Metabolomic and Genomic Approach to Study Defense Induction by Nesidiocoris tenuis against Tuta absoluta and Tetranychus urticae in Tomato Plants
Abstract
:1. Introduction
2. Results
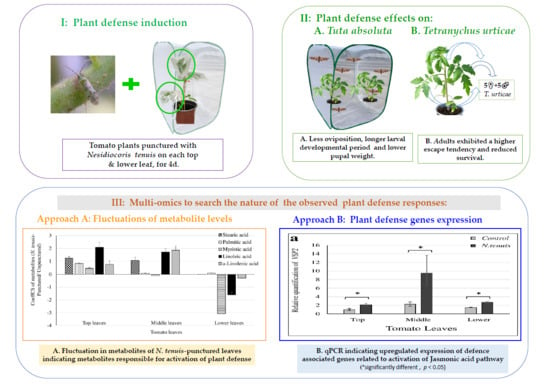
2.1. Oviposition Preference of T. absoluta
2.2. Effects of N. tenuis-Punctured Tomato Plants on Larval Development and Pupal Weight of T. absoluta
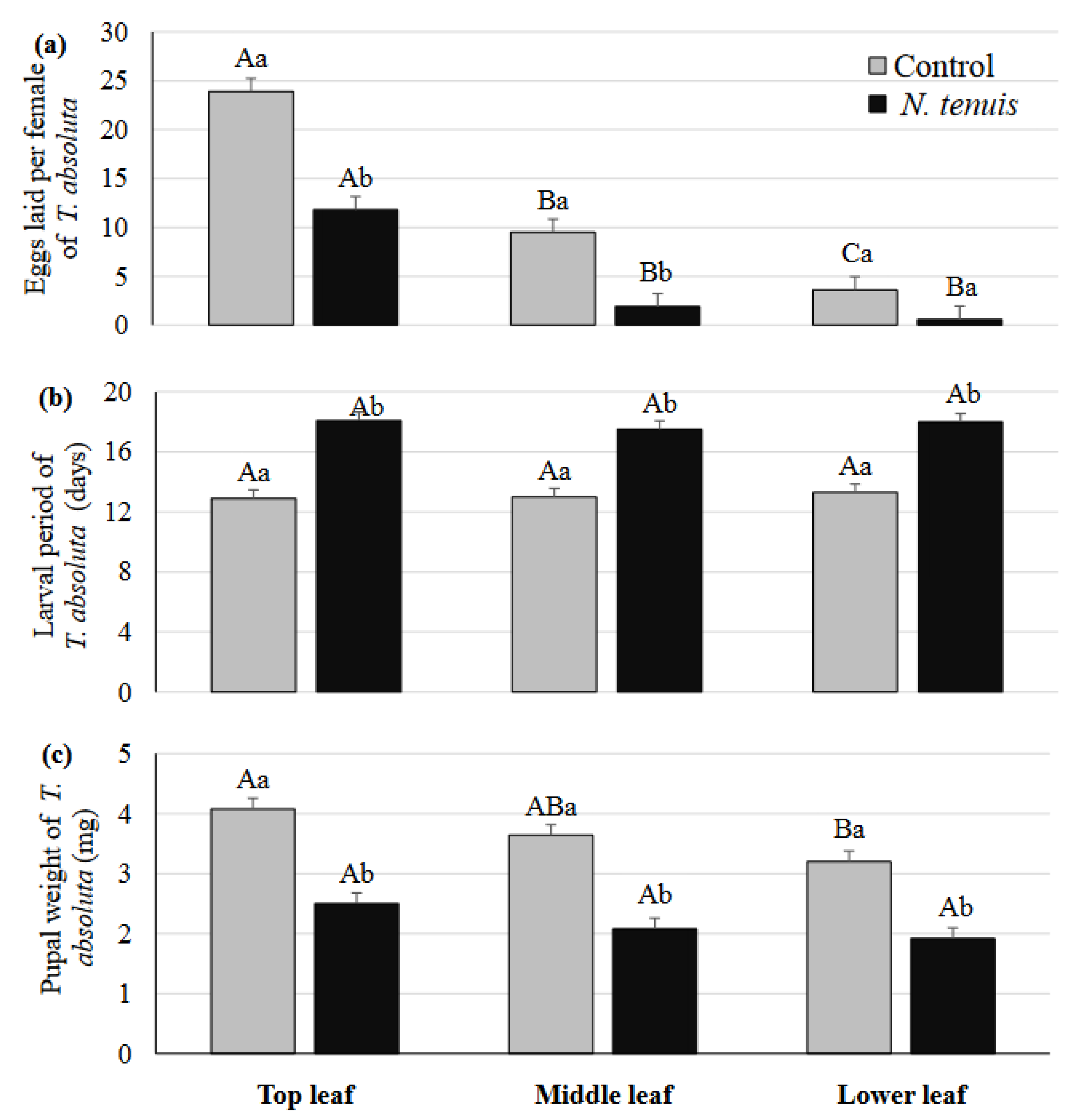
2.3. N. tenuis-Punctured Tomato Plants Induced Escape Tendency of T. urticae
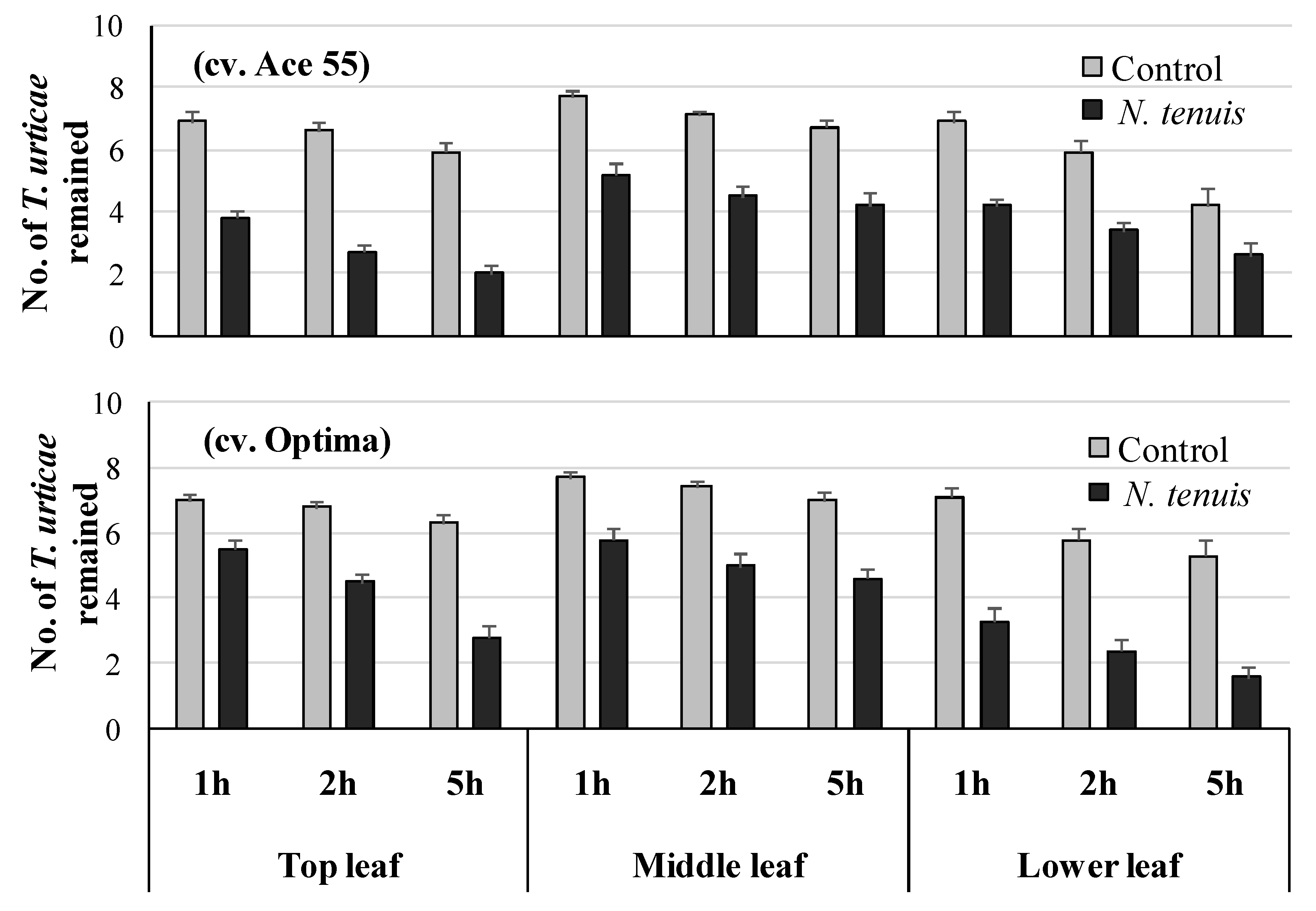
2.4. N. tenuis-Punctured Tomato Plants Affect Survival of T. urticae
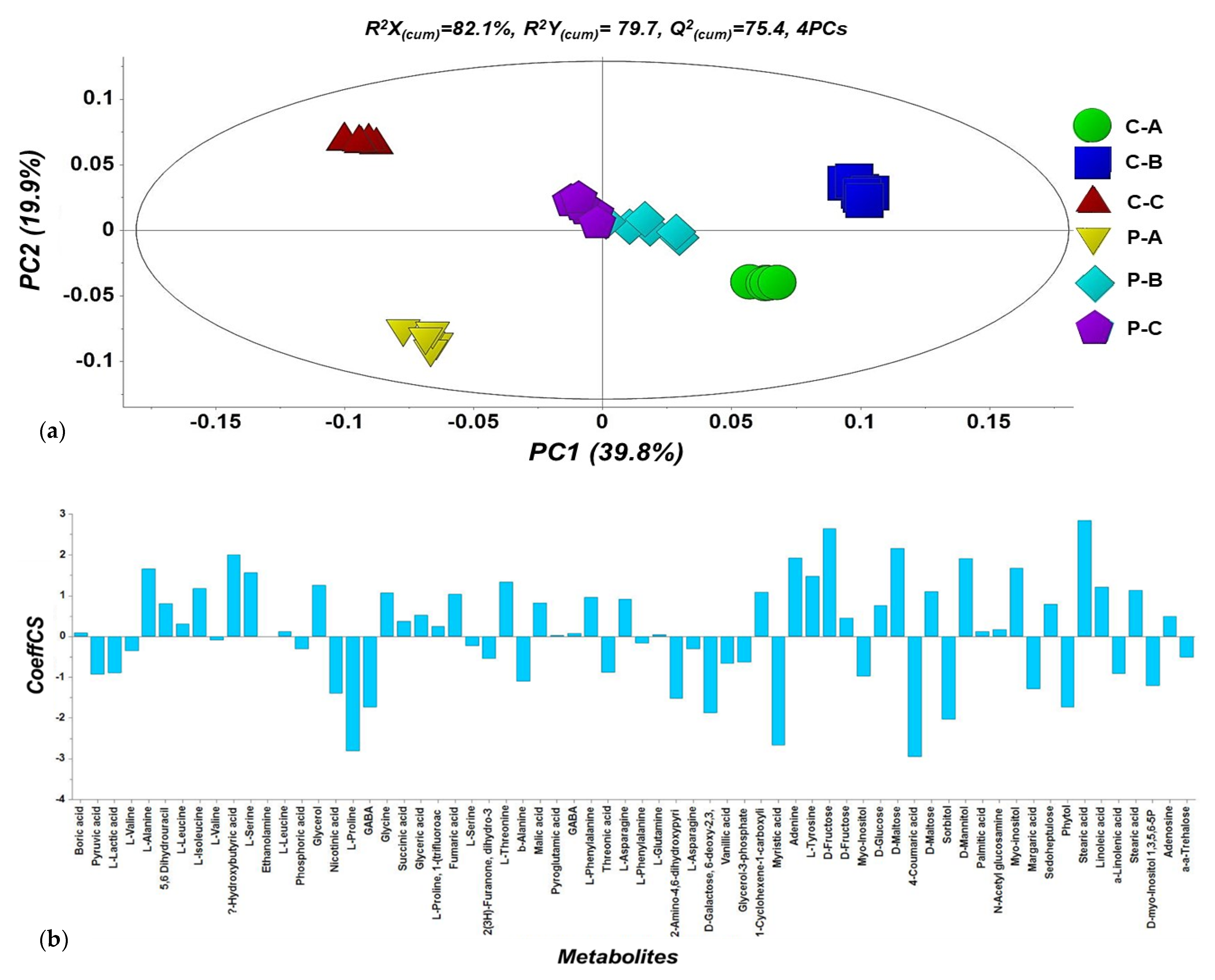
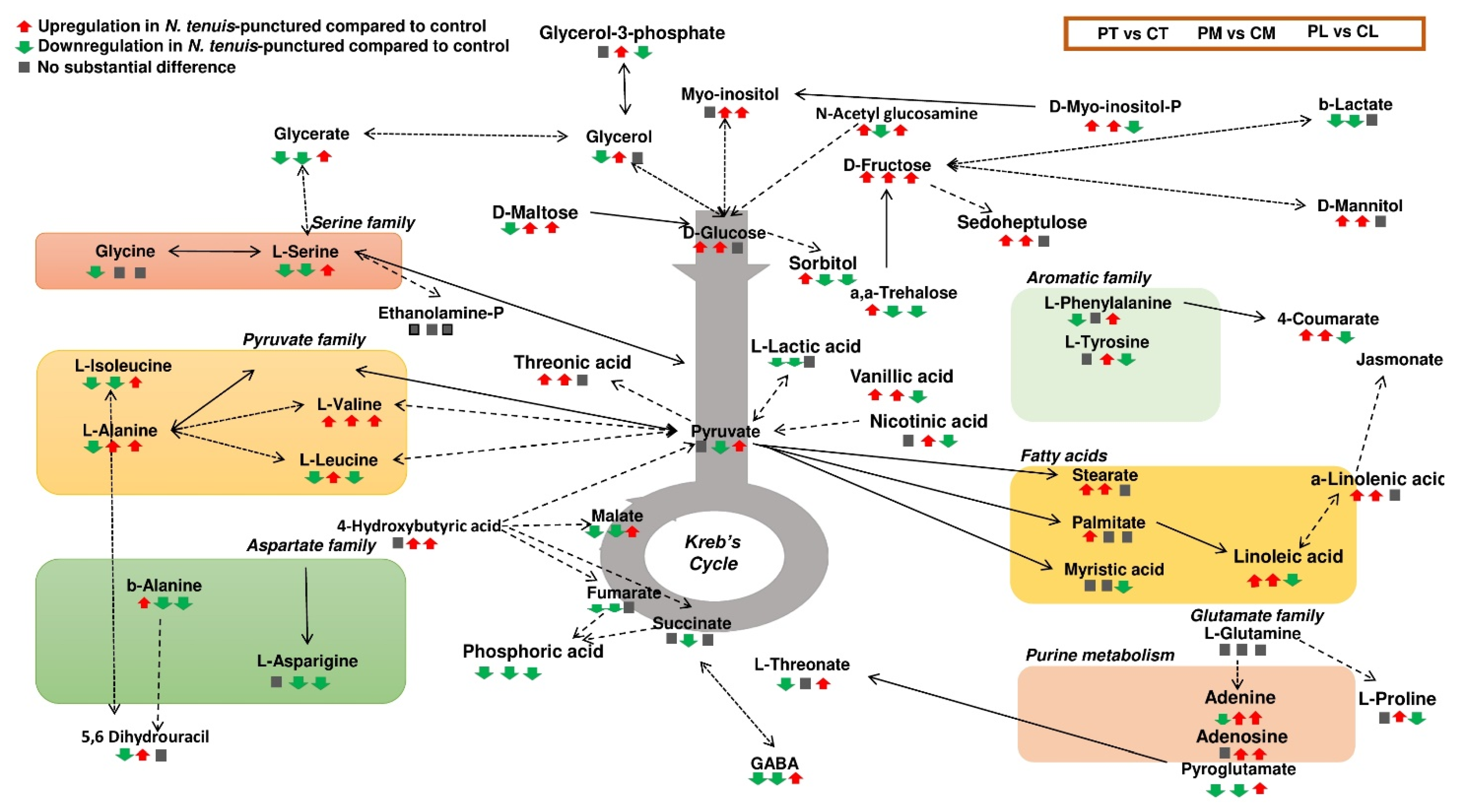
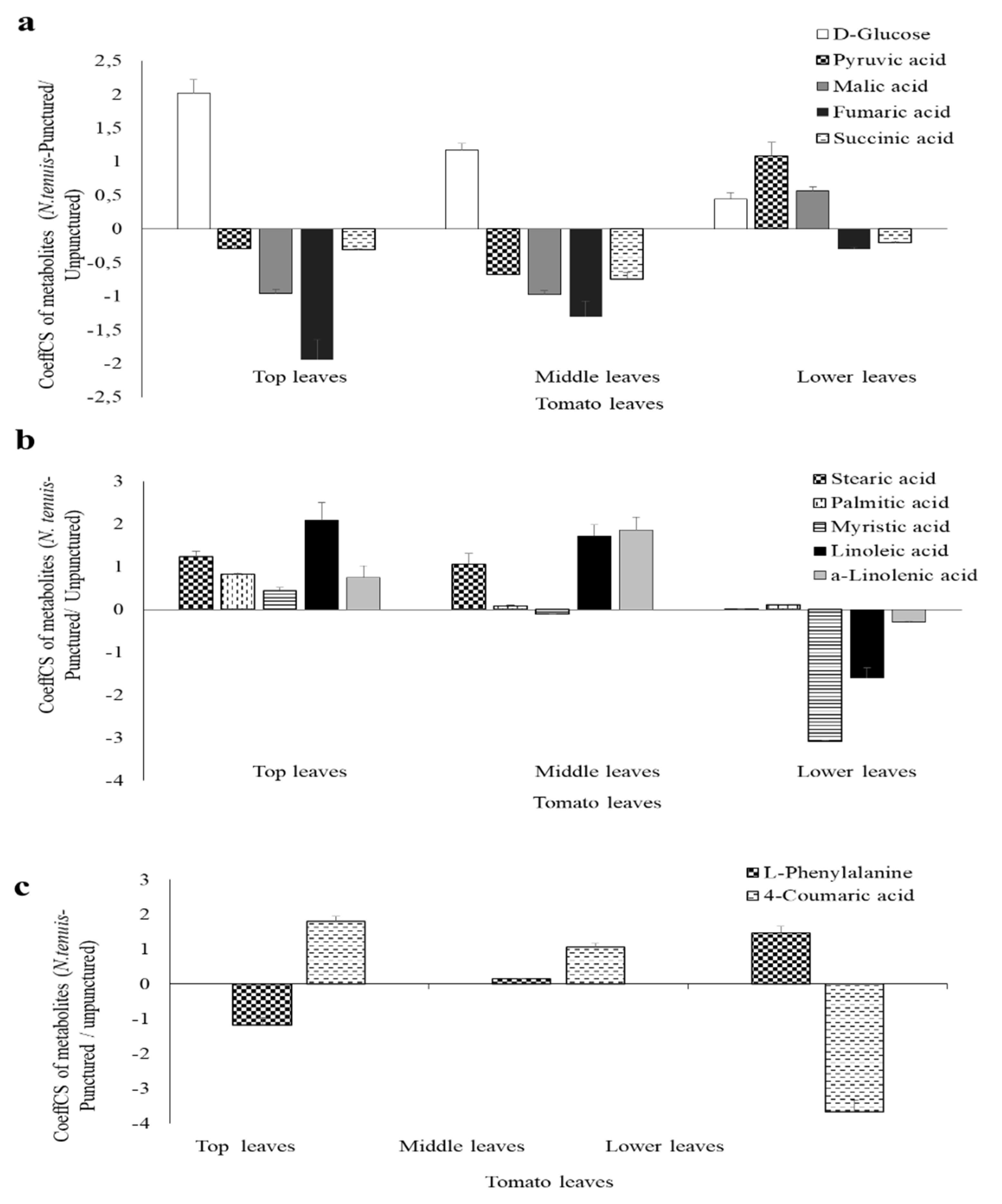
2.5. Overview of the Metabolomics Analyses of N. tenuis-Punctured Tomato Plants
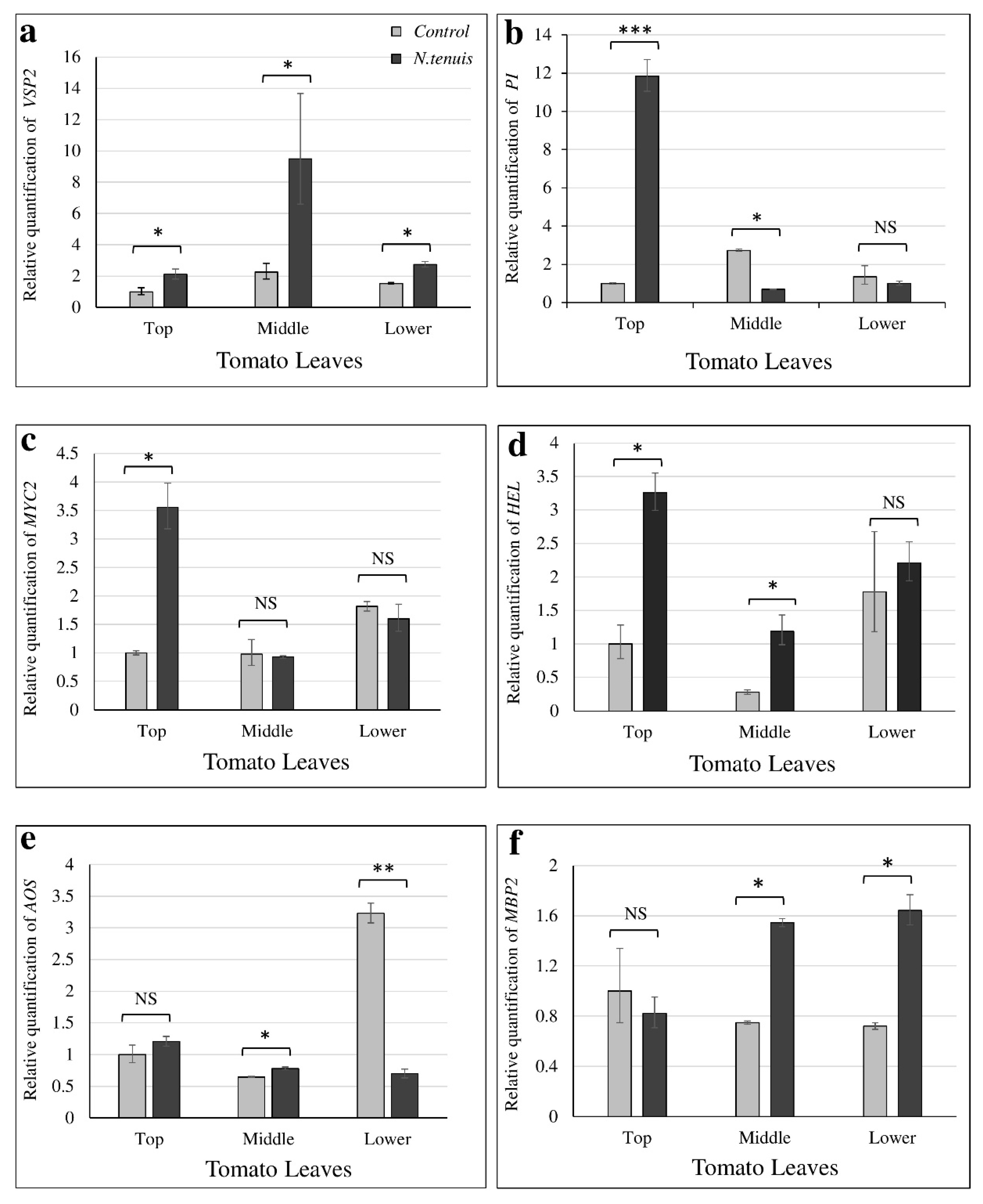
2.6. Plant Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Exposure of Tomato Plants to N. tenuis
4.3. Effects on Oviposition, Larval Development Period, and Pupal Weight of T. absoluta due to Exposure to N. tenuis
4.4. Effects on Escape Tendency and Survival of T. urticae due to Exposure to N. tenuis
4.5. Sampling and Sample Preparation for GC/EI/MS Metabolomics
4.6. GC/EI/MS Metabolite Profiling of Tomato Leaves and Data Pre-Processing
4.7. RNA Extraction and Plant Gene Expression Analysis
4.8. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.G.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Catalán Ruescas, D.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Biondi, A.; Guedes, R.; Wan, F.; Desneux, N. Ecology, worldwide spread, and management of the invasive South American Tomato Pinworm, Tuta absoluta: Past, present, and future. Annu. Rev. Entomol. 2018, 63, 239–258. [Google Scholar] [CrossRef]
- Roditakis, E.; Steinbach, D.; Moritz, G.; Vasakis, E.; Stavrakaki, M.; Ilias, A.; García-Vidal, L.; del Rosario Martinez-Aguirre, M.; Bielza, P.; Morou, E.; et al. Ryanodine receptor point mutations confer diamide insecticide resistance in tomato leafminer, Tuta absoluta (Lepidoptera: Gelechiidae). Insect Biochem. Mol. Biol. 2017, 80, 11–20. [Google Scholar] [CrossRef]
- Guedes, R.N.C.; Roditakis, E.; Campos, M.R.; Haddi, K.; Bielza, P.; Siqueira, H.A.A.; Tsagkarakou, A.; Vontas, J.; Nauen, R. Insecticide resistance in the tomato pinworm Tuta absoluta: Patterns, spread, mechanisms, management and outlook. J. Pest Sci. 2019, 92, 1329–1342. [Google Scholar] [CrossRef]
- Richardson, E.B.; Troczka, B.J.; Gutbrod, O.; Davies, T.G.; Nauen, R. Diamide resistance: 10 years of lessons from lepidopteran pests. J. Pest Sci. 2020, 93, 911–928. [Google Scholar] [CrossRef]
- Feraccini, C.; Bueno, V.H.; Dindo, M.L.; Ingegno, B.L.; Luna, M.G.; Salas Gervassio, N.G.; Sánchez, N.E.; Siscaro, G.; van Lenteren, J.C.; Zappalà, L.; et al. Natural enemies of Tuta absoluta in the Mediterranean basin, Europe and South America. Biocontrol Sci. Technol. 2019, 29, 578–609. [Google Scholar] [CrossRef]
- van Lenteren, J.C.; Alomar, O.; Ravensberg, W.J.; Urbaneja, A. Biological control agents for control of pests in greenhouses. In Integrated Pest and Disease Management in Greenhouse Crops; Gullino, M.L., Nicot, P.C., Albajes, R., Eds.; Springer Nature: Zug, Switzerland, 2020; pp. 409–439. [Google Scholar]
- Perdikis, D.; Lykouressis, D. Effects of various items, host plants and temperatures on the development and survival of Macrolophus pygmaeus Rambur (Hemiptera: Miridae). Biol. Control 2000, 17, 55–60. [Google Scholar] [CrossRef]
- Perdikis, D.; Fantinou, A.; Lykouressis, D. Enhancing pest control in annual crops by conservation of predatory Heteroptera. Biol. Control 2011, 59, 13–21. [Google Scholar] [CrossRef]
- Sarmah, N.; Devee, A.; Perdikis, D. Macrolophus pygmaeus (Hemiptera: Miridae) foraging on tomato leaves from different plant strata. Phytoparasitica 2019, 47, 663–670. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Urbaneja-Bernat, P.; Jaques, J.A.; Flors, V.; Urbaneja, A. Defensive plant responses induced by Nesidiocoris tenuis (Hemiptera: Miridae) on tomato plants. J. Pest Sci. 2015, 88, 543–554. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Hedo, M.; Bouagga, S.; Jaques, J.A.; Flors, V.; Urbaneja, A. Tomato plant responses to feeding behavior of three zoophytophagous predators (Hemiptera: Miridae). Biol. Control 2015, 86, 46–51. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Riahi, C.; Urbaneja, A. Use of zoophytophagous mirid bugs in horticultural crops: Current challenges and future perspectives. Pest Manag. Sci. 2021, 77, 33–42. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Arias-Sanguino, Á.M.; Urbaneja, A. Induced tomato plant resistance against Tetranychus urticae triggered by the phytophagy of Nesidiocoris tenuis. Front. Plant Sci. 2018, 9, 1419. [Google Scholar] [CrossRef]
- Pappas, M.L.; Steppuhn, A.; Geuss, D.; Topalidou, N.; Zografou, A.; Sabelis, M.W.; Broufas, G.D. Beyond predation: The zoophytophagous predator Macrolophus pygmaeus induces tomato resistance against spider mites. PLoS ONE 2015, 10, 0127251. [Google Scholar] [CrossRef]
- Zhang, N.X.; Messelink, G.J.; Alba, J.M.; Schuurink, R.C.; Kant, M.R.; Janssen, A. Phytophagy of omnivorous predator Macrolophus pygmaeus affects performance of herbivores through induced plant defenses. Oecologia 2018, 186, 101–113. [Google Scholar] [CrossRef]
- Bouagga, S.; Urbaneja, A.; Rambla, J.L.; Flors, V.; Granell, A.; Jaques, J.A.; Pérez-Hedo, M. Zoophytophagous mirids provide pest control by inducing direct defenses, antixenosis and attraction to parasitoids in sweet pepper plants. Pest Manag. Sci. 2018, 74, 1286–1296. [Google Scholar] [CrossRef]
- War, A.R.; Taggar, G.K.; Hussain, B.; Taggar, M.S.; Nair, R.M.; Sharma, H.C. Plant defense against herbivory and insect adaptations. AoB Plants 2018, 10, 037. [Google Scholar] [CrossRef]
- Stintzi, A.; Weber, H.; Reymond, P.; Browse, J.; Farmer, E.E. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 2001, 98, 12837–12842. [Google Scholar] [CrossRef]
- Wasternack, C. How jasmonates earned their laurels: Past and present. J. Plant Growth Regul. 2015, 34, 761–794. [Google Scholar] [CrossRef]
- Van Dam, N.; van der Meijimden, E. A role for metabolomics in plant ecology. Annu. Plant Rev. 2011, 43, 87–107. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites—Pathways, transcription factors and applied aspects—A brief review. Nat. Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef]
- Reymond, P.; Weber, H.; Damond, M.; Farmer, E.E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 2000, 12, 707–719. [Google Scholar] [CrossRef]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Sing. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Schittko, U.; Preston, C.; Baldwin, I. Eating the evidence? Manduca sexta larvae cannot disrupt specific jasmonate induction in Nicotiana attenuata by rapid consumption. Planta 2000, 210, 343–346. [Google Scholar] [CrossRef]
- Karban, R.; Baldwin, I.T. Induced Responses to Herbivory; Chicago Univ. Press: Chicago, IL, USA, 1997. [Google Scholar]
- Agrawal, A.A. Induced responses to herbivory and increased plant performance. Science 1998, 279, 1201–1202. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Ku, K.M.; Becker, T.M.; Juvik, J.A. Transcriptome and metabolome analyses of glucosinolates in two broccoli cultivars following jasmonate treatment for the induction of glucosinolate defense to Trichoplusia ni (Hübner). Int. J. Mol. Sci. 2016, 17, 1135. [Google Scholar] [CrossRef]
- de Falco, B.; Manzo, D.; Incerti, G.; Garonna, A.P.; Ercolano, M.; Lanzotti, V. Metabolomics approach based on NMR spectroscopy and multivariate data analysis to explore the interaction between the leafminer Tuta absoluta and tomato (Solanum lycopersicum). Phytochem. Anal. 2019, 30, 556–563. [Google Scholar] [CrossRef]
- Han, P.; Desneux, N.; Becker, C.; Romain, L.; Bot, J.L.; Adamowicz, S.; Zhang, J.; Lavoir, A.V. Bottom-up effects of irrigation, fertilization and plant resistance on Tuta absoluta: Implications for integrated pest management. J. Pest Sci. 2018, 92, 1359–1370. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, S.; Wang, S.; Shan, W.; Wang, X.; Lin, Y.; Su, F.; Yang, Z.; Yu, X. Defensive responses of tea plants (Camellia sinensis) against tea green leafhopper attack: A multi-omics study. Front. Plant. Sci. 2020, 10, 1705. [Google Scholar] [CrossRef]
- Strapasson, P.; Pinto-Zevallos, D.M.; Paudel, S.; Rajotte, E.G.; Felton, G.W.; Zarbin, P.H. Enhancing plant resistance at the seed stage: Low concentrations of methyl jasmonate reduce the performance of the leaf miner Tuta absoluta but do not alter the behavior of its predator Chrysoperla externa. J. Chem. Ecol. 2014, 40, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Esmaeily, S.; Amin, S.M.; Izadi, H. Induced eggplant resistance against Trialeurodes vaporariorum triggered by jasmonic acid, abscisic acid, and Nesidiocoris tenuis feeding. Bull. Entomol. Res. 2020, 110, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Rambla, J.L.; Granell, A.; Urbaneja, A. Biological activity and specificity of Miridae-induced plant volatiles. Biol. Control 2018, 63, 203–213. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, L.; Xu, L.; Yan, D.; Min, L.; Zhang, X. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nature Comm. 2014, 5, 5372. [Google Scholar] [CrossRef]
- Chanda, B.; Xia, Y.; Mandal, M.K.; Yu, K.; Sekine, K.T.; Gao, Q.M.; Selote, D.; Hu, Y.; Stromberg, A.; Navarre, D.; et al. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 2011, 43, 421–427. [Google Scholar] [CrossRef]
- Sanchez, J.A.; del Amor, F.M.; Flores, P.; López-Gallego, E. Nutritional variations at Nesidiocoris tenuis feeding sites and reciprocal interactions between the mirid and tomato plants. J. Appl. Entom. 2016, 140, 161–173. [Google Scholar] [CrossRef]
- van Dam, N.M.; Horn, M.; Mareš, M.; Baldwin, I.T. Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J. Chem. Ecol. 2001, 27, 547–568. [Google Scholar] [CrossRef]
- Anderson, P.; Agrell, J. Within-plant variation in induced defense in developing leaves of cotton plants. Oecologia 2005, 144, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Liu, Y.; Ahn, J.E.; Datta, S.; Salzman, R.A.; Moon, J.; Huyghues-Despointes, B.; Pittendrigh, B.; Murdock, L.L.; Koiwa, H.; Zhu-Salzman, K. Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol. 2005, 139, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Isman, M.; Akhtar, Y. Plant natural products as a source for developing environmentally acceptable insecticides. In Insecticides Design Using Advanced Technologies Springer: Berlin/Heidelberg, Germany, 2007; pp. 235–248. [CrossRef]
- Ulrich-Merzenich, G.; Zeitler, H.; Jobst, D.; Panek, D.; Vetter, H.; Wagner, H. Application of the ‘‘-Omic-’’ technologies in phytomedicine. Phytomedicine 2007, 14, 70–82. [Google Scholar] [CrossRef]
- Naselli, M.; Urbaneja, A.; Siscaro, G.; Jaques, J.A.; Zappala, L.; Flors, V.; Perez-Hedo, M. Stage-related defense response induction in tomato plants by Nesidiocoris tenuis. Int. J. Mol. Sci. 2016, 17, 1210. [Google Scholar] [CrossRef] [PubMed]
- De Puysseleyr, V.; Hofte, M.; De Clercq, P. Ovipositing Orius laevigatus increase tomato resistance against Frankliniella occidentalis feeding by inducing the wound response. Arth-Plant Int. 2011, 5, 71–80. [Google Scholar] [CrossRef]
- Kostopoulou, S.; Ntatsi, G.; Arapis, G.; Aliferis, K.A. Assessment of the effects of metribuzin, glyphosate, and their mixtures on the metabolism of the model plant Lemna minor L. applying metabolomics. Chemosphere 2020, 239, 124582. [Google Scholar] [CrossRef]
- Kalampokis, I.F.; Kapetanakis, G.C.; Aliferis, K.A.; Diallinas, G. Multiple nucleobase transporters contribute to boscalid sensitivity in Aspergillus nidulans. Fungal Genet. Biol. 2018, 115, 52–63. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Mitsuhiro, K.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Meth. 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, K.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2017, 15, 53. [Google Scholar] [CrossRef]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dormann, P.; Weckwerth, W.; Gibon, Y.; Willmitzer, L.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Aliferis, K.A.; Cubeta, M.A.; Jabaji, S. Chemotaxonomy of fungi in the Rhizoctonia solani species complex performing GC/MS metabolite profiling. Metabolomics 2013, 9, 159–169. [Google Scholar] [CrossRef]
- Yoo, B.C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.j.; Lucas, W.J. A systemic small RNA signaling system in plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2004, 3, 1101–1108. [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef]
- Bosch, M.; Wright, L.P.; Gershenzon, J.; Wasternack, C.; Hause, B.; Schaller, A.; Stintzi, A. Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 2014, 166, 396–410. [Google Scholar] [CrossRef] [Green Version]
- SAS Institute JMP Version 14.0; SAS Institute Inc.: Cary, NC, USA, 2016.
| Gene Name | Criteria for Selection | Primers Forward (F) and Reverse (R) | Refs | |
|---|---|---|---|---|
| Proteinase inhibitor II | PI-II | Induced by the JA pathway, harmful for the digestive system of insects | F: GGATATGCCCAGGTTCAGAAGGAA R: AATAGCAACCCTTGTACCCTGTGC | [59] |
| Vegetative storage protein 2 | VSP2 | Acid phosphatase and anti-insect activity, specific to JA, induced by MYC2 | F: CTGGTTATGCAGTCCCACAAT R: ACGTCGATATTGTTTGCCAAG | This study |
| Myrosinase-binding protein 2 | MBP2 | May contribute to the production of toxins protecting against herbivory | F: CACAAACATCAGAGGCCATTT R: TGCACCATGTTTTACTGACCA | This study |
| Allene oxide synthase | AOS | Component of the JA-biosynthesis pathway, coi1-dependent | F: GATTTCGTTGTGATGGTTTCG R: TCGACGTTGAGTGTACCGTAA | This study |
| Hevein-like peptide | HEL | Antimicrobial peptide, highly induced by herbivory | F: TGTTGATTATATCCGCGATTG R: TTGGAAGGTGAACAAAATTCG | This study |
| Myelocytomatosis oncogene transcription factor 2 | MYC2 | Major transcription factor, component of the JA pathway, anti-insect activity | F: GATGATCCAACAAGCCACAGT R: CGATGTCAACGCTACCCTAAG | This study |
| TAP4 interacting protein of 41 kDa | TIP41 | Housekeeping gene | F: GCTGCGTTTCTGGCTTAGG R: ATGGAGTTTTTGAGTCTTCTGC | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmah, N.; Kaldis, A.; Kalampokis, I.; Aliferis, K.A.; Voloudakis, A.; Perdikis, D. Metabolomic and Genomic Approach to Study Defense Induction by Nesidiocoris tenuis against Tuta absoluta and Tetranychus urticae in Tomato Plants. Metabolites 2022, 12, 838. https://doi.org/10.3390/metabo12090838
Sarmah N, Kaldis A, Kalampokis I, Aliferis KA, Voloudakis A, Perdikis D. Metabolomic and Genomic Approach to Study Defense Induction by Nesidiocoris tenuis against Tuta absoluta and Tetranychus urticae in Tomato Plants. Metabolites. 2022; 12(9):838. https://doi.org/10.3390/metabo12090838
Chicago/Turabian StyleSarmah, Nomi, Athanasios Kaldis, Ioannis Kalampokis, Konstantinos A. Aliferis, Andreas Voloudakis, and Dionysios Perdikis. 2022. "Metabolomic and Genomic Approach to Study Defense Induction by Nesidiocoris tenuis against Tuta absoluta and Tetranychus urticae in Tomato Plants" Metabolites 12, no. 9: 838. https://doi.org/10.3390/metabo12090838
APA StyleSarmah, N., Kaldis, A., Kalampokis, I., Aliferis, K. A., Voloudakis, A., & Perdikis, D. (2022). Metabolomic and Genomic Approach to Study Defense Induction by Nesidiocoris tenuis against Tuta absoluta and Tetranychus urticae in Tomato Plants. Metabolites, 12(9), 838. https://doi.org/10.3390/metabo12090838