The New Era of Salivaomics in Dentistry: Frontiers and Facts in the Early Diagnosis and Prevention of Oral Diseases and Cancer
Abstract
:1. Introduction
2. Salivaomics in Dentistry
2.1. Proteomics
2.2. Transcriptomics
2.3. Metabolome
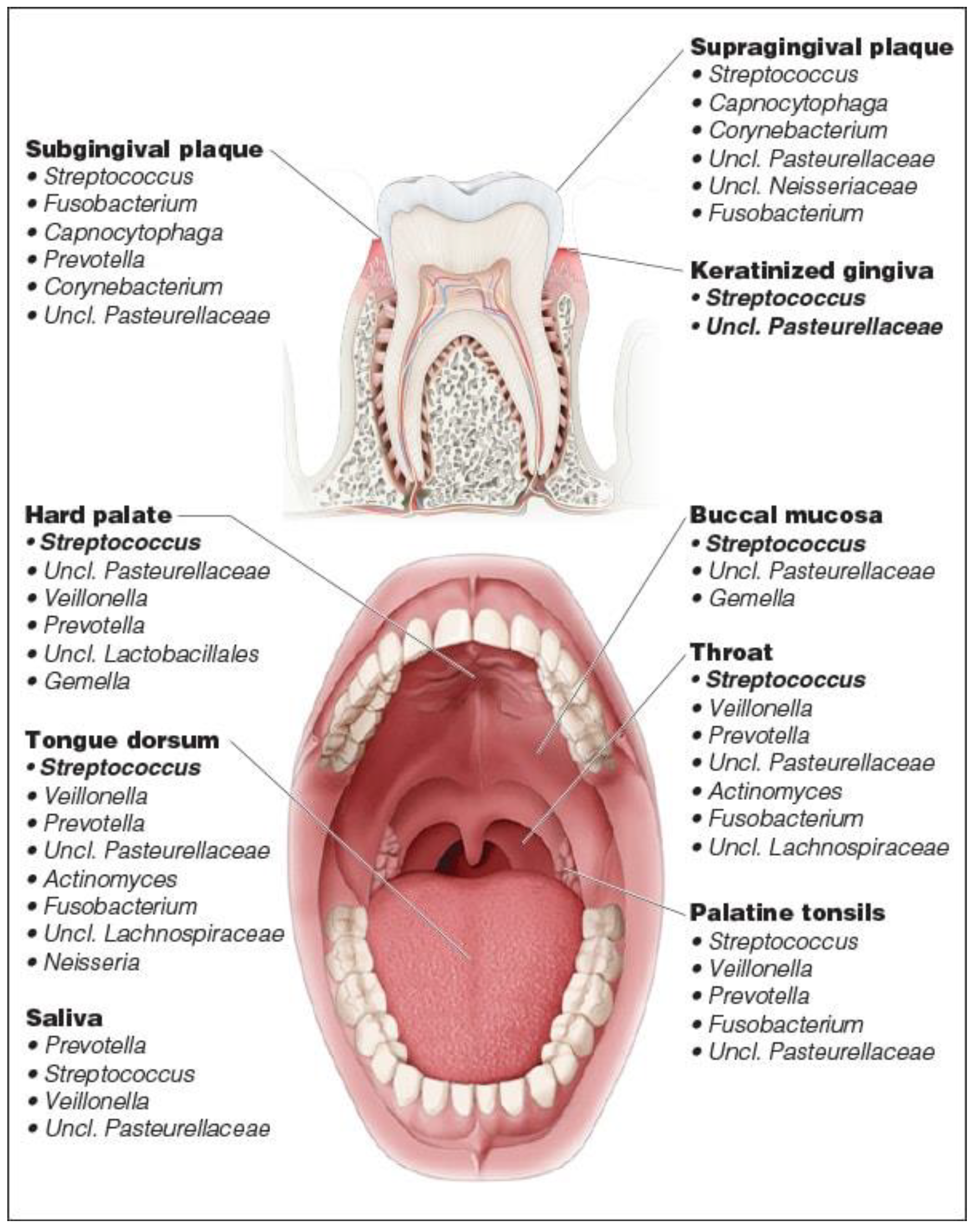
2.4. Microbiome
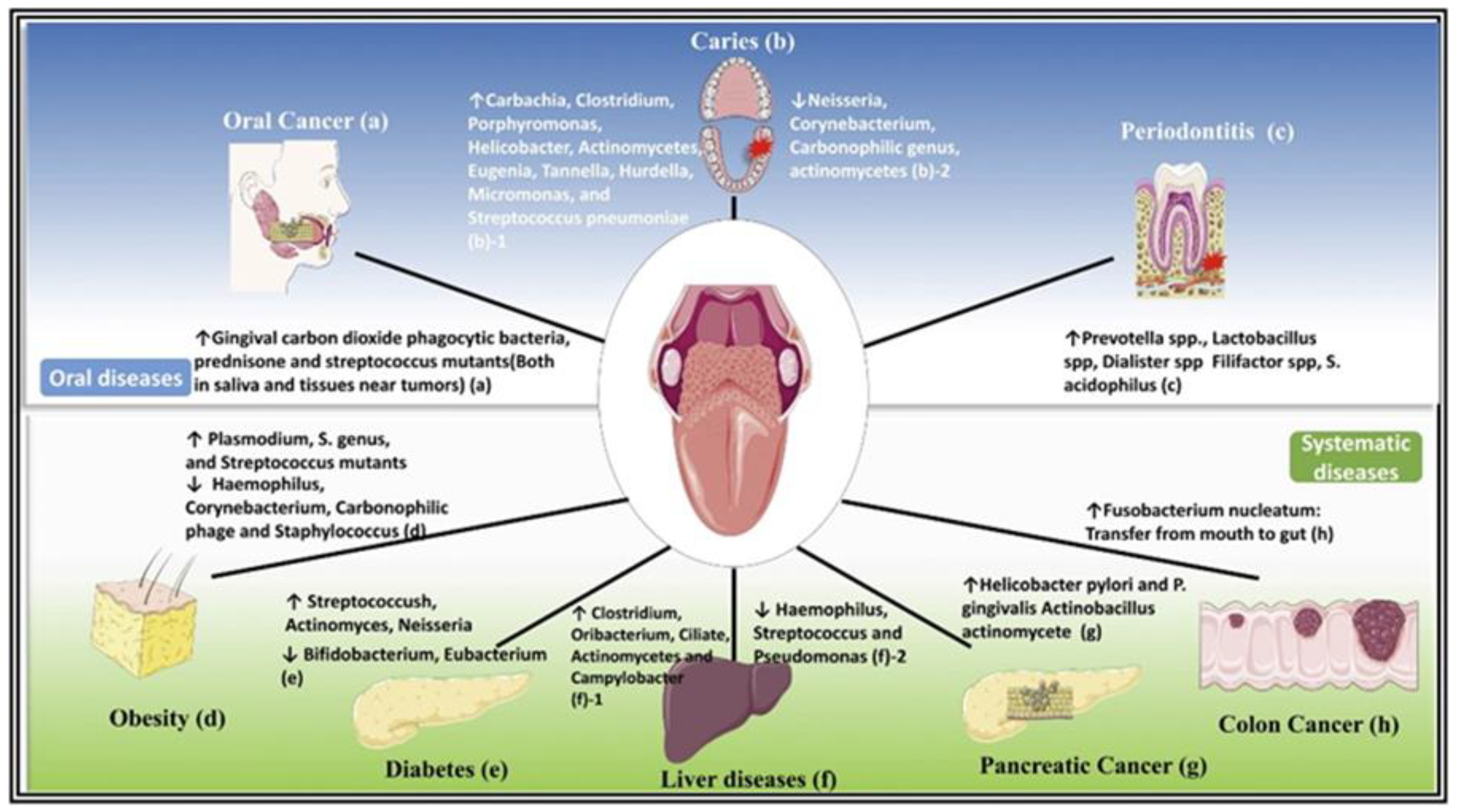
3. Salivaomics and Oral Microbiome
4. Periodontal Disease and Its Impact on Salivaomics
5. Salivaomics and Premalignant and OSCC
5.1. Oral Lichen Planus
5.2. Leukoplakia
| Disease | Biomarker | Changes | |
|---|---|---|---|
| OSCC | IL-1 | ↑ | [81] |
| OSCC | IL-6 | ↑ | [89] |
| OSCC | IL-8 | ↑ | [90] |
| OSCC | TNF-α | ↑ | [89] |
| OLP | IL-6 | ↑ | [99,100] |
| OLP | IL-8 | ↑ | [99,100] |
| OLP | TNF-α | ↑ | [99,100] |
| OLP | cortisol | ↑ | [98] |
| OLP | Nitric Oxide (NO) | ↑ | [98] |
| OLP | Reactive Oxygen Species (ROS) | ↑ | [98] |
| Leukoplakia | TNF-α | ↑ | [103] |
| Leukoplakia | IL-8 | ↑ | [104] |
| Leukoplakia | IL-6 | ↑ | [104] |
6. Discussion
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Malamud, D. Saliva as a diagnostic fluid. Dent. Clin. N. Am. 2011, 55, 159–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javaid, M.A.; Ahmed, A.S.; Durand, R.; Tran, S.D. Saliva as a diagnostic tool for oral and systemic diseases. J. Oral. Biol. Craniofac. Res. 2016, 6, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, E.; Lamster, I.B. The diagnostic applications of saliva—A review. Crit. Rev. Oral. Biol. Med. 2002, 13, 197–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sindhu, S.; Jagannathan, N. Saliva: A Cutting Edge in Diagnostic Procedures. J. Oral. Dis. 2014, 2014, 168584. [Google Scholar] [CrossRef] [Green Version]
- Davatzikos, C.; Rathore, S.; Bakas, S.; Pati, S.; Bergman, M.; Kalarot, R.; Sridharan, P.; Gastounioti, A.; Jahani, N.; Cohen, E.; et al. Cancer imaging phenomics toolkit: Quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 2018, 5, 011018. [Google Scholar] [CrossRef] [PubMed]
- Rathnayake, N.; Gieselmann, D.-R.; Heikkinen, A.M.; Tervahartiala, T.; Sorsa, T. Salivary Diagnostics—Point-of-Care diagnostics of MMP-8 in dentistry and medicine. Diagnostics 2017, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Goldoni, R.; Scolaro, A.; Boccalari, E.; Dolci, C.; Scarano, A.; Inchingolo, F.; Ravazzani, P.; Muti, P.; Tartaglia, G. Malignancies and Biosensors: A Focus on Oral Cancer Detection through Salivary Biomarkers. Biosensors 2021, 11, 396. [Google Scholar] [CrossRef]
- Shah, S. Salivaomics: The current scenario. J. Oral. Maxillofac. Pathol. 2018, 22, 375–381. [Google Scholar] [CrossRef]
- Wong, D.T. Salivaomics. J. Am. Dent. Assoc. 2012, 143, 19s–24s. [Google Scholar] [CrossRef]
- Koneru, S.; Tanikonda, R. Salivaomics—A promising future in early diagnosis of dental diseases. Dent. Res. J. 2014, 11, 11–15. [Google Scholar]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of Salivary Biomarkers in Oral Cancer Detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis 1998, 19, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Kaczor-Urbanowicz, K.E.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsani, K.R.; Sakellari, D. Saliva proteomics updates in biomedicine. J. Biol Res. 2019, 26, 17. [Google Scholar] [CrossRef] [Green Version]
- Pennington, S.R.; Dunn, M.J. Proteomics: From Protein Sequence to Function; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Uzozie, A.C.; Aebersold, R. Advancing translational research and precision medicine with targeted proteomics. J. Proteom. 2018, 189, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Esteves, C.V.; Campos, W.G.d.; Souza, M.M.d.; Lourenço, S.V.; Siqueira, W.L.; Lemos-Júnior, C.A. Diagnostic potential of saliva proteome analysis: A review and guide to clinical practice. Braz. Oral Res. 2019, 33, e043. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Govila, V.; Saini, A. Proteomics—The research frontier in periodontics. J. Oral Biol. Craniofacial Res. 2015, 5, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Amado, F.M.; Ferreira, R.P.; Vitorino, R. One decade of salivary proteomics: Current approaches and outstanding challenges. Clin. Biochem. 2013, 46, 506–517. [Google Scholar] [CrossRef]
- Xiao, H.; Wong, D.T. Method development for proteome stabilization in human saliva. Anal. Chim. Acta 2012, 722, 63–69. [Google Scholar] [CrossRef]
- Hortin, G.L.; Sviridov, D. The dynamic range problem in the analysis of the plasma proteome. J. Proteom. 2010, 73, 629–636. [Google Scholar] [CrossRef]
- Wik, L.; Nordberg, N.; Broberg, J.; Björkesten, J.; Assarsson, E.; Henriksson, S.; Grundberg, I.; Pettersson, E.; Westerberg, C.; Liljeroth, E.; et al. Proximity Extension Assay in Combination with Next-Generation Sequencing for High-throughput Proteome-wide Analysis. Mol. Cell Proteom. 2021, 20, 100168. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, M.; Eriksson, A.; Tran, B.; Assarsson, E.; Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic. Acids Res. 2011, 39, e102. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Bucht Thorsen, S.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.J.; Fang, X.; Tan, W. Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun. 2002, 292, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Santonocito, S.; Polizzi, A.; Palazzo, G.; Isola, G. The Emerging Role of microRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives. Int. J. Mol. Sci. 2021, 22, 5456. [Google Scholar] [CrossRef]
- Kagiya, T. MicroRNAs: Potential biomarkers and therapeutic targets for alveolar bone loss in periodontal disease. Int. J. Mol. Sci. 2016, 17, 1317. [Google Scholar] [CrossRef] [Green Version]
- Kebschull, M.; Papapanou, P.N. Mini but mighty: Micro RNA s in the pathobiology of periodontal disease. Periodontology 2000 2015, 69, 201–220. [Google Scholar] [CrossRef] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Cheng, Y.; Dong, L.; Zhang, J.; Zhao, Y.; Li, Z. Recent advances in microRNA detection. Analyst 2018, 143, 1758–1774. [Google Scholar] [CrossRef]
- Castoldi, M.; Schmidt, S.; Benes, V.; Hentze, M.W.; Muckenthaler, M.U. miChip: An array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat. Protoc. 2008, 3, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Karbiener, M.; Scheideler, M. Microarray analysis of small non-coding RNAs. In Small Non-Coding RNAs; Springer: Berlin/Heidelberg, Germany, 2015; pp. 161–171. [Google Scholar]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Busk, P.K. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. 2014, 15, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkonen, J.J.; Singh, S.P.; Herrala, M.; Lappalainen, R.; Myllymaa, S.; Kullaa, A.M. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J. Periodontal. Res. 2016, 51, 431–437. [Google Scholar] [CrossRef]
- Ar, P.; Gulati, A.; Mehta, D.; Sugandhan, S. Diagnostic applications of saliva in dentistry. Int. J. Clin. Pediatr. Dent. 2009, 2, 7–13. [Google Scholar] [CrossRef]
- Martina, E.; Campanati, A.; Diotallevi, F.; Offidani, A. Saliva and Oral Diseases. J. Clin. Med. 2020, 9, 466. [Google Scholar] [CrossRef] [Green Version]
- Lederberg, J.; McCray, A.T. Ome Sweet Omics—A genealogical treasury of words. Scientist 2001, 15, 8. [Google Scholar]
- Human Oral Microbiome Database. 2016. Available online: https://www.homd.org/ (accessed on 10 April 2022).
- Takahashi, N. Microbial ecosystem in the oral cavity: Metabolic diversity in an ecological niche and its relationship with oral diseases. Int. Congr. Ser. 2005, 1284, 103–112. [Google Scholar] [CrossRef]
- Belstrøm, D. The salivary microbiota in health and disease. J. Oral Microbiol. 2020, 12, 1723975. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.; Totsika, M.; Morrison, M.; Punyadeera, C. Oral Microbiome: A New Biomarker Reservoir for Oral and Oropharyngeal Cancers. Theranostics 2017, 7, 4313–4321. [Google Scholar] [CrossRef]
- Buszewski, B.; Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V. Identification of microorganisms by modern analytical techniques. J. AOAC Int. 2017, 100, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Aro, K.; Kaczor-Urbanowicz, K.; Carreras-Presas, C.M. Salivaomics in oral cancer. Curr. Opin. Otolaryngol. Head Neck Surg. 2019, 27, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Rodriguez, R.; Trinh, M.; Gunsolley, J.; Xu, P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS ONE 2013, 8, e65520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burne, R.; Zeng, L.; Ahn, S.; Palmer, S.; Liu, Y.; Lefebure, T.; Stanhope, M.; Nascimento, M. Progress dissecting the oral microbiome in caries and health. Adv. Dent. Res. 2012, 24, 77–80. [Google Scholar] [CrossRef]
- Lynge Pedersen, A.M.; Belstrøm, D. The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent. 2019, 80, S3–S12. [Google Scholar] [CrossRef]
- Sampaio-Maia, B.; Caldas, I.M.; Pereira, M.L.; Pérez-Mongiovi, D.; Araujo, R. The Oral Microbiome in Health and Its Implication in Oral and Systemic Diseases. Adv. Appl. Microbiol. 2016, 97, 171–210. [Google Scholar] [CrossRef]
- Wirth, R.; Pap, B.; Maróti, G.; Vályi, P.; Komlósi, L.; Barta, N.; Strang, O.; Minárovits, J.; Kovács, K.L. Toward Personalized Oral Diagnosis: Distinct Microbiome Clusters in Periodontitis Biofilms. Front. Cell Infect. Microbiol. 2021, 11, 747814. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef] [Green Version]
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Sedghi, L.; DiMassa, V.; Harrington, A.; Lynch, S.V.; Kapila, Y.L. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontology 2000 2021, 87, 107–131. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Porsch, M.; Grosse, I.; Hoffmann, K.; Schaller, H.G.; Reichert, S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral. Biol. 2019, 99, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Plachokova, A.S.; Andreu-Sánchez, S.; Noz, M.P.; Fu, J.; Riksen, N.P. Oral Microbiome in Relation to Periodontitis Severity and Systemic Inflammation. Int. J. Mol. Sci. 2021, 22, 5876. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.; Haffajee, A.; Devlin, P.; Norris, C.; Posner, M.; Goodson, J. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Perera, M.; Al-Hebshi, N.N.; Speicher, D.J.; Perera, I.; Johnson, N.W. Emerging role of bacteria in oral carcinogenesis: A review with special reference to perio-pathogenic bacteria. J. Oral Microbiol. 2016, 8, 32762. [Google Scholar] [CrossRef] [Green Version]
- Bandara, H.; Panduwawala, C.; Samaranayake, L. Biodiversity of the Human Oral Mycobiome in Health and Disease. Oral Dis. 2018, 25, 363–371. [Google Scholar] [CrossRef]
- Loos, B.G.; Papantonopoulos, G.; Jepsen, S.; Laine, M.L. What is the Contribution of Genetics to Periodontal Risk? Dent. Clin. N. Am. 2015, 59, 761–780. [Google Scholar] [CrossRef] [Green Version]
- Petersen, P.E. The World Oral Health Report 2003: Continuous improvement of oral health in the 21st century—The approach of the WHO Global Oral Health Programme. Community Dent. Oral Epidemiol. 2003, 31, 3–23. [Google Scholar] [CrossRef]
- Sanz, M.; Del Castillo, A.M.; Jepsen, S.; Gonzalez-Juanatey, J.R.; D’Aiuto, F.; Bouchard, P.; Chapple, I.; Dietrich, T.; Gotsman, I.; Graziani, F.; et al. Periodontitis and Cardiovascular Diseases. Consensus Report. Glob. Heart 2020, 15, 1. [Google Scholar] [CrossRef] [Green Version]
- Ji, S.; Choi, Y. Point-of-care diagnosis of periodontitis using saliva: Technically feasible but still a challenge. Front. Cell Infect. Microbiol. 2015, 5, 65. [Google Scholar] [CrossRef]
- WHO. International Program on Chemical Safety. Biomarkers in Risk Asessment: Validity and Validation; Environmental Health Criteria 222; WHO: Geneva, Switzerland, 2001. [Google Scholar]
- Hirtz, C.; O’Flynn, R.; Voisin, P.M.; Deville de Périère, D.; Lehmann, S.; Guedes, S.; Amado, F.; Ferreira, R.; Trindade, F.; Vitorino, R. The potential impact of salivary peptides in periodontitis. Crit. Rev. Clin. Lab. Sci. 2021, 58, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Gröschl, M. Saliva: A reliable sample matrix in bioanalytics. Bioanalysis 2017, 9, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Giannobile, W.V.; Beikler, T.; Kinney, J.S.; Ramseier, C.A.; Morelli, T.; Wong, D.T. Saliva as a diagnostic tool for periodontal disease: Current state and future directions. Periodontology 2000 2009, 50, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Pouso, A.I.; Pérez-Sayáns, M.; Bravo, S.B.; López-Jornet, P.; García-Vence, M.; Alonso-Sampedro, M.; Carballo, J.; García-García, A. Protein-Based Salivary Profiles as Novel Biomarkers for Oral Diseases. Dis. Markers 2018, 2018, 6141845. [Google Scholar] [CrossRef]
- Tasoulas, J.; Patsouris, E.; Giaginis, C.; Theocharis, S. Salivaomics for oral diseases biomarkers detection. Expert Rev. Mol. Diagn. 2016, 16, 285–295. [Google Scholar] [CrossRef]
- Shin, M.S.; Kim, Y.G.; Shin, Y.J.; Ko, B.J.; Kim, S.; Kim, H.D. Deep sequencing salivary proteins for periodontitis using proteomics. Clin. Oral Investig. 2019, 23, 3571–3580. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Kc, S.; Wang, X.Z.; Gallagher, J.E. Diagnostic sensitivity and specificity of host-derived salivary biomarkers in periodontal disease amongst adults: Systematic review. J. Clin. Periodontol. 2020, 47, 289–308. [Google Scholar] [CrossRef]
- Buzalaf, M.A.R.; Ortiz, A.C.; Carvalho, T.S.; Fideles, S.O.M.; Araújo, T.T.; Moraes, S.M.; Buzalaf, N.R.; Reis, F.N. Saliva as a diagnostic tool for dental caries, periodontal disease and cancer: Is there a need for more biomarkers? Expert Rev. Mol. Diagn. 2020, 20, 543–555. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.J.; Song, Y.; Lee, H.A.; Kim, S.; Chung, J. Metabolic phenotyping of saliva to identify possible biomarkers of periodontitis using proton nuclear magnetic resonance. J. Clin. Periodontol. 2021, 48, 1240–1249. [Google Scholar] [CrossRef]
- Schmalz, G.; Li, S.; Burkhardt, R.; Rinke, S.; Krause, F.; Haak, R.; Ziebolz, D. MicroRNAs as Salivary Markers for Periodontal Diseases: A New Diagnostic Approach? Biomed. Res. Int. 2016, 2016, 1027525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, S.K.; Bhardwaj, A.; Leavesley, S.J.; Grizzle, W.E.; Singh, S.; Singh, A.P. MicroRNAs as potential clinical biomarkers: Emerging approaches for their detection. Biotech. Histochem. 2013, 88, 373–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuevas-González, M.V.; Suaste-Olmos, F.; García-Calderón, A.G.; Tovar-Carrillo, K.L.; Espinosa-Cristóbal, L.F.; Nava-Martínez, S.D.; Cuevas-González, J.C.; Zambrano-Galván, G.; Saucedo-Acuña, R.A.; Donohue-Cornejo, A. Expression of MicroRNAs in Periodontal Disease: A Systematic Review. Biomed. Res. Int. 2021, 2021, 2069410. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Fujimori, K.; Yoneda, T.; Tomofuji, T.; Ekuni, D.; Azuma, T.; Maruyama, T.; Mizuno, H.; Sugiura, Y.; Morita, M. Detection of Salivary miRNAs Reflecting Chronic Periodontitis: A Pilot Study. Molecules 2019, 24, 1034. [Google Scholar] [CrossRef] [Green Version]
- Kang, L.; Li, N.; Wang, L. The Expression of miR-23a and miR-146a in the Saliva of Patients with Periodontitis and Its Clinical Significance. Biomed. Res. Int. 2021, 2021, 5135278. [Google Scholar] [CrossRef]
- Yakob, M.; Fuentes, L.; Wang, M.B.; Abemayor, E.; Wong, D.T. Salivary biomarkers for detection of oral squamous cell carcinoma—Current state and recent advances. Curr. Oral. Health Rep. 2014, 1, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Societa Italiana di Chirurgia Orale ed Implantologia. SICOI—Manuale di Chirurgia Orale; Masson, E., Ed.; Societa Italiana di Chirurgia Orale ed Implantologia: Firenze, Italy, 2011; p. 924. [Google Scholar]
- Bagan, J.; Sarrion, G.; Jimenez, Y. Oral cancer: Clinical features. Oral Oncol. 2010, 46, 414–417. [Google Scholar] [CrossRef]
- Bigler, L.R.; Streckfus, C.F.; Dubinsky, W.P. Salivary biomarkers for the detection of malignant tumors that are remote from the oral cavity. Clin. Lab. Med. 2009, 29, 71–85. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Panta, P.; Venna, V.R. Salivary RNA signatures in oral cancer detection. Anal. Cell Pathol. 2014, 2014, 450629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radhika, T.; Jeddy, N.; Nithya, S.; Muthumeenakshi, R.M. Salivary biomarkers in oral squamous cell carcinoma—An insight. J. Oral Biol. Craniofac. Res. 2016, 6, S51–S54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chundru, V.N.S.; Nirmal, R.M.; Srikanth, B.; Bojji, M.; Midhun, N.; Lakshmi, B.J. Salivaomics for Oral Cancer Detection: An Insight. J. Pharm. Bioallied Sci. 2021, 13, S52–S56. [Google Scholar] [CrossRef] [PubMed]
- Nagler, R.M.; Barak, M.; Peled, M.; Ben-Aryeh, H.; Filatov, M.; Laufer, D. Early diagnosis and treatment monitoring roles of tumor markers Cyfra 2-11 and TPS in oral squamous cell carcinoma. Cancer 1999, 85, 1018–1025. [Google Scholar] [CrossRef]
- Govindraju, P.; Kumar, T. Genomic Alphabets of Saliva as a Biomarker in Oral Cancer. J. Indian Acad. Oral Med. Radiol. 2017, 29, 300. [Google Scholar] [CrossRef]
- Li, Y.; St John, M.A.; Zhou, X.; Kim, Y.; Sinha, U.; Jordan, R.C.; Eisele, D.; Abemayor, E.; Elashoff, D.; Park, N.H.; et al. Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.Z.; Cheng, X.Q.; Li, J.Y.; Zhang, P.; Yi, P.; Xu, X.; Zhou, X.D. Saliva in the diagnosis of diseases. Int. J. Oral Sci. 2016, 8, 133–137. [Google Scholar] [CrossRef] [Green Version]
- Triantos, D.; Horefti, E.; Paximadi, E.; Kyriakopoulou, Z.; Karakassiliotis, G.; Papanastasiou, K.; Lelekis, M.; Panos, G.; Donta-Bakoyianni, C.; Rapidis, A.; et al. Presence of human herpes virus-8 in saliva and non-lesional oral mucosa in HIV-infected and oncologic immunocompromised patients. Oral Microbiol. Immunol. 2004, 19, 201–204. [Google Scholar] [CrossRef]
- Yete, S.; Saranath, D. MicroRNAs in oral cancer: Biomarkers with clinical potential. Oral Oncol. 2020, 110, 105002. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [Green Version]
- Scott, G.K.; Goga, A.; Bhaumik, D.; Berger, C.E.; Sullivan, C.S.; Benz, C.C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007, 282, 1479–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczor-Urbanowicz, K.E.; Wei, F.; Rao, S.L.; Kim, J.; Shin, H.; Cheng, J.; Tu, M.; Wong, D.T.; Kim, Y. Clinical validity of saliva and novel technology for cancer detection. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2019, 1872, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Melguizo-Rodríguez, L.; Costela-Ruiz, V.J.; Manzano-Moreno, F.J.; Ruiz, C.; Illescas-Montes, R. Salivary Biomarkers and Their Application in the Diagnosis and Monitoring of the Most Common Oral Pathologies. Int. J. Mol. Sci. 2020, 21, 5173. [Google Scholar] [CrossRef]
- Humberto, J.S.M.; Pavanin, J.V.; Rocha, M.; Motta, A.C.F. Cytokines, cortisol, and nitric oxide as salivary biomarkers in oral lichen planus: A systematic review. Braz. Oral Res. 2018, 32, e82. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhang, J.; Sun, W.; Du, G.; Zhou, G. Inflammation-related cytokines in oral lichen planus: An overview. J. Oral Pathol. Med. 2015, 44, 1–14. [Google Scholar] [CrossRef]
- Liu, F.; Wu, J.; Ye, F. Expression of miRNA-155 and miRNA-146a in peripheral blood mononuclear cells and plasma of oral lichen planus patients. Zhonghua Kou Qiang Yi Xue Za Zhi = Zhonghua Kouqiang Yixue Zazhi= Chin. J. Stomatol. 2015, 50, 23–27. [Google Scholar]
- Mello, F.W.; Miguel, A.F.P.; Dutra, K.L.; Porporatti, A.L.; Warnakulasuriya, S.; Guerra, E.N.S.; Rivero, E.R.C. Prevalence of oral potentially malignant disorders: A systematic review and meta-analysis. J. Oral Pathol. Med. 2018, 47, 633–640. [Google Scholar] [CrossRef]
- Deepthi, G.; Nandan, S.R.; Kulkarni, P.G. Salivary Tumour Necrosis Factor-α as a Biomarker in Oral Leukoplakia and Oral Squamous Cell Carcinoma. Asian Pac. J. Cancer Prev. 2019, 20, 2087–2093. [Google Scholar] [CrossRef] [Green Version]
- Kaur, J.; Jacobs, R. Proinflammatory cytokine levels in oral lichen planus, oral leukoplakia, and oral submucous fibrosis. J. Korean Assoc. Oral Maxillofac. Surg. 2015, 41, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-D.; Karna, S.; Shin, Y.; Vu, H.; Cho, H.-J.; Kim, S. S100A8 and S100A9 in saliva, blood and gingival crevicular fluid for screening established periodontitis: A cross-sectional study. BMC Oral Health 2021, 21, 388. [Google Scholar] [CrossRef]
- Gstaiger, M.; Aebersold, R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 2009, 10, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Chiappin, S.; Antonelli, G.; Gatti, R.; De Palo, E.F. Saliva specimen: A new laboratory tool for diagnostic and basic investigation. Clin. Chim. Acta 2007, 383, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.R.; Kim, H.-R.; Chae, H.-J. Compliance with Saliva Collection Protocol in Healthy Volunteers: Strategies for Managing Risk and Errors. Int. J. Med. Sci. 2018, 15, 823–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.H.; Boland, B.; Daureeawoo, Y.; Donaldson, E.; Small, K.; Tuomainen, J. Effect of aging on stimulated salivary flow in adults. J. Am. Geriatr Soc. 2013, 61, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Shakeeb, N.; Varkey, P.; Ajit, A. Human Saliva as a Diagnostic Specimen for Early Detection of Inflammatory Biomarkers by Real-Time RT-PCR. Inflammation 2021, 44, 1713–1723. [Google Scholar] [CrossRef]
- Garewal, D.J.; Garewal, D.R. Saliva: A Diagnostic Marker in Health and Disease. Med. Dent. Sci. 2022, 1, 3–6. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zohaib, S.; Najeeb, S.; Zafar, M.S.; Slowey, P.D.; Almas, K. Human Saliva Collection Devices for Proteomics: An Update. Int. J. Mol. Sci. 2016, 17, 846. [Google Scholar] [CrossRef] [Green Version]
- Isola, G. Saliva biotechnology as a diagnostic tool for periodontal diseases: New challenges for clinical practice. Front. Biosci. Elite 2022, 14, 9. [Google Scholar] [CrossRef]


| Disease | Biomarker | Changes | Mechanism | |
|---|---|---|---|---|
| Periodontitis | IL-1β | ↑ | support bone destruction | [66] |
| Periodontitis | IgA | ↑ | Inhibition of bacterial adhesion and activity | [66] |
| Periodontitis | MMP-8 | ↑ | destruction of the supporting tissues | [67] |
| Periodontitis | Nitric Oxide (NO) | ↓ | Regulation of immune and inflammatory cell cellular mediating | [68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papale, F.; Santonocito, S.; Polizzi, A.; Giudice, A.L.; Capodiferro, S.; Favia, G.; Isola, G. The New Era of Salivaomics in Dentistry: Frontiers and Facts in the Early Diagnosis and Prevention of Oral Diseases and Cancer. Metabolites 2022, 12, 638. https://doi.org/10.3390/metabo12070638
Papale F, Santonocito S, Polizzi A, Giudice AL, Capodiferro S, Favia G, Isola G. The New Era of Salivaomics in Dentistry: Frontiers and Facts in the Early Diagnosis and Prevention of Oral Diseases and Cancer. Metabolites. 2022; 12(7):638. https://doi.org/10.3390/metabo12070638
Chicago/Turabian StylePapale, Flavia, Simona Santonocito, Alessandro Polizzi, Antonino Lo Giudice, Saverio Capodiferro, Gianfranco Favia, and Gaetano Isola. 2022. "The New Era of Salivaomics in Dentistry: Frontiers and Facts in the Early Diagnosis and Prevention of Oral Diseases and Cancer" Metabolites 12, no. 7: 638. https://doi.org/10.3390/metabo12070638
APA StylePapale, F., Santonocito, S., Polizzi, A., Giudice, A. L., Capodiferro, S., Favia, G., & Isola, G. (2022). The New Era of Salivaomics in Dentistry: Frontiers and Facts in the Early Diagnosis and Prevention of Oral Diseases and Cancer. Metabolites, 12(7), 638. https://doi.org/10.3390/metabo12070638










