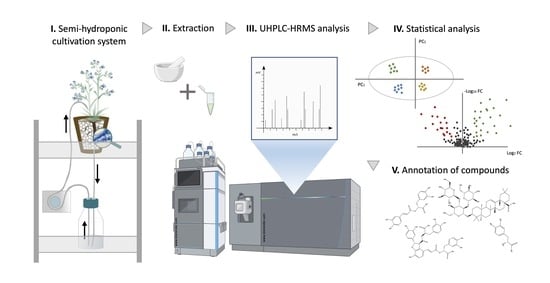
The Metabolic Profile of Anchusa officinalis L. Differs According to Its Associated Arbuscular Mycorrhizal Fungi
Abstract
1. Introduction
2. Results
2.1. Root Colonization by AMF and Plant Total Fresh Weight
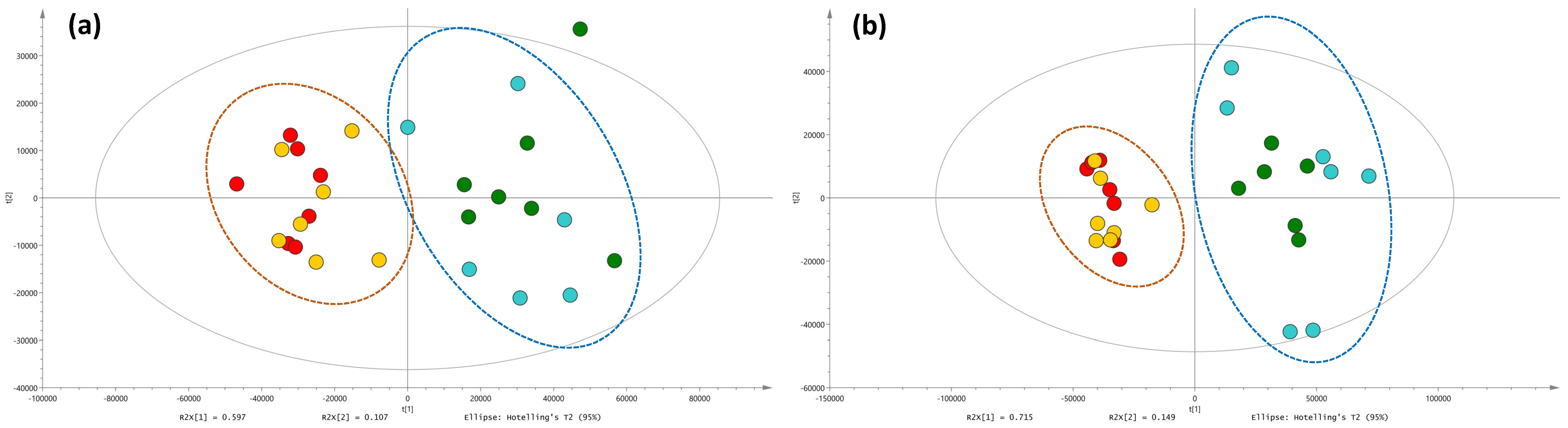
2.2. Metabolic Profiles and Metabolomic Analysis of A. officinalis Plants
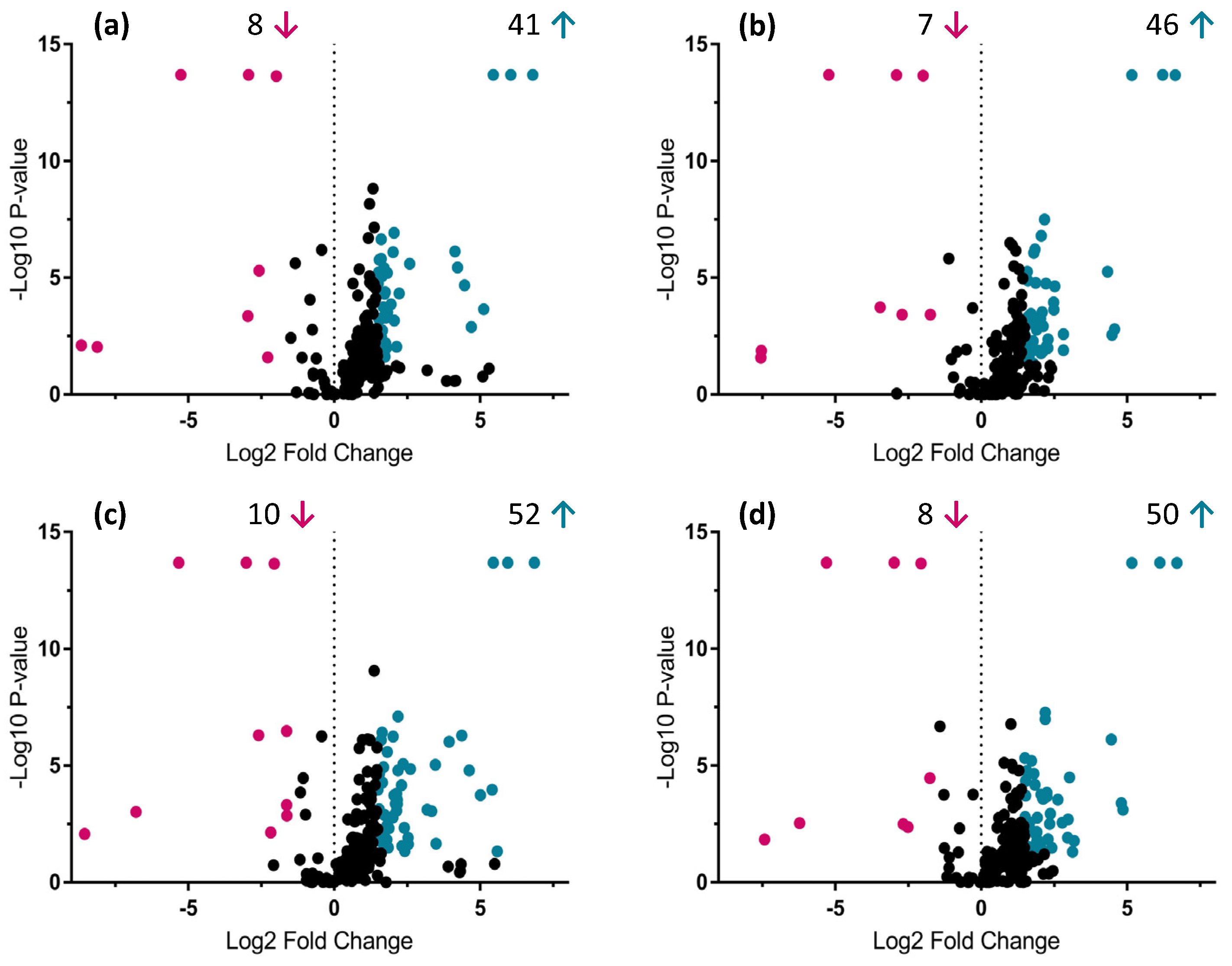
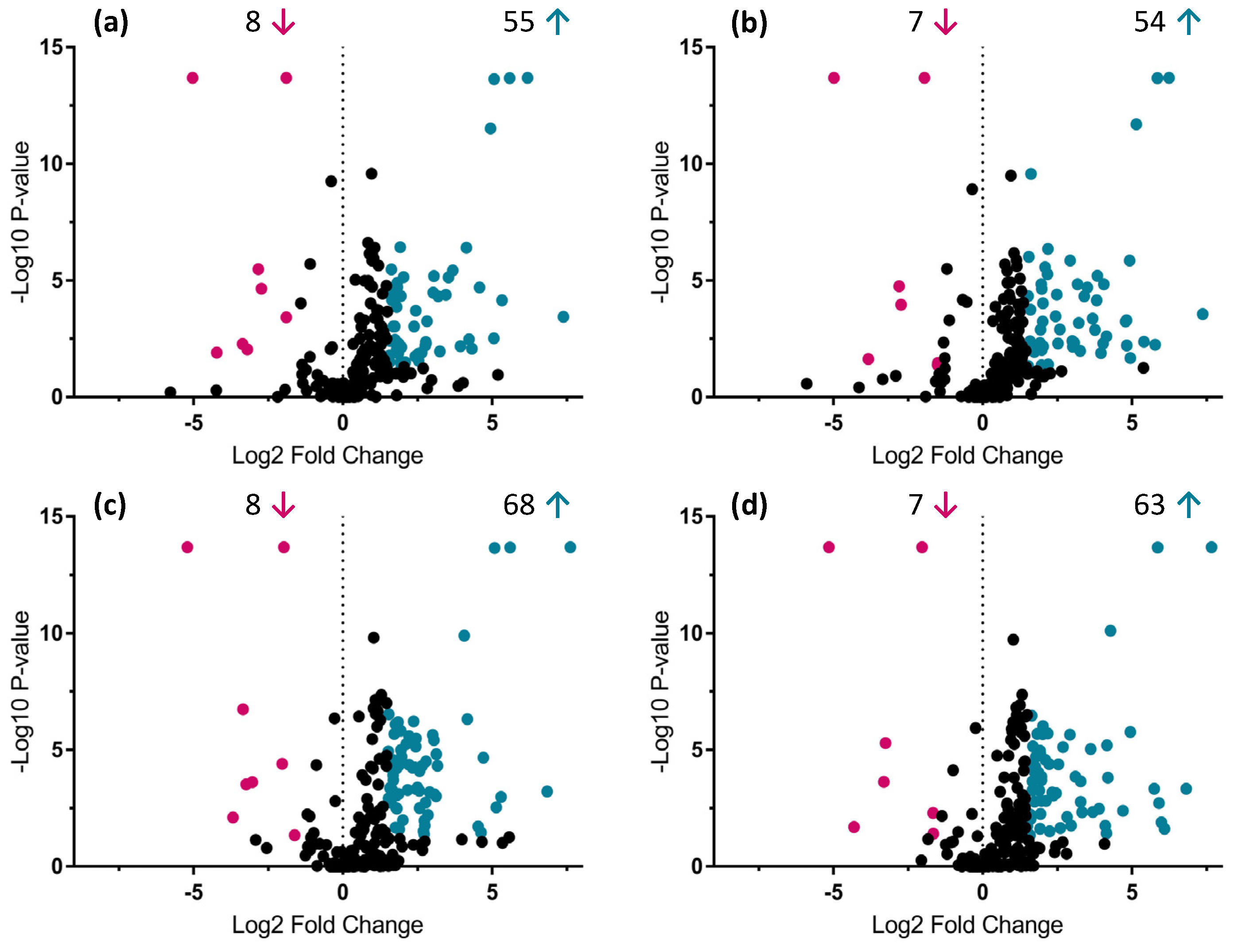
2.3. Identification of Primary and Secondary Metabolites Affected by AMF
3. Discussion
3.1. Impact of AMF Species on Primary and Secondary Metabolites in Roots and Shoots of Anchusa officinalis
3.1.1. Impact on Primary Metabolism
3.1.2. Impact on Secondary Metabolism
4. Material and Methods
4.1. Chemicals
4.2. Biological Material
4.3. Anchusa officinalis Colonization
4.4. Experimental Setup
4.5. Plant Harvest and AMF Roots Colonization
4.6. Analysis of Primary and Secondary Metabolites in Roots and Shoots of A. officinalis
4.6.1. Samples Preparation
4.6.2. UHPLC-HRMS Analysis and Untargeted Metabolomics Data Processing
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kilam, D.; Sharma, P.; Agnihotri, A.; Kharkwal, A.; Varma, A. Microbial Symbiosis and Bioactive Ingredients of Medicinal Plants. In Mycorrhiza-Eco-Physiology, Secondary Metabolites, Nanomaterials; Varma, A., Prasad, R., Tuteja, N., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 283–302. ISBN 978-3-319-57849-1. [Google Scholar]
- Kaur, S.; Suseela, V. Unraveling Arbuscular Mycorrhiza-Induced Changes in Plant Primary and Secondary Metabolome. Metabolites 2020, 10, 335. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Jakobsen, I.; Grønlund, M.; Smith, F.A. Roles of Arbuscular Mycorrhizas in Plant Phosphorus Nutrition: Interactions between Pathways of Phosphorus Uptake in Arbuscular Mycorrhizal Roots Have Important Implications for Understanding and Manipulating Plant Phosphorus Acquisition. Plant Physiol. 2011, 156, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Arato, M.; Borghi, L.; Nouri, E.; Reinhardt, D. Beneficial Services of Arbuscular Mycorrhizal Fungi–From Ecology to Application. Front. Plant Sci. 2018, 9, 1270. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of Arbuscular Mycorrhizal Fungi in Plant Growth Regulation: Implications in Abiotic Stress Tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Guo, L.-P.; Chen, B.; Hao, Z.; Wang, J.-Y.; Huang, L.-Q.; Yang, G.; Cui, X.-M.; Yang, L.; Wu, Z.-X. Arbuscular mycorrhizal symbiosis and active ingredients of medicinal plants: Current research status and prospectives. Mycorrhiza 2013, 23, 253–265. [Google Scholar] [CrossRef]
- Schliemann, W.; Ammer, C.; Strack, D. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 2008, 69, 112–146. [Google Scholar] [CrossRef]
- Hill, E.M.; Robinson, L.A.; Abdul-Sada, A.; Vanbergen, A.J.; Hodge, A.; Hartley, S.E. Arbuscular Mycorrhizal Fungi and Plant Chemical Defence: Effects of Colonisation on Aboveground and Belowground Metabolomes. J. Chem. Ecol. 2018, 44, 198–208. [Google Scholar] [CrossRef]
- Rivero, J.; Gamir, J.; Aroca, R.; Pozo, M.J.; Flors, V. Metabolic transition in mycorrhizal tomato roots. Front. Microbiol. 2015, 6, 598. [Google Scholar] [CrossRef]
- Schweiger, R.; Baier, M.C.; Persicke, M.; Müller, C. High specificity in plant leaf metabolic responses to arbuscular mycorrhiza. Nat. Commun. 2014, 5, 3886. [Google Scholar] [CrossRef]
- Bernardo, L.; Carletti, P.; Badeck, F.W.; Rizza, F.; Morcia, C.; Ghizzoni, R.; Rouphael, Y.; Colla, G.; Terzi, V.; Lucini, L. Metabolomic responses triggered by arbuscular mycorrhiza enhance tolerance to water stress in wheat cultivars. Plant Physiol. Biochem. 2019, 137, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.-L.; Belval, L.; Martin, I.R.; Roth, L.; Laloue, H.; Deglène-Benbrahim, L.; Valat, L.; Bertsch, C.; Chong, J. Arbuscular Mycorrhizal Symbiosis Triggers Major Changes in Primary Metabolism Together with Modification of Defense Responses and Signaling in Both Roots and Leaves of Vitis vinifera. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Cartabia, A.; Tsiokanos, E.; Tsafantakis, N.; Lalaymia, I.; Termentzi, A.; Miguel, M.; Fokialakis, N.; Declerck, S. The Arbuscular Mycorrhizal Fungus Rhizophagus irregularis MUCL 41833 Modulates Metabolites Production of Anchusa officinalis L. Under Semi-Hydroponic Cultivation. Front. Plant Sci. 2021, 12, 1766. [Google Scholar] [CrossRef] [PubMed]
- Zubek, S.; Błaszkowski, J.; Mleczko, P. Arbuscular mycorrhizal and dark septate endophyte associations of medicinal plants. Acta Soc. Bot. Pol. 2011, 80, 285–292. [Google Scholar] [CrossRef][Green Version]
- Giovannini, L.; Palla, M.; Agnolucci, M.; Avio, L.; Sbrana, C.; Turrini, A.; Giovannetti, M. Arbuscular Mycorrhizal Fungi and Associated Microbiota as Plant Biostimulants: Research Strategies for the Selection of the Best Performing Inocula. Agronomy 2020, 10, 106. [Google Scholar] [CrossRef]
- Declerck, S.; Strullu, D.G.; Fortin, A. Vitro Culture of Mycorrhizas; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2005; ISBN 978-3-540-24027-3. [Google Scholar]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frey, N.F.D.; Gianinazzi-Pearson, V. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, 20117–20122. [Google Scholar] [CrossRef]
- Zhang, N.; Luo, J.; Bhattacharya, D. Chapter Eight—Advances in Fungal Phylogenomics and Their Impact on Fungal Systematics. In Advances in Genetics; Fungal Phylogenetics and Phylogenomics; Townsend, J.P., Wang, Z., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 100, pp. 309–328. [Google Scholar]
- Brieudes, V.; Angelis, A.; Vougogiannopoulou, K.; Pratsinis, H.; Kletsas, D.; Mitakou, S.; Halabalaki, M.; Skaltsounis, L.A. Phytochemical Analysis and Antioxidant Potential of the Phytonutrient-Rich Decoction of Cichorium spinosum and C. intybus. Planta Medica 2016, 82, 1070–1078. [Google Scholar] [CrossRef]
- Li, X.; Yu, C.; Sun, W.; Liu, G.; Jia, J.; Wang, Y. Simultaneous determination of magnesium lithospermate B, rosmarinic acid, and lithospermic acid in beagle dog serum by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2878–2882. [Google Scholar] [CrossRef]
- Shi, X.; Yang, Y.; Ren, H.; Sun, S.; Mu, L.T.; Chen, X.; Wang, Y.; Zhang, Y.; Wang, L.H.; Sun, C. Identification of multiple components in deep eutectic solvent extract of Acanthopanax senticosus root by ultra-high-performance liquid chromatography with quadrupole orbitrap mass spectrometry. Phytochem. Lett. 2019, 35, 175–185. [Google Scholar] [CrossRef]
- Dresler, S.; Szymczak, G.; Wójcik, M. Comparison of some secondary metabolite content in the seventeen species of the Boraginaceae family. Pharm. Biol. 2017, 55, 691–695. [Google Scholar] [CrossRef]
- Liu, A.-H.; Guo, H.; Ye, M.; Lin, Y.-H.; Sun, J.-H.; Xu, M.; Guo, D.-A. Detection, characterization and identification of phenolic acids in Danshen using high-performance liquid chromatography with diode array detection and electrospray ionization mass spectrometry. J. Chromatogr. A 2007, 1161, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Miyase, T.; Kuroyanagi, M.; Umehara, K.; Noguchi, H. Oligosaccharide polyesters from roots of Polygala glomerata. Phytochemistry 1998, 47, 45–52. [Google Scholar] [CrossRef]
- Nomoto, Y.; Sugimoto, S.; Matsunami, K.; Otsuka, H. Hirtionosides A–C, gallates of megastigmane glucosides, 3-hydroxyoctanoic acid glucosides and a phenylpropanoid glucoside from the whole plants of Euphorbia hirta. J. Nat. Med. 2012, 67, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Abu-Reidah, I.M.; Arraez-Roman, D.; Carretero, A.S.; Gutierrez, A.F. Extensive characterisation of bioactive phenolic constituents from globe artichoke (Cynara scolymus L.) by HPLC–DAD-ESI-QTOF-MS. Food Chem. 2013, 141, 2269–2277. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-G.; Cha, B.-J.; Seo, W.-D.; Jeong, R.-H.; Shrestha, S.; Kim, J.-Y.; Kang, H.-C.; Baek, N.-I. Feruloyl Sucrose Esters from Oryza sativa Roots and Their Tyrosinase Inhibition Activity. Chem. Nat. Compd. 2015, 51, 1094–1098. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Ludwig, I.A.; Polyviou, T.; Malkova, D.; García, A.; Moreno-Rojas, J.M.; Crozier, A. Identification of Plasma and Urinary Metabolites and Catabolites Derived from Orange Juice (Poly)phenols: Analysis by High-Performance Liquid Chromatography–High-Resolution Mass Spectrometry. J. Agric. Food Chem. 2016, 64, 5724–5735. [Google Scholar] [CrossRef]
- Abu-Reidah, I.M.; Arráez-Román, D.; Al-Nuri, M.; Warad, I.; Segura-Carretero, A. Untargeted metabolite profiling and phytochemical analysis of Micromeria fruticosa L. (Lamiaceae) leaves. Food Chem. 2018, 279, 128–143. [Google Scholar] [CrossRef]
- Romussi, G.; Falsone, G.; Wendisch, D.; Parodi, B. Inhaltsstoffe von Boraginaceae, 7. Anchusosid-8 und -9: Zwei neue Triterpensaponine aus Anchusa officinalis L. Eur. J. Org. Chem. 1984, 1984, 1869–1872. [Google Scholar] [CrossRef]
- Barros, L.; Dueñas, M.; Dias, M.I.; Sousa, M.J.; Santos-Buelga, C.; Ferreira, I.C.F.R. Phenolic Profiles of Cultivated, in Vitro Cultured and Commercial Samples of Melissa Officinalis L. Infusions. Food Chem. 2013, 136, 1–8. [Google Scholar] [CrossRef]
- Bandara, B.M.R.; Jayasinghe, L.; Karunaratne, V.; Wannigama, G.; Kraus, W.; Bokel, M.; Sotheeswaran, S. Diploclisin, a bidesmosidic triterpenoid saponin from Diploclisia glaucescens. Phytochemistry 1989, 28, 2783–2785. [Google Scholar] [CrossRef]
- Sharma, Y.; Velamuri, R.; Fagan, J.; Schaefer, J. Full-Spectrum Analysis of Bioactive Compounds in Rosemary (Rosmarinus officinalis L.) as Influenced by Different Extraction Methods. Molecules 2020, 25, 4599. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.-W.; Shimizu, N.; Dou, D.-Q.; Takeda, T.; Fu, R.; Pei, Y.-H.; Chen, Y.-J. Five New Triterpenoid Saponins from the Roots of Platycodon grandiflorum. Chem. Pharm. Bull. 2006, 54, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Krzyżanowska-Kowalczyk, J.; Pecio, Ł.; Mołdoch, J.; Ludwiczuk, A.; Kowalczyk, M. Novel Phenolic Constituents of Pulmo-naria Officinalis L. LC-MS/MS Comparison of Spring and Autumn Metabolite Profiles. Molecules 2018, 23, 2277. [Google Scholar] [CrossRef] [PubMed]
- Lehbili, M.; Magid, A.A.; Kabouche, A.; Voutquenne-Nazabadioko, L.; Abedini, A.; Morjani, H.; Sarazin, T.; Gangloff, S.C.; Kabouche, Z. Oleanane-type triterpene saponins from Calendula stellata. Phytochemistry 2017, 144, 33–42. [Google Scholar] [CrossRef]
- Romussi, G.; Ciarallo, G.; Falsone, G.; Schneider, C. Inhaltsstoffe von Boraginaceae Triterpensaponine aus Anchusa officinalis L. Eur. J. Org. Chem. 1979, 1979, 2028–2035. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, J.; Zhang, W.; Wang, Q.; Du, Y.; Sun, Q.; Li, C.; Xu, H. Characterisation of hederacoside C metabolites using ultrahigh-performance liquid chromatography quadrupole Orbitrap mass spectrometry based on automatic fragment ion search. Phytochem. Anal. 2020, 31, 395–407. [Google Scholar] [CrossRef]
- Basiru, S.; Mwanza, H.P.; Hijri, M. Analysis of Arbuscular Mycorrhizal Fungal Inoculant Benchmarks. Microorganisms 2020, 9, 81. [Google Scholar] [CrossRef]
- Xie, M.-M.; Chen, S.-M.; Zou, Y.-N.; Srivastava, A.K.; Rahman, M.M.; Wu, Q.-S.; Kuča, K. Effects of Rhizophagus intraradices and Rhizobium trifolii on growth and N assimilation of white clover. Plant Growth Regul. 2021, 93, 311–318. [Google Scholar] [CrossRef]
- Jiménez-Arias, D.; Machado, F.J.G.; Morales-Sierra, S.; Luis, J.C.; Suarez, E.; Hernández, M.; Valdés, F.; Borges, A.A. Lettuce plants treated with L-pyroglutamic acid increase yield under water deficit stress. Environ. Exp. Bot. 2018, 158, 215–222. [Google Scholar] [CrossRef]
- Canellas, N.O.A.; Olivares, F.L.; Canellas, L.P. Metabolite fingerprints of maize and sugarcane seedlings: Searching for markers after inoculation with plant growth-promoting bacteria in humic acids. Chem. Biol. Technol. Agric. 2019, 6. [Google Scholar] [CrossRef]
- Sun, L.; Fan, K.; Wang, L.; Ma, D.; Wang, Y.; Kong, X.; Li, H.; Ren, Y.; Ding, Z. Correlation among Metabolic Changes in Tea Plant Camellia sinensis (L.) Shoots, Green Tea Quality and the Application of Cow Manure to Tea Plantation Soils. Molecules 2021, 26, 6180. [Google Scholar] [CrossRef]
- Helsper, J.P.; Loewus, F.A. Metabolism of l-Threonic Acid in Rumex x acutus L. and Pelargonium crispum (L.) L’Hér. Plant Physiol. 1982, 69, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Boskovic, I.; Đukić, D.A.; Mašković, P.; Mandić, L.; Perovic, S. Phytochemical composition and antimicrobial, antioxidant and cytotoxic activities of Anchusa officinalis L. extracts. Biologia 2018, 73, 1035–1041. [Google Scholar] [CrossRef]
- Mugford, S.T.; Osbourn, A. Saponin Synthesis and Function. In Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches; Bach, T.J., Rohmer, M., Eds.; Springer: New York, NY, USA, 2013; pp. 405–424. ISBN 978-1-4614-4063-5. [Google Scholar]
- Hussain, M.; Qasim, M.; Bamisile, B.S.; Wang, L. Role of Saponins in Plant Defense against the Diamondback Moth, Plutella Xylostella (L.). Preprints 2017, 2017060035. [Google Scholar] [CrossRef]
- Xie, W.; Hao, Z.; Zhou, X.; Jiang, X.; Xu, L.; Wu, S.; Zhao, A.; Zhang, X.; Chen, B. Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza 2018, 28, 285–300. [Google Scholar] [CrossRef]
- Chen, K.-K.; Xie, Z.-J.; Dai, W.; Wang, Q. A new oleanolic-type triterpene glycoside from Anchusa italica. Nat. Prod. Res. 2016, 31, 959–965. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Maeda, T.; Yamaguchi, K.; Kameoka, H.; Tanaka, S.; Ezawa, T.; Shigenobu, S.; Kawaguchi, M. The genome of Rhizophagus clarus HR1 reveals a common genetic basis for auxotrophy among arbuscular mycorrhizal fungi. BMC Genom. 2018, 19, 465. [Google Scholar] [CrossRef]
- Maeda, T.; Kobayashi, Y.; Kameoka, H.; Okuma, N.; Takeda, N.; Yamaguchi, K.; Bino, T.; Shigenobu, S.; Kawaguchi, M. Evidence of non-tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis. Commun. Biol. 2018, 1, 87. [Google Scholar] [CrossRef]
- Walker, C.; Schüßler, A.; Vincent, B.; Cranenbrouck, S.; Declerck, S. Anchoring the Species Rhizophagus Intraradices (Formerly Glomus Intraradices). Fungal Syst. Evol. 2021, 8, 179–197. [Google Scholar] [CrossRef]
- Luthfiana, N.; Inamura, N.; Tantriani; Sato, T.; Saito, K.; Oikawa, A.; Chen, W.; Tawaraya, K. Metabolite profiling of the hyphal exudates of Rhizophagus clarus and Rhizophagus irregularis under phosphorus deficiency. Mycorrhiza 2021, 31, 403–412. [Google Scholar] [CrossRef]
- Yang, Y.; Ou, X.; Yang, G.; Xia, Y.; Chen, M.; Guo, L.; Liu, D. Arbuscular Mycorrhizal Fungi Regulate the Growth and Phyto-Active Compound of Salvia miltiorrhiza Seedlings. Appl. Sci. 2017, 7, 68. [Google Scholar] [CrossRef]
- Garcés-Ruiz, M.; Calonne, M.; Plouznikoff, K.; Misson, C.; Navarrete-Mier, M.; Cranenbrouck, S.; Declerck, S. Dynamics of Short-Term Phosphorus Uptake by Intact Mycorrhizal and Non-mycorrhizal Maize Plants Grown in a Circulatory Semi-Hydroponic Cultivation System. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Walker, C. A Simple Blue Staining Technique for Arbuscular Mycorrhizal and Other Root-Inhabiting Fungi. Inoculum 2005, 56, 68–69. [Google Scholar]
- Pinheiro, J.C.; Bates, D.M. (Eds.) Linear Mixed-Effects Models: Basic Concepts and Examples. In Mixed-Effects Models in S and S-PLUS; Statistics and Computing; Springer: New York, NY, USA, 2000; pp. 3–56. ISBN 978-0-387-22747-4. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-projet.org (accessed on 17 June 2022).
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D. R Core Team Nlme Citation Info. Available online: https://cran.r-project.org/web/packages/nlme/citation.html (accessed on 26 October 2021).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. RStudio Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics; 2021. Available online: https://CRAN.R-project.org/package=ggplot2 (accessed on 17 June 2022).
- Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research; 2021. Available online: https://CRAN.R-project.org/package=agricolae (accessed on 17 June 2022).



| AMF Treatments | AMF Root Colonization (%) | Fresh Weight (g) | |
|---|---|---|---|
| TC | AC | ||
| R. irregularis (MUCL 41833) | 70 ± 2 a | 10 ± 3 a | 5.68 ± 1.5 a |
| R. intraradices (MUCL 49410) | 81 ± 2 b | 17 ± 3 a | 8.22 ± 1.5 a |
| R. clarus (MUCL 46238) | 74 ± 2 ab | 12 ± 3 a | 5.13 ± 1.5 a |
| R. aggregatus (MUCL 49408) | 77 ± 2 ab | 14 ± 3 a | 8.06 ± 1.5 a |
| Peak | Proposed Phytochemicals | Rt (min) | Precursor Ion—[M-H]− | m/z Calcd. | Δm (ppm) | MS/MS Fragment Ions (m/z) | Chemical Formula | Affected In | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) PMs | 1 | D-Glutamine | 1.43 | 145.0620 | 146.0686 | 1.13 | 146, 128, 102 | C5H10N2O3 | R | [20] |
| 2 | L-Aspartic acid | 1.44 | 132.0303 | 133.0370 | 0.67 | 132, 115, 88, 71 | C4H7NO4 | S, R | [20] | |
| 3 | L-Glutamic acid | 1.46 | 146.0660 | 147.0530 | 0.75 | 146, 128, 102 | C5H9NO4 | S | [20] | |
| 4 | L-Threonic acid | 1.51 | 135.0300 | 136.0366 | 1.06 | 135, 117, 89, 75, 61 | C4H8O5 | S | [20] | |
| 5 | DL-Malic acid | 1.59 | 133.0144 | 134.0210 | 0.93 | 133, 115, 89, 72, 71 | C4H6O5 | S, R | [21] | |
| 6 | DL-pyroglutamic acid | 1.65 | 128.0355 | 129.0420 | 1.28 | 128, 82, 62 | C5H7NO3 | S, R | [22] | |
| (b) SMs | 7 | Allantoin | 1.49 | 157.0359 | 158.0434 | −1.56 | 114, 97, 71, 59 | C4H6N4O3 | S | [23] |
| 8 | Danshensu | 3.53 | 197.0451 | 198.0523 | 0.64 | 179, 153, 135, 121, 73 | C9H10O5 | S | [24] | |
| 9 | Glomeratose A | 4.50 | 561.1837 | 562.1892 | 2.19 | 342, 240, 191, 163, 121, 59 | C24H34O15 | R | [25] | |
| 10 | Methyl dihydrosinapic acid glucoside | 4.90 | 401.1458 | 402.1520 | 1.15 | 208, 193, 175, 163, 121, 93, 71 | C18H26O10 | R | [26] | |
| 11 | Salicylic acid glucoside | 4.94 | 299.0776 | 300.0840 | 1.50 | 137, 93 | C13H16O8 | S | [27] | |
| 12 | 3-Feruloyl-6′acetyl sucrose | 5.05 | 559.1679 | 560.1736 | 2.16 | 193, 179, 161, 133 | C24H32O15 | R | [28] | |
| 13 | Methylsyringinoside | 5.22 | 547.2039 | 548.2100 | 1.26 | 219, 191, 176, 161, 121, 93, 71 | C24H36O14 | R | [14] | |
| 14 | Barlerin | 5.37 | 447.1514 | 448.1575 | 1.97 | 269, 161, 113, 101, 71 | C19H28O12 | S, R | - | |
| 15 | Dihydroferulic acid 4-O-glucuronide | 5.78 | 371.0990 | 372.1051 | 1.67 | 179, 163, 121, 73 | C16H20O10 | S | [29] | |
| 16 | Yunnaneic acid D | 5.79 | 539.1206 | 540.1262 | 1.82 | 297, 271, 197, 179, 161, 135, 109, 73 | C27H24O12 | S | [30] | |
| 17 | Lithospermic acid | 5.81 | 537.1050 | 538.1106 | 2.11 | 339, 295, 269, 197, 179, 161, 135, 109, 73 | C27H22O12 | S | [24] | |
| 18 | Isofraxidin | 5.98 | 221.0457 | 222.0523 | 1.30 | 177, 161, 145, 133, 123, 108, 95, 85, 67 | C11H10O5 | R | - | |
| 19 | Anchusoside-9 | 6.07 | 827.4449 | 828.4502 | 1.8 | 665, 503, 161, 113, 85, 71 | C42H68O16 | R | [31] | |
| 20 | Bayogenin triglycoside | 6.09 | 1001.4954 | 1002.5030 | 0.17 | 942, 797, 635 | C49H78O21 | S, R | - | |
| 21 | Rosmarinic acid glucoside | 6.14 | 521.1311 | 522.1368 | 2.13 | 359, 197, 179, 161, 135, 123, 73 | C24H26O13 | R | [32] | |
| 22 | Acetylanchusoside-9 | 6.22 | 869.4543 | 870.4608 | 0.33 | 707, 503, 161, 113, 85, 71 | C44H70O17 | R | [31] | |
| 23 | SA derivative I | 6.33 | 537.1049 | 538.1106 | 2.73 | 285, 185, 135, 109, 121 | C27H22O12 | R | - | |
| 24 | Methylsyringin | 6.40 | 385.1509 | 386.1571 | 1.57 | 207, 191, 176, 161, 121, 93, 71 | C18H26O9 | S, R | [14] | |
| 25 | Bayogenin diglycoside | 6.42 | 839.4435 | 840.4502 | 1.00 | 633, 423, 161, 113, 85, 71 | C43H68O16 | S | [33] | |
| 26 | Salvianolic acid (SA) A | 6.48 | 493.1150 | 494.1207 | 2.11 | 295, 267, 197, 185, 169, 135, 109, 73 | C26H22O10 | R | [34] | |
| 27 | Dihydroxybayogenin diglycoside | 6.49 | 843.4406 | 844.4451 | 2.58 | 621, 459, 161, 113, 101, 71 | C42H68O17 | S | [35] | |
| 28 | SA derivative II | 6.51 | 537.1046 | 538.1106 | 3.01 | 295, 185, 135, 109, 121 | C27H22O12 | R | - | |
| 29 | Rosmarinic acid (RA) | 6.53 | 359. 0779 | 360.0840 | 1.95 | 197, 179, 161, 135, 123, 73, 62 | C18H16O8 | S, R | [36] | |
| 30 | Salvianolic acid (SA) E | 6.70 | 717.1478 | 718.1528 | 1.72 | 339, 321, 295, 185, 161, 135, 109, 73 | C36H30O16 | R | [24] | |
| 31 | 6″-Acetyl-methyl syringin | 6.75 | 427.1616 | 428.1677 | 0.59 | 384, 219, 208, 191, 176, 161, 121, 93, 73 | C20H28O10 | R | - | |
| 32 | Clinopodic acid A | 6.98 | 343.0829 | 344.0891 | 0.59 | 197, 179, 145, 135, 123, 117, 89, 73 | C18H16O7 | R | - | |
| 33 | Dehydro SA B | 7.10 | 715.1324 | 716.1372 | 2.70 | 339, 295, 185, 135, 109, 72 | C36H28O16 | R | [24] | |
| 34 | Dehydro RA | 7.00 | 357.0622 | 358.0683 | 0.70 | 197, 179, 161, 133, 123, 73 | C18H14O8 | R | [14] | |
| 35 | Methyl RA | 7.06 | 373.0935 | 374.0996 | 0.65 | 197, 179, 161, 135, 123, 73 | C19H18O8 | S | [36] | |
| 36 | Citrinin | 7.75 | 249.0771 | 250.0836 | 0.22 | 205, 157, 143, 122, 104 | C13H14O5 | R | [22] | |
| 37 | Malonylanchusoside-2 | 8.15 | 1027.5135 | 1028.5187 | 1.38 | 779, 659, 617, 599, 455, 159, 129, 113, 101, 87 | C51H80O21 | S | [37] | |
| 38 | Hydroxy Malonyl anchusoside-7 | 8.30 | 1043.5081 | 1044.5136 | 2.21 | 795, 659, 617, 471, 159, 129, 113, 101, 87 | C51H80O22 | S | - | |
| 39 | Hydroxy Dimalonylanchusoside 2/7 | 8.43 | 1129.5087 | 1130.5140 | 2.24 | 659, 471, 455, 159, 111, 101, 87 | C54H82O25 | S | - | |
| 40 | Malonylanchusoside-7 | 9.26 | 1027.5138 | 1028.5187 | 1.86 | 779, 659, 617, 599, 455, 161, 113, 101, 89 | C51H80O21 | S | [38] | |
| 41 | Gingerol | 9.54 | 293.1662 | 294.1826 | 1.39 | 236, 221, 148, 127, 97, 72 | C17H26O4 | S, R | [22] | |
| 42 | Embellin | 10.07 | 293.1766 | 294.1826 | 2.51 | 249, 193, 177, 136, 97, 79 | C17H26O4 | S, R | - |
 | ||
|---|---|---|
| Comp. | [M-H]− | MS/MS (m/z) |
| 19 | 827.4449 | 665 (E1), 503 (E2), 161, 113, 85, 71 |
| 22 | 869.4543 | 707 (E1), 503 (E2), 161, 113, 85, 71 |
| 31 | 427.1616 | 384 (K), 219 (F), 208 (G), 191(G/I), 176 (G/H), 161 (G/H/I), 121(G/I/J), 93, 73 |
| 37 | 1027.5135 | 779 (A/C), 659 (B/A1/A2), 617 (A1/A2/C), 599 (A1/A2/D), 455 (A4), 159, 129, 113, 101, 87 |
| 40 | 1027.5138 | 779 (A/C), 659 (B/A2/A3), 617 (A2/A3/C), 599 (A2/A3/D), 455 (A4), 159, 129, 113, 101, 87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsiokanos, E.; Cartabia, A.; Tsafantakis, N.; Lalaymia, I.; Termentzi, A.; Miguel, M.; Declerck, S.; Fokialakis, N. The Metabolic Profile of Anchusa officinalis L. Differs According to Its Associated Arbuscular Mycorrhizal Fungi. Metabolites 2022, 12, 573. https://doi.org/10.3390/metabo12070573
Tsiokanos E, Cartabia A, Tsafantakis N, Lalaymia I, Termentzi A, Miguel M, Declerck S, Fokialakis N. The Metabolic Profile of Anchusa officinalis L. Differs According to Its Associated Arbuscular Mycorrhizal Fungi. Metabolites. 2022; 12(7):573. https://doi.org/10.3390/metabo12070573
Chicago/Turabian StyleTsiokanos, Evangelia, Annalisa Cartabia, Nikolaos Tsafantakis, Ismahen Lalaymia, Aikaterini Termentzi, Maria Miguel, Stéphane Declerck, and Nikolas Fokialakis. 2022. "The Metabolic Profile of Anchusa officinalis L. Differs According to Its Associated Arbuscular Mycorrhizal Fungi" Metabolites 12, no. 7: 573. https://doi.org/10.3390/metabo12070573
APA StyleTsiokanos, E., Cartabia, A., Tsafantakis, N., Lalaymia, I., Termentzi, A., Miguel, M., Declerck, S., & Fokialakis, N. (2022). The Metabolic Profile of Anchusa officinalis L. Differs According to Its Associated Arbuscular Mycorrhizal Fungi. Metabolites, 12(7), 573. https://doi.org/10.3390/metabo12070573