793–1803. [Google Scholar] [CrossRef]
- Yin, Y.; Yu, G.; Chen, Y.; Jiang, S.; Wang, M.; Jin, Y.; Lan, X.; Liang, Y.; Sun, H. Genome-Wide Transcriptome and Proteome Analysis on Different Developmental Stages of Cordyceps militaris. PLoS ONE 2012, 7, e51853. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, G.; Li, C.; Ling, J. Cordycepin and pentostatin biosynthesis gene identified through transcriptome and proteomics analysis of Cordyceps kyushuensis Kob. Microbiol. Res. 2018, 218, 12–21. [Google Scholar] [CrossRef]
- Yang, N.-N.; Jiang, N.; Ma, Q.-Y.; Kong, F.-D.; Xie, Q.-Y.; Zhou, L.-M.; Yu, Z.-F.; Zhao, Y.-X. Chemical study of the strain Cordyceps spp. from cell fusion between Cordyceps militaris and Cordyceps cicadae. J. Asian Nat. Prod. Res. 2018, 21, 449–455. [Google Scholar] [CrossRef]
- Zhao, C.; Bu, H.; Zhu, J.; Wang, Y.; Oliver, K.M.; Hu, F.; Huang, B.; Li, Z.; Peng, F. Integration of Untargeted Metabolomics with Transcriptomics Provides Insights into Beauvericin Biosynthesis in Cordyceps chanhua under H2O2-Induced Oxidative Stress. J. Fungi 2022, 8, 484. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Talbot, C.L.; Chandravanshi, B.; Ksiazek, A.; Sood, A.; Chowdhury, K.H.; Maschek, J.A.; Cox, J.; Babu, A.K.S.; Paz, H.A.; et al. Cordyceps inhibits ceramide biosynthesis and improves insulin resistance and hepatic steatosis. Sci. Rep. 2022, 12, 7273. [Google Scholar] [CrossRef]
- Yin, Y.; Chen, B.; Song, S.; Li, B.; Yang, X.; Wang, C. Production of Diverse Beauveriolide Analogs in Closely Related Fungi: A Rare Case of Fungal Chemodiversity. mSphere 2020, 5, e00667-20. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Q.; Weng, Q. Secondary metabolites (SMs) of Isaria cicadae and Isaria tenuipes. RSC Adv. 2018, 9, 172–184. [Google Scholar] [CrossRef]
- Tuli, H.S.; Sharma, A.K.; Sandhu, S.S.; Kashyap, D. Cordycepin: A bioactive metabolite with therapeutic potential. Life Sci. 2013, 93, 863–869. [Google Scholar] [CrossRef]
- Holbein, S.; Wengi, A.; Decourty, L.; Freimoser, F.M.; Jacquier, A.; Dichtl, B. Cordycepin interferes with 3′ end formation in yeast independently of its potential to terminate RNA chain elongation. RNA 2009, 15, 837–849. [Google Scholar] [CrossRef]
- Wen, T.; Jiang, C.; Shen, D.; Li, C.; Lu, Q.; Liu, X.; Kang, Y.; Chen, X.; Liu, B.; Qin, C.; et al. Evaluation of the Antitumor Activity by Cordycepin with Gold Nanoparticle. J. Nanosci. Nanotechnol. 2016, 16, 7134–7139. [Google Scholar] [CrossRef]
- Dalla Rosa, L.; da Silva, A.S.; Gressler, L.T.; Oliveira, C.B.; Dambrós, M.G.; Miletti, L.C.; França, R.T.; Lopes, S.T.; Samara, Y.N.; da Veiga, M.L.; et al. Cordycepin (3′-deoxyadenosine) pentostatin (deoxycoformycin) combination treatment of mice experimentally infected with Trypanosoma evansi. Parasitology 2013, 140, 663–671. [Google Scholar] [CrossRef]
- Sarvaria, A.; Topp, Z.; Saven, A. Current Therapy and New Directions in the Treatment of Hairy Cell Leukemia: A Review. JAMA Oncol. 2016, 2, 123–129. [Google Scholar] [CrossRef]
- Yoshikawa, N.; Nakamura, K.; Yamaguchi, Y.; Kagota, S.; Shinozuka, K.; Kunitomo, M. Reinforcement of antitumor effect of Cordyceps sinensis by 2′-deoxycoformycin, an adenosine deaminase inhibitor. In Vivo 2007, 21, 291–295. [Google Scholar]
- Feng, P.; Shang, Y.; Cen, K.; Wang, C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 11365–11370. [Google Scholar] [CrossRef]
- Russell, A.H.; Truman, A.W. Genome mining strategies for ribosomally synthesised and post-translationally modified pep-tides. Comput. Struct. Biotechnol. J. 2020, 18, 1838–1851. [Google Scholar] [CrossRef]
- Chu, L.; Huang, J.; Muhammad, M.; Deng, Z.; Gao, J. Genome mining as a biotechnological tool for the discovery of novel marine natural products. Crit. Rev. Biotechnol. 2020, 40, 571–589. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Blin, K.; Kim, H.U.; Medema, M.H.; Weber, T. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 2017, 20, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal Cordycepin Biosynthesis Is Coupled with the Production of the Safeguard Molecule Pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489.e4. [Google Scholar] [CrossRef]
- Wang, X.; Gao, Y.L.; Zhang, M.L.; Zhang, H.D.; Huang, J.Z.; Li, L. Genome mining and biosynthesis of the Acyl-CoA: Cho-lesterol acyltransferase inhibitor beauveriolide I and III in Cordyceps militaris. J. Biotechnol. 2020, 309, 85–91. [Google Scholar] [CrossRef]
- Gao, Y.-L.; Yu, C.; Li, L. Heterologous expression of a natural product biosynthetic gene cluster from Cordyceps militaris. J. Antibiot. 2021, 75, 16–20. [Google Scholar] [CrossRef]
- Wang, W.-J.; Vogel, H.; Yao, Y.-J.; Ping, L. The nonribosomal peptide and polyketide synthetic gene clusters in two strains of entomopathogenic fungi in Cordyceps. FEMS Microbiol. Lett. 2012, 336, 89–97. [Google Scholar] [CrossRef][Green Version]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; Sahu, J.; Iyer, S.V.; Khamari, L.; De Silva, N.; et al. PHI-base in 2022: A multi-species phenotype database for Pathogen–Host Interactions. Nucleic Acids Res. 2021, 50, D837–D847. [Google Scholar] [CrossRef] [PubMed]
- Garron, M.-L.; Henrissat, B. The continuing expansion of CAZymes and their families. Curr. Opin. Chem. Biol. 2019, 53, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Func-tions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-H.; Lin, C.-H.; Chung, K.-R. A nonribosomal peptide synthetase mediates siderophore production and virulence in the citrus fungal pathogen Alternaria alternata. Mol. Plant Pathol. 2013, 14, 497–505. [Google Scholar] [CrossRef]
- Jalal, M.A.F.; Love, S.K.; van der Helm, D. N alpha-dimethylcoprogens. Three novel trihydroxamate siderophores from pathogenic fungi. Biol. Met. 1988, 1, 4–8. [Google Scholar] [CrossRef]
- Hai, Y.; Tang, Y. Biosynthesis of Long-Chain N-Acyl Amide by a Truncated Polyketide Synthase–Nonribosomal Peptide Synthetase Hybrid Megasynthase in Fungi. J. Am. Chem. Soc. 2018, 140, 1271–1274. [Google Scholar] [CrossRef]
- Ahn, Y.J.; Park, S.J.; Lee, S.G.; Shin, S.C.; Choi, D.H. Cordycepin: Selective growth inhibitor derived from liquid culture of Cordyceps militaris against Clostridium spp. J. Agric. Food Chem. 2000, 48, 2744–2748. [Google Scholar] [CrossRef]
- Cacho, R.A.; Tang, Y.; Chooi, Y.-H. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front. Microbiol. 2015, 5, 774. [Google Scholar] [CrossRef][Green Version]
- Du, L.; Lou, L. PKS and NRPS release mechanisms. Nat. Prod. Rep. 2009, 27, 255–278. [Google Scholar] [CrossRef]
- Parsley, L.C.; Linneman, J.; Goode, A.M.; Becklund, K.; George, I.; Goodman, R.M.; Lopanik, N.B.; Liles, M.R. Polyketide synthase pathways identified from a metagenomic library are derived from soil Acidobacteria. FEMS Microbiol. Ecol. 2011, 78, 176–187. [Google Scholar] [CrossRef]
- Yuan, X.L.; Li, Y.Q.; Yi, W. The Diversity of Polyketide Synthase (PKS) Gene in Cordyceps militaris. J. West China For. Sci. 2019, 48, 97–103+113. [Google Scholar]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [PubMed]
- Awakawa, T.; Yang, X.-L.; Wakimoto, T.; Abe, I. Pyranonigrin E: A PKS-NRPS Hybrid Metabolite from Aspergillus niger Identified by Genome Mining. ChemBioChem 2013, 14, 2095–2099. [Google Scholar] [CrossRef]
- Theobald, S.; Vesth, T.C.; Andersen, M.R. Genus level analysis of PKS-NRPS and NRPS-PKS hybrids reveals their origin in Aspergilli. BMC Genom. 2019, 20, 847. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, S.; Guo, T.; Li, L.; Li, T.; Wang, A.; Zhang, D.; Wang, Y.; Sun, Y. Bioactive PKS–NRPS Alkaloids from the Plant-Derived Endophytic Fungus Xylaria arbuscula. Molecules 2021, 27, 136. [Google Scholar] [CrossRef]
- Seshime, Y.; Juvvadi, P.R.; Tokuoka, M.; Koyama, Y.; Kitamoto, K.; Ebizuka, Y.; Fujii, I. Functional expression of the Aspergillus flavus PKS–NRPS hybrid CpaA involved in the biosynthesis of cyclopiazonic acid. Bioorganic Med. Chem. Lett. 2009, 19, 3288–3292. [Google Scholar] [CrossRef]
- Nielsen, M.L.; Isbrandt, T.; Petersen, L.M.; Mortensen, U.H.; Andersen, M.R.; Hoof, J.B.; Larsen, T.O. Linker Flexibility Facil-itates Module Exchange in Fungal Hybrid PKS-NRPS Engineering. PLoS ONE 2016, 11, e0161199. [Google Scholar] [CrossRef]
- Zhong, Z.; Lin, L.; Zheng, H.; Bao, J.; Chen, M.; Zhang, L.; Tang, W.; Ebbole, D.J.; Wang, Z. Emergence of a hybrid PKS-NRPS secondary metabolite cluster in a clonal population of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2020, 22, 2709–2723. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, W.; Li, Z.; Li, H.; Geng, C.; Huang, X.; Lu, X. Discovery and Characterization of a PKS-NRPS Hybrid in As-pergillus terreus by Genome Mining. J. Nat. Prod. 2020, 83, 473–480. [Google Scholar] [CrossRef]
- Sigrist, R.; Luhavaya, H.; McKinnie, S.; Ferreira da Silva, A.; Jurberg, I.D.; Moore, B.S.; Gonzaga de Oliveira, L. Nonlinear Biosynthetic Assembly of Alpiniamide by a Hybrid cis/trans-AT PKS-NRPS. ACS Chem. Biol. 2020, 15, 1067–1077. [Google Scholar] [CrossRef]



| Item | Value | Item | Count | Percentage (%) |
|---|---|---|---|---|
| Total length (Mb) | 30.1 | NR | 8596 | 98.75 |
| Max length (bp) | 413,323 | SwissProt | 6206 | 71.29 |
| GC content (%) | 54.11 | KEGG | 3614 | 41.52 |
| Gene number | 8705 | GO | 5861 | 62.33 |
| Total gene number (bp) | 13,973,781 | EggNOg | 7877 | 90.49 |
| Average gene number (bp) | 1605.2 | P450 | 8480 | 97.42 |
| Gene/Genome (%) | 47.3169 | TCDB | 1358 | 15.60 |
| Contigs | 645 | |||
| Scaffolds | 527 | |||
| Contigs N50 | 101,518 | |||
| Scaffolds N50 | 131,856 | |||
| Contigs N90 | 27,054 | |||
| Scaffolds N90 | 32,698 |
| Item | C. pseudotenuipes This Study | C. tenuipes This Study | C. cicadae ASM296887v1 | C. militaris GCA_000225605.1 |
|---|---|---|---|---|
| Contigs | 645 | 384 | 1799 | 597 |
| Scaffolds | 527 | 285 | 595 | 32 |
| Total length (Mb) | 30.1 | 30.06 | 34.11 | 32.27 |
| GC content (%) | 54.11 | 53.72 | 52.70 | 51.40 |
| Scaffold N50 (bp) | 131,856 | 172,867 | 212,207 | 4,551,492 |
| Contig N50 (bp) | 101,518 | 140,681 | 47,316 | 10,818 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Wang, Y.; Yuan, X.; Huang, O.; Dong, Q.; Li, D.; Ding, S.; Ma, F.; Yu, H. Genomic Comparative Analysis of Cordyceps pseudotenuipes with Other Species from Cordyceps. Metabolites 2022, 12, 844. https://doi.org/10.3390/metabo12090844
Lu Y, Wang Y, Yuan X, Huang O, Dong Q, Li D, Ding S, Ma F, Yu H. Genomic Comparative Analysis of Cordyceps pseudotenuipes with Other Species from Cordyceps. Metabolites. 2022; 12(9):844. https://doi.org/10.3390/metabo12090844
Chicago/Turabian StyleLu, Yingling, Yi Wang, Xiaolong Yuan, Ou Huang, Quanying Dong, Dandan Li, Shujin Ding, Fuxian Ma, and Hong Yu. 2022. "Genomic Comparative Analysis of Cordyceps pseudotenuipes with Other Species from Cordyceps" Metabolites 12, no. 9: 844. https://doi.org/10.3390/metabo12090844
APA StyleLu, Y., Wang, Y., Yuan, X., Huang, O., Dong, Q., Li, D., Ding, S., Ma, F., & Yu, H. (2022). Genomic Comparative Analysis of Cordyceps pseudotenuipes with Other Species from Cordyceps. Metabolites, 12(9), 844. https://doi.org/10.3390/metabo12090844






