Influence of Genetic West African Ancestry on Metabolomics among Hypertensive Patients
Abstract
:1. Introduction
2. Results
2.1. Study Population
2.2. Data Processing and Quality Control of PEAR Metabolomics Data
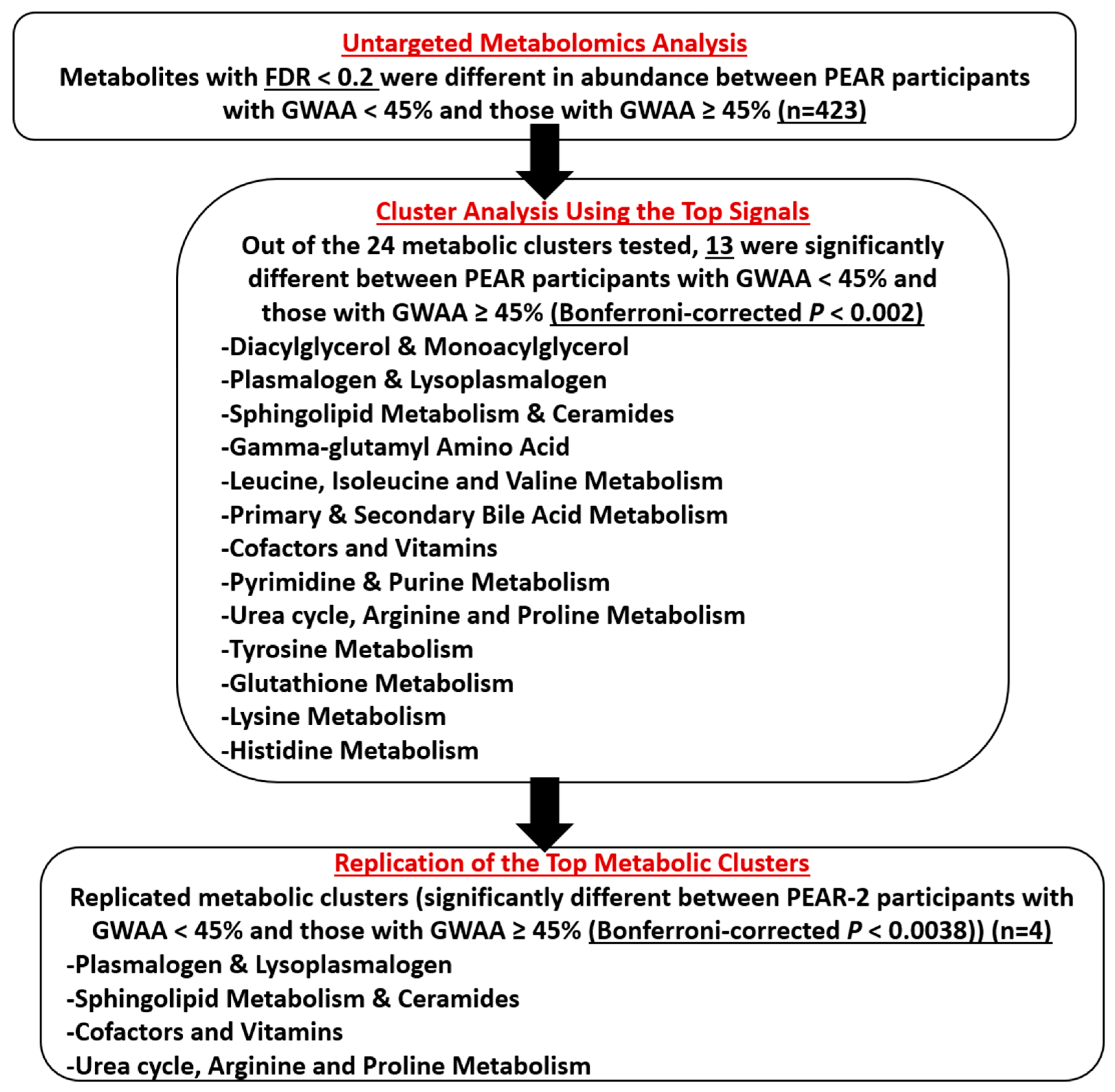
2.3. Untargeted Metabolomics Analysis
2.4. Cluster Analysis Using the Top Signals
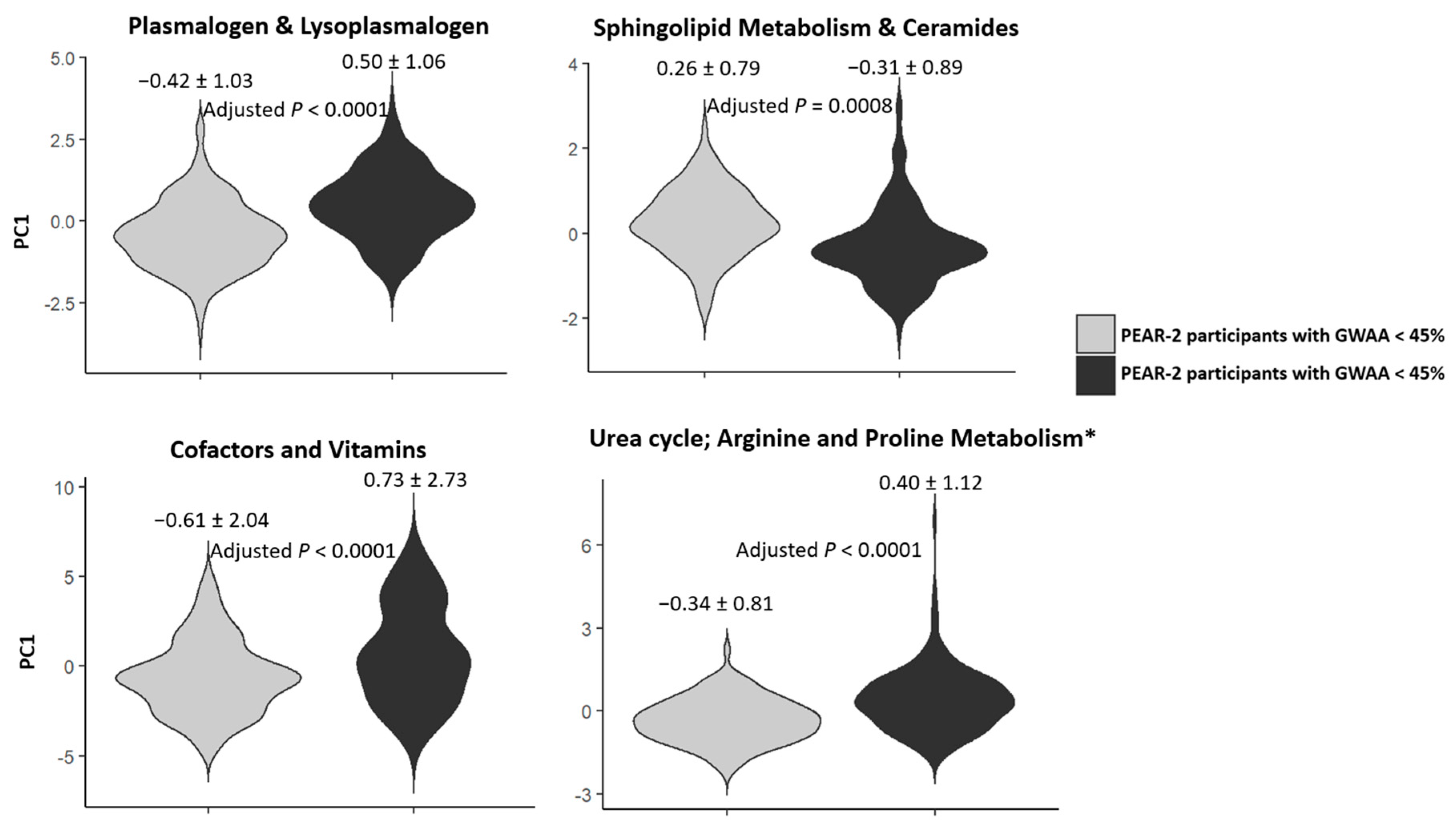
2.5. Replication of the Top Metabolic Clusters
3. Discussion
4. Materials and Methods
4.1. Study Design and Participants
4.2. Genotyping and Estimation of GWAA
4.3. Untargeted Metabolomics Profiling
4.4. Data Processing and QC of PEAR Metabolomics Data
4.5. Statistical Analyses
4.5.1. Untargeted Metabolomics Analysis (Screening Phase)
4.5.2. Cluster Analysis Using the Top Signal(s) (Discovery Phase)
4.5.3. Replication Phase of the Top Metabolic Cluster(s)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aggarwal, R.; Chiu, N.; Wadhera, R.K.; Moran, A.E.; Raber, I.; Shen, C.; Yeh, R.W.; Kazi, D.S. Racial/Ethnic Disparities in Hypertension Prevalence, Awareness, Treatment, and Control in the United States, 2013 to 2018. Hypertension 2021, 78, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control and Prevention, National Center for Health Statistics. National Vital Statistics System: Public Use Data File Documentation: Mortality Multiple Cause-of-Death Micro-Data Files. 2020. Available online: https://www.cdc.gov/nchs/data/dvs/Multiple-Cause-Record-Layout-2020.pdf (accessed on 15 June 2022).
- Brewster, L.M.; Seedat, Y.K. Why do hypertensive patients of African ancestry respond better to calcium blockers and diuretics than to ACE inhibitors and β-adrenergic blockers? A systematic review. BMC Med. 2013, 11, 141. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.G.; Coffman, T.M.; Wilcox, C.S. Pathophysiology of Hypertension: The Mosaic Theory and Beyond. Circ. Res. 2021, 128, 847–863. [Google Scholar] [CrossRef]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Pazoki, R.; Dehghan, A.; Evangelou, E.; Warren, H.; Gao, H.; Caulfield, M.; Elliott, P.; Tzoulaki, I. Genetic Predisposition to High Blood Pressure and Lifestyle Factors: Associations With Midlife Blood Pressure Levels and Cardiovascular Events. Circulation 2018, 137, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, C.P.; Ng, F.L.; Nicholls, H.L.; Gupta, A.; Barnes, M.R.; Munroe, P.B.; Caulfield, M.J. Over 1000 genetic loci influencing blood pressure with multiple systems and tissues implicated. Hum. Mol. Genet. 2019, 28, R151–R161. [Google Scholar] [CrossRef]
- Ng, F.L.; Warren, H.R.; Caulfield, M.J. Hypertension genomics and cardiovascular prevention. Ann. Transl. Med. 2018, 6, 291. [Google Scholar] [CrossRef]
- Everett, J.R.; Holmes, E.; Veselkov, K.A.; Lindon, J.C.; Nicholson, J.K. A Unified Conceptual Framework for Metabolic Phenotyping in Diagnosis and Prognosis. Trends Pharmacol. Sci. 2019, 40, 763–773. [Google Scholar] [CrossRef]
- Goïta, Y.; Chao de la Barca, J.M.; Keïta, A.; Diarra, M.B.; Dembélé, K.C.; Chabrun, F.; Dramé, B.S.I.; Kassogué, Y.; Diakité, M.; Mirebeau-Prunier, D.; et al. Sexual Dimorphism of Metabolomic Profile in Arterial Hypertension. Sci. Rep. 2020, 10, 7517. [Google Scholar] [CrossRef]
- Øvrehus, M.A.; Bruheim, P.; Ju, W.; Zelnick, L.R.; Langlo, K.A.; Sharma, K.; de Boer, I.H.; Hallan, S.I. Gene Expression Studies and Targeted Metabolomics Reveal Disturbed Serine, Methionine, and Tyrosine Metabolism in Early Hypertensive Nephrosclerosis. Kidney Int. Rep. 2019, 4, 321–333. [Google Scholar] [CrossRef]
- Ke, C.; Zhu, X.; Zhang, Y.; Shen, Y. Metabolomic characterization of hypertension and dyslipidemia. Metabolomics 2018, 14, 117. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, Y.; Li, Z.; Song, Y.; Cai, X.; Zhang, T.; Yang, L.; Li, L.; Gao, S.; Li, Y.; et al. Identification of essential hypertension biomarkers in human urine by non-targeted metabolomics based on UPLC-Q-TOF/MS. Clin. Chim. Acta 2018, 486, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Ameta, K.; Gupta, A.; Kumar, S.; Sethi, R.; Kumar, D.; Mahdi, A.A. Essential hypertension: A filtered serum based metabolomics study. Sci. Rep. 2017, 7, 2153. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yu, Z.; Deng, S.; Chen, X.; Chen, L.; Guo, Z.; Zheng, H.; Cai, D.; Wen, B.; Wu, Q.; et al. A Targeted Metabolomics MRM-MS Study on Identifying Potential Hypertension Biomarkers in Human Plasma and Evaluating Acupuncture Effects. Sci. Rep. 2016, 6, 25871. [Google Scholar] [CrossRef]
- Hao, Y.; Wang, Y.; Xi, L.; Li, G.; Zhao, F.; Qi, Y.; Liu, J.; Zhao, D. A Nested Case-Control Study of Association between Metabolome and Hypertension Risk. Biomed. Res. Int. 2016, 2016, 7646979. [Google Scholar] [CrossRef]
- Wang, L.; Hou, E.; Wang, Y.; Yang, L.; Zheng, X.; Xie, G.; Sun, Q.; Liang, M.; Tian, Z. Reconstruction and analysis of correlation networks based on GC-MS metabolomics data for young hypertensive men. Anal. Chim. Acta 2015, 854, 95–105. [Google Scholar] [CrossRef]
- van Deventer, C.A.; Lindeque, J.Z.; van Rensburg, P.J.; Malan, L.; van der Westhuizen, F.H.; Louw, R. Use of metabolomics to elucidate the metabolic perturbation associated with hypertension in a black South African male cohort: The SABPA study. J. Am. Soc. Hypertens. 2015, 9, 104–114. [Google Scholar] [CrossRef]
- Altmaier, E.; Fobo, G.; Heier, M.; Thorand, B.; Meisinger, C.; Römisch-Margl, W.; Waldenberger, M.; Gieger, C.; Illig, T.; Adamski, J.; et al. Metabolomics approach reveals effects of antihypertensives and lipid-lowering drugs on the human metabolism. Eur. J. Epidemiol. 2014, 29, 325–336. [Google Scholar] [CrossRef]
- Hiltunen, T.P.; Rimpelä, J.M.; Mohney, R.P.; Stirdivant, S.M.; Kontula, K.K. Effects of four different antihypertensive drugs on plasma metabolomic profiles in patients with essential hypertension. PLoS ONE 2017, 12, e0187729. [Google Scholar] [CrossRef]
- Huang, C.C.; Huang, Y.L.; Lin, C.H.; Chen, J.W. Plasma metabolomic profiles associated with hypertension and blood pressure in response to thiazide diuretics. Hypertens. Res. 2022, 45, 464–473. [Google Scholar] [CrossRef]
- Brocker, C.N.; Velenosi, T.; Flaten, H.K.; McWilliams, G.; McDaniel, K.; Shelton, S.K.; Saben, J.; Krausz, K.W.; Gonzalez, F.J.; Monte, A.A. Metabolomic profiling of metoprolol hypertension treatment reveals altered gut microbiota-derived urinary metabolites. Hum. Genom. 2020, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Rotroff, D.M.; Shahin, M.H.; Gurley, S.B.; Zhu, H.; Motsinger-Reif, A.; Meisner, M.; Beitelshees, A.L.; Fiehn, O.; Johnson, J.A.; Elbadawi-Sidhu, M.; et al. Pharmacometabolomic Assessments of Atenolol and Hydrochlorothiazide Treatment Reveal Novel Drug Response Phenotypes. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 669–679. [Google Scholar] [CrossRef] [PubMed]
- He, W.J.; Li, C.; Mi, X.; Shi, M.; Gu, X.; Bazzano, L.A.; Razavi, A.C.; Nierenberg, J.L.; Dorans, K.; He, H.; et al. An untargeted metabolomics study of blood pressure: Findings from the Bogalusa Heart Study. J. Hypertens. 2020, 38, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Walejko, J.M.; Kim, S.; Goel, R.; Handberg, E.M.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. Gut microbiota and serum metabolite differences in African Americans and White Americans with high blood pressure. Int. J. Cardiol. 2018, 271, 336–339. [Google Scholar] [CrossRef]
- Mels, C.M.; Delles, C.; Louw, R.; Schutte, A.E. Central systolic pressure and a nonessential amino acid metabolomics profile: The African Prospective study on the Early Detection and Identification of Cardiovascular disease and Hypertension. J. Hypertens. 2019, 37, 1157–1166. [Google Scholar] [CrossRef]
- Patel, M.J.; Batch, B.C.; Svetkey, L.P.; Bain, J.R.; Turer, C.B.; Haynes, C.; Muehlbauer, M.J.; Stevens, R.D.; Newgard, C.B.; Shah, S.H. Race and sex differences in small-molecule metabolites and metabolic hormones in overweight and obese adults. OMICS 2013, 17, 627–635. [Google Scholar] [CrossRef]
- Duggan, C.P.; Sunguya, B.; Stanford, F.C. Reporting of Race and Ethnicity in Medical and Scientific Journals. JAMA 2021, 326, 673. [Google Scholar] [CrossRef]
- Mensah, G.A.; Jaquish, C.; Srinivas, P.; Papanicolaou, G.J.; Wei, G.S.; Redmond, N.; Roberts, M.C.; Nelson, C.; Aviles-Santa, L.; Puggal, M.; et al. Emerging Concepts in Precision Medicine and Cardiovascular Diseases in Racial and Ethnic Minority Populations. Circ. Res. 2019, 125, 7–13. [Google Scholar] [CrossRef]
- Rao, S.; Segar, M.W.; Bress, A.P.; Arora, P.; Vongpatanasin, W.; Agusala, V.; Essien, U.R.; Correa, A.; Morris, A.A.; de Lemos, J.A.; et al. Association of Genetic West African Ancestry, Blood Pressure Response to Therapy, and Cardiovascular Risk Among Self-Reported Black Individuals in the Systolic Blood Pressure Reduction Intervention Trial (SPRINT). JAMA Cardiol. 2020, 6, 388–398. [Google Scholar] [CrossRef]
- Lindhorst, J.; Alexander, N.; Blignaut, J.; Rayner, B. Differences in hypertension between blacks and whites: An overview. Cardiovasc. J. Afr. 2007, 18, 241–247. [Google Scholar]
- Sagnella, G.A. Why is plasma renin activity lower in populations of African origin? J. Hum. Hypertens. 2001, 15, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2021 Update: A Report From the American Heart Association. Circulation 2021, 143, e254–e743. [Google Scholar] [CrossRef] [PubMed]
- Bozelli, J.C.; Azher, S.; Epand, R.M. Plasmalogens and Chronic Inflammatory Diseases. Front. Physiol. 2021, 12, 730829. [Google Scholar] [CrossRef] [PubMed]
- Rebholz, C.M.; Lichtenstein, A.H.; Zheng, Z.; Appel, L.J.; Coresh, J. Serum untargeted metabolomic profile of the Dietary Approaches to Stop Hypertension (DASH) dietary pattern. Am. J. Clin. Nutr. 2018, 108, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Spears, L.D.; Adak, S.; Dong, G.; Wei, X.; Spyropoulos, G.; Zhang, Q.; Yin, L.; Feng, C.; Hu, D.; Lodhi, I.J.; et al. Endothelial ether lipids link the vasculature to blood pressure, behavior, and neurodegeneration. J. Lipid Res. 2021, 62, 100079. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Howard, A.G.; Tsilimigras, M.C.B.; Avery, C.L.; Meyer, K.A.; Sha, W.; Sun, S.; Zhang, J.; Su, C.; et al. Gut Microbiota and Host Plasma Metabolites in Association with Blood Pressure in Chinese Adults. Hypertension 2021, 77, 706–717. [Google Scholar] [CrossRef]
- Hammad, S.M.; Hardin, J.R.; Wilson, D.A.; Twal, W.O.; Nietert, P.J.; Oates, J.C. Race disparity in blood sphingolipidomics associated with lupus cardiovascular comorbidity. PLoS ONE 2019, 14, e0224496. [Google Scholar] [CrossRef]
- Spijkers, L.J.; van den Akker, R.F.; Janssen, B.J.; Debets, J.J.; De Mey, J.G.; Stroes, E.S.; van den Born, B.J.; Wijesinghe, D.S.; Chalfant, C.E.; MacAleese, L.; et al. Hypertension is associated with marked alterations in sphingolipid biology: A potential role for ceramide. PLoS ONE 2011, 6, e21817. [Google Scholar] [CrossRef]
- Spijkers, L.J.; Alewijnse, A.E.; Peters, S.L. Sphingolipids and the orchestration of endothelium-derived vasoactive factors: When endothelial function demands greasing. Mol. Cells 2010, 29, 105–111. [Google Scholar] [CrossRef]
- Wawrzyniak, R.; Mpanga, A.Y.; Struck-Lewicka, W.; Kordalewska, M.; Polonis, K.; Patejko, M.; Mironiuk, M.; Szyndler, A.; Chrostowska, M.; Hoffmann, M.; et al. Untargeted Metabolomics Provides Insight into the Mechanisms Underlying Resistant Hypertension. Curr. Med. Chem. 2019, 26, 232–243. [Google Scholar] [CrossRef]
- Shahin, M.H.; Gong, Y.; Frye, R.F.; Rotroff, D.M.; Beitelshees, A.L.; Baillie, R.A.; Chapman, A.B.; Gums, J.G.; Turner, S.T.; Boerwinkle, E.; et al. Sphingolipid Metabolic Pathway Impacts Thiazide Diuretics Blood Pressure Response: Insights From Genomics, Metabolomics, and Lipidomics. J. Am. Heart Assoc. 2017, 7, e006656. [Google Scholar] [CrossRef]
- Mehanna, M.; McDonough, C.W.; Smith, S.M.; Gong, Y.; Gums, J.G.; Chapman, A.B.; Johnson, J.A.; McIntyre, L.; Cooper-DeHoff, R.M. Metabolomics Signature of Plasma Renin Activity and Linkage with Blood Pressure Response to Beta Blockers and Thiazide Diuretics in Hypertensive European American Patients. Metabolites 2021, 11, 645. [Google Scholar] [CrossRef]
- Ford, E.S.; Schleicher, R.L.; Mokdad, A.H.; Ajani, U.A.; Liu, S. Distribution of serum concentrations of alpha-tocopherol and gamma-tocopherol in the US population. Am. J. Clin. Nutr. 2006, 84, 375–383. [Google Scholar] [CrossRef]
- Boshtam, M.; Rafiei, M.; Sadeghi, K.; Sarraf-Zadegan, N. Vitamin E can reduce blood pressure in mild hypertensives. Int. J. Vitam. Nutr. Res. 2002, 72, 309–314. [Google Scholar] [CrossRef]
- Huang, J.; Weinstein, S.J.; Yu, K.; Männistö, S.; Albanes, D. Relationship Between Serum Alpha-Tocopherol and Overall and Cause-Specific Mortality. Circ. Res. 2019, 125, 29–40. [Google Scholar] [CrossRef]
- Loo, R.L.; Zou, X.; Appel, L.J.; Nicholson, J.K.; Holmes, E. Characterization of metabolic responses to healthy diets and association with blood pressure: Application to the Optimal Macronutrient Intake Trial for Heart Health (OmniHeart), a randomized controlled study. Am. J. Clin. Nutr. 2018, 107, 323–334. [Google Scholar] [CrossRef]
- Huynh, P.K.; Wilder, J.; Hiller, S.; Hagaman, J.; Takahashi, N.; Maeda-Smithies, N.; Li, F. Beneficial effects of nicotinamide on hypertensive mice with impaired endothelial nitric oxide function. J. Exp. Nephrol. 2020, 1, 1–8. [Google Scholar]
- Kane, A.E.; Sinclair, D.A. Sirtuins and NAD. Circ. Res. 2018, 123, 868–885. [Google Scholar] [CrossRef]
- Lecompte, S.; Szabo de Edelenyi, F.; Goumidi, L.; Maiani, G.; Moschonis, G.; Widhalm, K.; Molnár, D.; Kafatos, A.; Spinneker, A.; Breidenassel, C.; et al. Polymorphisms in the CD36/FAT gene are associated with plasma vitamin E concentrations in humans. Am. J. Clin. Nutr. 2011, 93, 644–651. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Major, J.M.; Yu, K.; Wheeler, W.; Zhang, H.; Cornelis, M.C.; Wright, M.E.; Yeager, M.; Snyder, K.; Weinstein, S.J.; Mondul, A.; et al. Genome-wide association study identifies common variants associated with circulating vitamin E levels. Hum. Mol. Genet. 2011, 20, 3876–3883. [Google Scholar] [CrossRef]
- Glyn, M.C.; Anderssohn, M.; Lüneburg, N.; Van Rooyen, J.M.; Schutte, R.; Huisman, H.W.; Fourie, C.M.; Smith, W.; Malan, L.; Malan, N.T.; et al. Ethnicity-specific differences in L-arginine status in South African men. J. Hum. Hypertens. 2012, 26, 737–743. [Google Scholar] [CrossRef]
- Atzler, D.; Gore, M.O.; Ayers, C.R.; Choe, C.U.; Böger, R.H.; de Lemos, J.A.; McGuire, D.K.; Schwedhelm, E. Homoarginine and cardiovascular outcome in the population-based Dallas Heart Study. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2501–2507. [Google Scholar] [CrossRef]
- Craig, A.; M C Mels, C.; Tsikas, D.; Boeger, R.H.; Schwedhelm, E.; Schutte, A.E.; Kruger, R. Central systolic blood pressure relates inversely to nitric oxide synthesis in young black adults: The African-PREDICT study. J. Hum. Hypertens. 2021, 35, 985–993. [Google Scholar] [CrossRef]
- Dong, J.Y.; Qin, L.Q.; Zhang, Z.; Zhao, Y.; Wang, J.; Arigoni, F.; Zhang, W. Effect of oral L-arginine supplementation on blood pressure: A meta-analysis of randomized, double-blind, placebo-controlled trials. Am. Heart J. 2011, 162, 959–965. [Google Scholar] [CrossRef]
- Monti, L.D.; Casiraghi, M.C.; Setola, E.; Galluccio, E.; Pagani, M.A.; Quaglia, L.; Bosi, E.; Piatti, P. L-arginine enriched biscuits improve endothelial function and glucose metabolism: A pilot study in healthy subjects and a cross-over study in subjects with impaired glucose tolerance and metabolic syndrome. Metabolism 2013, 62, 255–264. [Google Scholar] [CrossRef]
- Jeremy, R.W.; McCarron, H.; Sullivan, D. Effects of dietary L-arginine on atherosclerosis and endothelium-dependent vasodilatation in the hypercholesterolemic rabbit. Response according to treatment duration, anatomic site, and sex. Circulation 1996, 94, 498–506. [Google Scholar] [CrossRef]
- Blum, A.; Hathaway, L.; Mincemoyer, R.; Schenke, W.H.; Kirby, M.; Csako, G.; Waclawiw, M.A.; Panza, J.A.; Cannon, R.O. Oral L-arginine in patients with coronary artery disease on medical management. Circulation 2000, 101, 2160–2164. [Google Scholar] [CrossRef]
- van der Zwan, L.P.; Davids, M.; Scheffer, P.G.; Dekker, J.M.; Stehouwer, C.D.; Teerlink, T. L-Homoarginine and L-arginine are antagonistically related to blood pressure in an elderly population: The Hoorn study. J. Hypertens. 2013, 31, 1114–1123. [Google Scholar] [CrossRef]
- Kazmi, N.; Elliott, H.R.; Burrows, K.; Tillin, T.; Hughes, A.D.; Chaturvedi, N.; Gaunt, T.R.; Relton, C.L. Associations between high blood pressure and DNA methylation. PLoS ONE 2020, 15, e0227728. [Google Scholar] [CrossRef]
- Yuswan, M.H.; A Jalil, N.H.; Mohamad, H.; Keso, S.; Mohamad, N.A.; Tengku Md Yusoff, T.S.; Ismail, N.F.; Abdul Manaf, Y.N.; Mohd Hashim, A.; Mohd Desa, M.N.; et al. Hydroxyproline determination for initial detection of halal-critical food ingredients (gelatin and collagen). Food Chem. 2021, 337, 127762. [Google Scholar] [CrossRef]
- Querejeta, R.; López, B.; González, A.; Sánchez, E.; Larman, M.; Martínez Ubago, J.L.; Díez, J. Increased collagen type I synthesis in patients with heart failure of hypertensive origin: Relation to myocardial fibrosis. Circulation 2004, 110, 1263–1268. [Google Scholar] [CrossRef]
- Jaisson, S.; Pietrement, C.; Gillery, P. Carbamylation-derived products: Bioactive compounds and potential biomarkers in chronic renal failure and atherosclerosis. Clin. Chem. 2011, 57, 1499–1505. [Google Scholar] [CrossRef]
- Jaisson, S.; Kerkeni, M.; Santos-Weiss, I.C.; Addad, F.; Hammami, M.; Gillery, P. Increased serum homocitrulline concentrations are associated with the severity of coronary artery disease. Clin. Chem. Lab. Med. 2015, 53, 103–110. [Google Scholar] [CrossRef]
- Kraus, L.M.; Gaber, L.; Handorf, C.R.; Marti, H.P.; Kraus, A.P. Carbamoylation of glomerular and tubular proteins in patients with kidney failure: A potential mechanism of ongoing renal damage. Swiss Med. Wkly. 2001, 131, 139–145. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Frye, R.F.; Zhu, H.; Gong, Y.; Boyle, S.; Churchill, E.; Cooper-Dehoff, R.M.; Beitelshees, A.L.; Chapman, A.B.; Fiehn, O.; et al. Pharmacometabolomics reveals racial differences in response to atenolol treatment. PLoS ONE 2013, 8, e57639. [Google Scholar] [CrossRef]
- Johnson, J.A.; Boerwinkle, E.; Zineh, I.; Chapman, A.B.; Bailey, K.; Cooper-DeHoff, R.M.; Gums, J.; Curry, R.W.; Gong, Y.; Beitelshees, A.L.; et al. Pharmacogenomics of antihypertensive drugs: Rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am. Heart J. 2009, 157, 442–449. [Google Scholar] [CrossRef]
- Mehanna, M.; Gong, Y.; McDonough, C.W.; Beitelshees, A.L.; Gums, J.G.; Chapman, A.B.; Schwartz, G.L.; Johnson, J.A.; Turner, S.T.; Cooper-DeHoff, R.M. Blood pressure response to metoprolol and chlorthalidone in European and African Americans with hypertension. J. Clin. Hypertens. 2017, 19, 1301–1308. [Google Scholar] [CrossRef]
- Singh, S.; McDonough, C.W.; Gong, Y.; Alghamdi, W.A.; Arwood, M.J.; Bargal, S.A.; Dumeny, L.; Li, W.Y.; Mehanna, M.; Stockard, B.; et al. Genome Wide Association Study Identifies the HMGCS2 Locus to be Associated With Chlorthalidone Induced Glucose Increase in Hypertensive Patients. J. Am. Heart. Assoc. 2018, 7, e007339. [Google Scholar] [CrossRef]
- Gong, Y.; Wang, Z.; Beitelshees, A.L.; McDonough, C.W.; Langaee, T.Y.; Hall, K.; Schmidt, S.O.; Curry, R.W.; Gums, J.G.; Bailey, K.R.; et al. Pharmacogenomic Genome-Wide Meta-Analysis of Blood Pressure Response to β-Blockers in Hypertensive African Americans. Hypertension 2016, 67, 556–563. [Google Scholar] [CrossRef]
- Kosoy, R.; Nassir, R.; Tian, C.; White, P.A.; Butler, L.M.; Silva, G.; Kittles, R.; Alarcon-Riquelme, M.E.; Gregersen, P.K.; Belmont, J.W.; et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum. Mutat. 2009, 30, 69–78. [Google Scholar] [CrossRef]
- Keene, K.L.; Mychaleckyj, J.C.; Leak, T.S.; Smith, S.G.; Perlegas, P.S.; Divers, J.; Langefeld, C.D.; Freedman, B.I.; Bowden, D.W.; Sale, M.M. Exploration of the utility of ancestry informative markers for genetic association studies of African Americans with type 2 diabetes and end stage renal disease. Hum. Genet. 2008, 124, 147–154. [Google Scholar] [CrossRef]
- Collins-Schramm, H.E.; Phillips, C.M.; Operario, D.J.; Lee, J.S.; Weber, J.L.; Hanson, R.L.; Knowler, W.C.; Cooper, R.; Li, H.; Seldin, M.F. Ethnic-difference markers for use in mapping by admixture linkage disequilibrium. Am. J. Hum. Genet. 2002, 70, 737–750. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Liu, Y.; Nyunoya, T.; Leng, S.; Belinsky, S.A.; Tesfaigzi, Y.; Bruse, S. Softwares and methods for estimating genetic ancestry in human populations. Hum. Genom. 2013, 7, 1. [Google Scholar] [CrossRef]
- Evans, A.M.; Br, B.; Liu, Q.; Mitchell, M.W.; Rj, R.; Dai, H.; Sj, S.; DeHaven, C.D.; Lad, M. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High- Throughput Profiling Metabolomics. Metabolomics 2014, 4, 2. [Google Scholar]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [Google Scholar] [CrossRef]
- Bryc, K.; Durand, E.Y.; Macpherson, J.M.; Reich, D.; Mountain, J.L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015, 96, 37–53. [Google Scholar] [CrossRef]
- Stone, E.A.; Ayroles, J.F. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009, 5, e1000479. [Google Scholar] [CrossRef] [PubMed]
- Knief, U.; Forstmeier, W. Violating the normality assumption may be the lesser of two evils. Behav. Res. Methods 2021, 53, 2576–2590. [Google Scholar] [CrossRef]

| PEAR | PEAR-2 | |||||
|---|---|---|---|---|---|---|
| Variable | Participants with GWAA < 45% (n = 383) | Participants with GWAA ≥ 45% (n = 250) | p-Value | Participants with GWAA < 45% (n = 224) | Participants with GWAA ≥ 45% (n = 187) | p-Value |
| Age, years | 50.0 ± 9.8 | 47.3 ± 8.9 | 0.0005 | 50.8 ± 9.0 | 50.2 ± 9.1 | 0.5 |
| Females, N (%) | 163 (42.6%) | 165 (66%) | <0.0001 | 98 (43.8%) | 100 (53.5%) | 0.05 |
| BMI, kg/m2 | 30.5 ± 5.4 | 31.5 ± 6.0 | 0.02 | 30.8 ± 5.0 | 31.1 ± 5.4 | 0.5 |
| GWAA (%) | 1.6% (1–3.4%) | 85.4% (78.8–90.6%) | <0.0001 | 1.5% (0.9–3.6%) | 84.8% (79.2–90.5%) | <0.0001 |
| Baseline PRA, ng/mL/h | 0.9 (0.5–1.5) | 0.4 (0.2–0.6) | <0.0001 | 0.9 (0.5–1.8) | 0.4 (0.2–0.7) | <0.0001 |
| Baseline SBP, mmHg | 151.2 ± 12.2 | 151.4 ± 12.9 | 0.9 | 149.9 ± 12.4 | 150.6 ± 13.1 | 0.5 |
| Baseline DBP, mmHg | 98.0 ± 5.7 | 99.1 ± 6.7 | 0.03 | 98.0 ± 5.3 | 98.7 ± 6.0 | 0.2 |
| Metabolic Cluster | Number of PEAR Metabolites Identified from Screening Phase | % Variability Explained by PC1 | Regression p (Comparing PC1 Values between PEAR Participants with GWAA < 45% and Those with GWAA ≥ 45%) |
|---|---|---|---|
| Diacylglycerol and monoacylglycerol | 19 | 54.7% | 0.0013 * |
| Plasmalogen and lysoplasmalogen | 17 | 44.3% | <0.0001 * |
| PC, PE, PI, lysophospholipid and phospholipid metabolism | 48 | 27.4% ⁰ | 0.0034 |
| Sphingolipid metabolism and ceramides | 33 | 30.6% | <0.0001 * |
| Steroid and sterol | 12 | 34.6% | 0.036 |
| Gamma-glutamyl amino acid | 9 | 44.3% | 0.0017 * |
| Leucine, isoleucine and valine metabolism | 14 | 42.5% ⁰ | 0.0007 * |
| Primary and secondary bile acid metabolism | 24 | 21.6% | <0.0001 * |
| Cofactors and vitamins | 23 | 22.6% | <0.0001 * |
| Pyrimidine and purine metabolism | 14 | 27.4% | <0.0001 * |
| Urea cycle; arginine and proline metabolism | 12 | 37.1% ⁰ | <0.0001 * |
| Tyrosine metabolism | 7 | 64.8% ⁰ | <0.0001 * |
| Glutamate metabolism | 5 | 43.4% | 0.039 |
| Glycine, serine and threonine metabolism | 9 | 40.4% | 0.11 |
| Glutathione metabolism | 5 | 59.6% | 0.0014 * |
| Methionine, cysteine, SAM and taurine metabolism | 7 | 37.9% | 0.0026 |
| Lysine metabolism | 8 | 47.4% ⁰ | <0.0001 * |
| Alanine and aspartate metabolism | 4 | 50.3% | 0.03 |
| Histidine metabolism | 4 | 52.9% | <0.0001 * |
| Carbohydrate | 5 | 41.9% | 0.0043 |
| Creatine metabolism | 3 | 54.0% | 0.07 |
| Fatty acid | 41 | 23.9% | 0.16 |
| Fatty acid metabolism (acyl carnitine) | 21 | 31.8% | 0.0086 |
| Tryptophan metabolism | 9 | 45.3% ⁰ | 0.36 |
| Metabolic Cluster | Number of Metabolites in PEAR-2 | % Variability Explained by PC1 | Regression p (Comparing PC1 Values between PEAR-2 Participants with GWAA < 45% and Those with GWAA ≥ 45%) |
|---|---|---|---|
| Diacylglycerol and monoacylglycerol | 5 | 58.8% | 0.05 |
| Plasmalogen and lysoplasmalogen | 15 | 47.2% | <0.0001 * |
| Sphingolipid metabolism and ceramides | 14 | 42.3% | 0.0008 * |
| Gamma-glutamyl amino acid | 7 | 64.0% ⁰ | 0.19 |
| Leucine, isoleucine and valine metabolism | 12 | 56.2% | 0.57 |
| Primary and secondary bile acid metabolism | 22 | 26.6% | 0.05 |
| Cofactors and vitamins | 22 | 28.0% | <0.0001 * |
| Pyrimidine and purine metabolism | 13 | 27.4% | 0.28 |
| Urea cycle; arginine and proline metabolism | 12 | 37.5% ⁰ | <0.0001 * |
| Tyrosine metabolism | 5 | 57.6% | 0.009 |
| Glutathione metabolism | 4 | 59.1% | 0.6 |
| Lysine metabolism | 7 | 53.8% | 0.23 |
| Histidine metabolism | 4 | 46.3% | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehanna, M.; McDonough, C.W.; Smith, S.M.; Gong, Y.; Gums, J.G.; Chapman, A.B.; Johnson, J.A.; Cooper-DeHoff, R.M. Influence of Genetic West African Ancestry on Metabolomics among Hypertensive Patients. Metabolites 2022, 12, 783. https://doi.org/10.3390/metabo12090783
Mehanna M, McDonough CW, Smith SM, Gong Y, Gums JG, Chapman AB, Johnson JA, Cooper-DeHoff RM. Influence of Genetic West African Ancestry on Metabolomics among Hypertensive Patients. Metabolites. 2022; 12(9):783. https://doi.org/10.3390/metabo12090783
Chicago/Turabian StyleMehanna, Mai, Caitrin W. McDonough, Steven M. Smith, Yan Gong, John G. Gums, Arlene B. Chapman, Julie A. Johnson, and Rhonda M. Cooper-DeHoff. 2022. "Influence of Genetic West African Ancestry on Metabolomics among Hypertensive Patients" Metabolites 12, no. 9: 783. https://doi.org/10.3390/metabo12090783
APA StyleMehanna, M., McDonough, C. W., Smith, S. M., Gong, Y., Gums, J. G., Chapman, A. B., Johnson, J. A., & Cooper-DeHoff, R. M. (2022). Influence of Genetic West African Ancestry on Metabolomics among Hypertensive Patients. Metabolites, 12(9), 783. https://doi.org/10.3390/metabo12090783






