UHPLC-MS Metabolomic Fingerprinting, Antioxidant, and Enzyme Inhibition Activities of Himantormia lugubris from Antarctica
Abstract
:1. Introduction
2. Results and Discussion
2.1. UHPLC Chromatographic Analysis of Himantormia lugubris Extracts
2.1.1. Aromatic Derivatives
2.1.2. Carbohydrates or Derivatives
2.1.3. Fatty Acids
2.1.4. Depsides
2.1.5. Dibenzofurans
2.2. Quantitative Analysis
2.3. Total Phenolic Contents and Antioxidant and Enzymatic Inhibitory Activity
2.4. Docking Assays
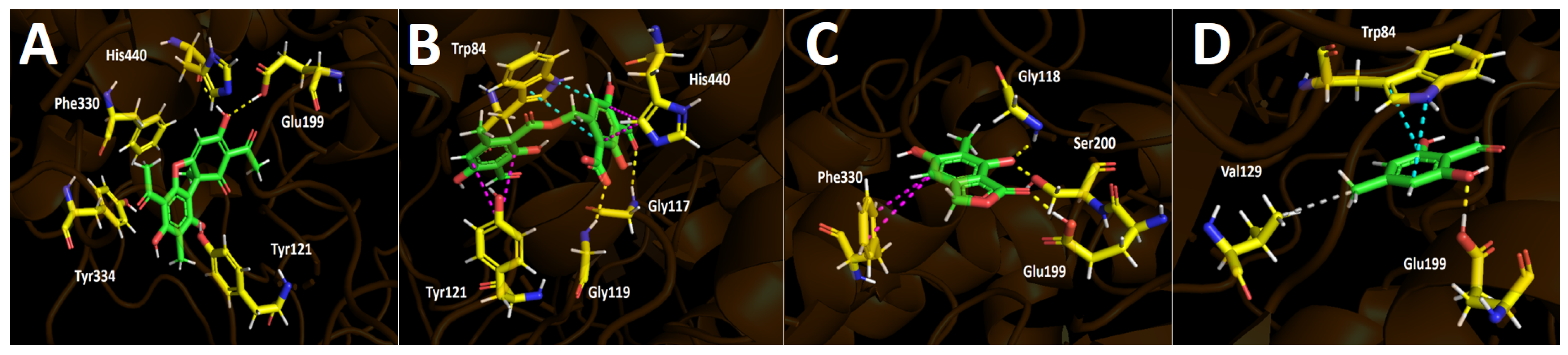
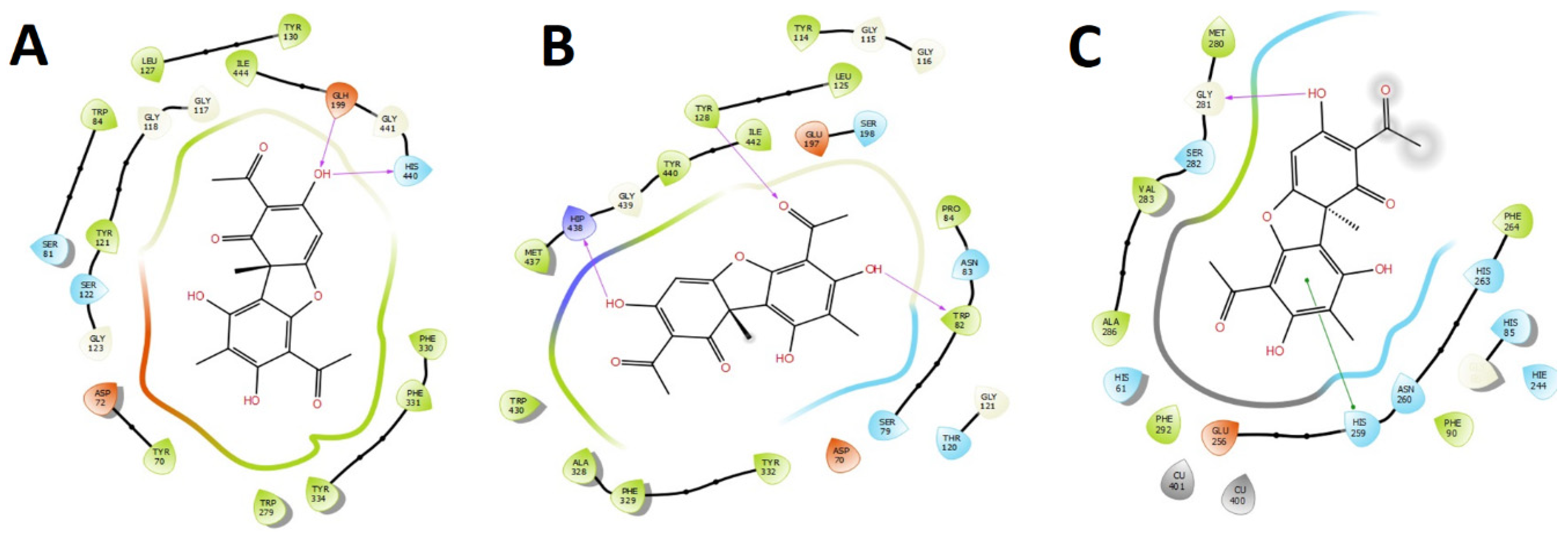
2.4.1. Acetylcholinesterase (TcAChE) Docking Results
2.4.2. Butyrylcholinesterase (hBuChE) Docking Results
2.4.3. Tyrosinase (Tyr) Docking Results
3. Materials and Methods
3.1. Chemicals
3.2. Plant Material
3.3. Extraction and Isolation Procedure
3.3.1. Extraction
3.3.2. Isolation
3.4. UHPLC–DAD–MS Instrument
3.5. LC Parameters and MS Parameters
3.6. HPLC Quantitative Analysis
3.7. Total Phenolic (TP) Content
3.8. Antioxidant Activity
3.8.1. DPPH Scavenging Activity
3.8.2. Ferric-Reducing Antioxidant Power Assay (FRAP)
3.8.3. Oxygen Radical Absorbance Capacity (ORAC) Assay
3.9. Determination of Cholinesterase Inhibition
3.10. Tyrosinase Inhibition Assay
3.11. Docking Simulations
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siddiqi, K. Noncommunicable diseases. In Public Health: An Action Guide to Improving Health; Walley, J., Wright, J., Eds.; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Rodrigues, S.A.; Terrón-Alfonso, A.; Elix, J.A.; Pérez-Ortega, S.; Tønsberg, T.; Fernández-Salegui, A.B.; Soares, A.M.V.M. Lecanora sorediomarginata, a new epiphytic lichen species discovered along the Portuguese coast. Lichenologist 2011, 43, 99–111. [Google Scholar] [CrossRef]
- Popovici, V.; Bucur, L.; Vochita, G.; Gherghel, D.; Mihai, C.T.; Rambu, D.; Calcan, S.I.; Costache, T.; Cucolea, I.E.; Matei, E.; et al. In Vitro anticancer activity and oxidative stress biomarkers status determined by Usnea barbata (L.) f.h. wigg. dry extracts. Antioxidants 2021, 10, 1141. [Google Scholar] [CrossRef]
- Okuyama, E.; Umeyama, K.; Yamazaki, M.; Kinoshita, Y.; Yamamoto, Y. Usnic acid and diffractaic acid as analgesic and antipyretic components of Usnea diffracta. Planta Med. 1995, 61, 113–115. [Google Scholar] [CrossRef]
- Furmanek, Ł.; Czarnota, P.; Seaward, M.R.D. Antifungal activity of lichen compounds against dermatophytes: A review. J. Appl. Microbiol. 2019, 127, 308–325. [Google Scholar] [CrossRef]
- Rowe, J.G.; Gimenezb, M.D.G.; Saenz Rodriguezb, M.T. Some Lichen Products Have Antimicrobial Activity. Z. Für Nat. C 1999, 54, 605–609. [Google Scholar] [CrossRef]
- Mitrović, T.; Stamenković, S.; Cvetković, V.; Tošić, S.; Stanković, M.; Radojević, I.; Stefanović, O.; Čomić, L.; Đačić, D.; Ćurčić, M.; et al. Antioxidant, Antimicrobial and Antiproliferative Activities of Five Lichen Species. Int. J. Mol. Sci. 2011, 12, 5428–5448. [Google Scholar] [CrossRef]
- Carocho, M.; Ferreira, I.C.F.R. A review on antioxidants, prooxidants and related controversy: Natural and synthetic compounds, screening and analysis methodologies and future perspectives. Food Chem. Toxicol. 2013, 51, 15–25. [Google Scholar] [CrossRef]
- Fernández-Moriano, C.; Divakar, P.K.; Crespo, A.; Gómez-Serranillos, M.P. In Vitro neuroprotective potential of lichen metabolite fumarprotocetraric acid via intracellular redox modulation. Toxicol. Appl. Pharmacol. 2017, 316, 83–94. [Google Scholar] [CrossRef]
- Kekuda, P.T.; Rao, P. Lichens as promising resources of enzyme inhibitors: A review. Prashith Kekuda J. Drug Deliv. Ther. 2019, 9, 665–676. [Google Scholar] [CrossRef]
- Sancho, L.; de los Ríos, A.; Pintado, A.; Colesie, C.; Raggio, J.; Ascaso, C.; Green, A. Himantormia lugubris, an Antarctic endemic on the edge of the lichen symbiosis. Symbiosis 2020, 82, 49–58. [Google Scholar] [CrossRef]
- González, C.; Cartagena, C.; Caballero, L.; Melo, F.; Areche, C.; Cornejo, A.; De Filippis, B.; Eckert, G.P. The Fumarprotocetraric Acid Inhibits Tau Covalently, Avoiding Cytotoxicity of Aggregates in Cells. Molecules 2021, 26, 3760. [Google Scholar] [CrossRef]
- Cetin Cakmak, K.; Gülçin, İ. Anticholinergic and antioxidant activities of usnic acid-an activity-structure insight. Toxicol. Rep. 2019, 6, 1273–1280. [Google Scholar] [CrossRef]
- Malaspina, P.; Catellani, E.; Burlando, B.; Brignole, D.; Cornara, L.; Bazzicalupo, M.; Candiani, S.; Obino, V.; De Feo, V.; Caputo, L.; et al. Depigmenting potential of lichen extracts evaluated by in vitro and in vivo tests. PeerJ 2020, 2020, e9150. [Google Scholar] [CrossRef]
- Nagatsu, T.; Nakashima, A.; Watanabe, H.; Ito, S.; Wakamatsu, K. Neuromelanin in Parkinson’s Disease: Tyrosine Hydroxylase and Tyrosinase. Int. J. Mol. Sci. 2022, 23, 4176. [Google Scholar] [CrossRef]
- Li, Q.; Mo, J.; Xiong, B.; Liao, Q.; Chen, Y.; Wang, Y.; Xing, S.; He, S.; Lyu, W.; Zhang, N.; et al. Discovery of Resorcinol-Based Polycyclic Structures as Tyrosinase Inhibitors for Treatment of Parkinson’s Disease. ACS Chem. Neurosci. 2022, 13, 81–96. [Google Scholar] [CrossRef]
- Lopes, T.I.B.; Coelho, R.G.; Honda, N.K. Inhibition of mushroom tyrosinase activity by orsellinates. Chem. Pharm. Bull. 2018, 66, 61–64. [Google Scholar] [CrossRef]
- Rocchetti, G.; Chiodelli, G.; Giuberti, G.; Ghisoni, S.; Baccolo, G.; Blasi, F.; Montesano, D.; Trevisan, M.; Lucini, L. UHPLC-ESI-QTOF-MS profile of polyphenols in Goji berries (Lycium barbarum L.) and its dynamics during in vitro gastrointestinal digestion and fermentation. J. Funct. Foods 2018, 40, 564–572. [Google Scholar] [CrossRef]
- Liu, S.; Marsol-Vall, A.; Laaksonen, O.; Kortesniemi, M.; Yang, B. Characterization and Quantification of Nonanthocyanin Phenolic Compounds in White and Blue Bilberry (Vaccinium myrtillus) Juices and Wines Using UHPLC-DAD-ESI-QTOF-MS and UHPLC-DAD. J. Agric. Food Chem. 2020, 68, 7734–7744. [Google Scholar] [CrossRef]
- Barrientos, R.; Fernández-Galleguillos, C.; Pastene, E.; Simirgiotis, M.; Romero-Parra, J.; Ahmed, S.; Echeverría, J. Metabolomic Analysis, Fast Isolation of Phenolic Compounds, and Evaluation of Biological Activities of the Bark From Weinmannia trichosperma Cav. (Cunoniaceae). Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]
- Barrientos, R.E.; Ahmed, S.; Cortés, C.; Fernández-Galleguillos, C.; Romero-Parra, J.; Simirgiotis, M.J.; Echeverría, J. Chemical Fingerprinting and Biological Evaluation of the Endemic Chilean Fruit Greigia sphacelata (Ruiz and Pav.) Regel (Bromeliaceae) by UHPLC-PDA-Orbitrap-Mass Spectrometry. Molecules 2020, 25, 3750. [Google Scholar] [CrossRef]
- Castro, O.N.; Benites, J.; Rodilla, J.; Santiago, J.C.; Simirgiotis, M.; Sepulveda, B.; Areche, C. Metabolomic Analysis of the Lichen Everniopsis trulla Using Ultra High Performance Liquid Chromatography-Quadrupole-Orbitrap Mass Spectrometry (UHPLC-Q-OT-MS). Chromatographia 2017, 80, 967–973. [Google Scholar] [CrossRef]
- Cornejo, A.; Salgado, F.; Caballero, J.; Vargas, R.; Simirgiotis, M.; Areche, C. Secondary Metabolites in Ramalina terebrata Detected by UHPLC/ESI/MS/MS and Identification of Parietin as Tau Protein Inhibitor. Int. J. Mol. Sci. 2016, 17, 1303. [Google Scholar] [CrossRef]
- Torres-Benítez, A.; Rivera-Montalvo, M.; Sepúlveda, B.; Castro, O.N.; Nagles, E.; Simirgiotis, M.J.; Garciá-Beltrán, O.; Areche, C. Metabolomic Analysis of Two Parmotrema Lichens: P. robustum (Degel.) Hale and P. andinum (Mull. rg.) Hale Using UHPLC-ESI-OT-MS-MS. Molecules 2017, 22, 1861. [Google Scholar] [CrossRef]
- Salgado, F.; Albornoz, L.; Cortéz, C.; Stashenko, E.; Urrea-Vallejo, K.; Nagles, E.; Galicia-Virviescas, C.; Cornejo, A.; Ardiles, A.; Simirgiotis, M.; et al. Secondary metabolite profiling of species of the genus usnea by UHPLC-ESI-OT-MS-MS. Molecules 2018, 23, 54. [Google Scholar] [CrossRef]
- Chae, H.-J.; Kim, G.-J.; Deshar, B.; Kim, H.-J.; Shin, M.-J.; Kwon, H.; Youn, U.-J.; Nam, J.-W.; Kim, S.-H.; Choi, H.; et al. Anticancer activity of 2-O-caffeoyl alphitolic acid extracted from the Lichen, Usnea barbata 2017-KL-10. Molecules 2021, 26, 3937. [Google Scholar] [CrossRef]
- Alahmadi, A.A. Usnic acid biological activity: History, evaluation and usage. Int. J. Basic Clin. Pharmacol. 2017, 6, 2752–2759. [Google Scholar] [CrossRef]
- Prateeksha; Paliya, B.S.; Bajpai, R.; Jadaun, V.; Kumar, J.; Kumar, S.; Upreti, D.K.; Singh, B.R.; Nayaka, S.; Joshi, Y.; et al. The genus Usnea: A potent phytomedicine with multifarious ethnobotany, phytochemistry and pharmacology. RSC Adv. 2016, 6, 21672–21696. [Google Scholar] [CrossRef]
- Duman, D.C.; Aras, S.; Atakol, O. Determination of Usnic Acid Content in Some Lichen Species Found in Anatolia. J. Appl. Biol. Sci. 2019, 2, 41–44. [Google Scholar]
- Ji, X.; Khan, I.A. Quantitative determination of usnic acid in Usnea lichen and its products by reversed-phase liquid chromatography with photodiode array detector. J. AOAC Int. 2005, 88, 1265–1268. [Google Scholar] [CrossRef]
- Cansaran, D.; Çetin, D.; Halici, M.G.; Atakol, O. Determination of usnic acid in some Rhizoplaca species from middle Anatolia and their antimicrobial activities. Z. Naturforsch.-Sect. C J. Biosci. 2006, 61, 47–51. [Google Scholar] [CrossRef]
- Popovici, V.; Bucur, L.; Popescu, A.; Schröder, V.; Costache, T.; Rambu, D.; Cucolea, I.E.; Gîrd, C.E.; Caraiane, A.; Gherghel, D.; et al. Antioxidant and Cytotoxic Activities of Usnea barbata (L.) F.H. Wigg. Dry Extracts in Different Solvents. Plants 2021, 10, 909. [Google Scholar] [CrossRef]
- Popovici, V.; Bucur, L.A.; Schröder, V.; Gherghel, D.; Mihai, C.T.; Caraiane, A.; Badea, F.C.; Vochita, G.; Badea, V. Evaluation of the Cytotoxic Activity of the Usnea barbata (L.) F.H. Wigg Dry Extract. Molecules 2020, 25, 1865. [Google Scholar] [CrossRef]
- Tang, J.Y.; Wu, K.H.; Wang, Y.Y.; Farooqi, A.A.; Huang, H.W.; Yuan, S.S.F.; Jian, R.I.; Tsao, L.Y.; Chen, P.A.; Chang, F.R.; et al. Methanol extract of usnea barbata induces cell killing, apoptosis, and dna damage against oral cancer cells through oxidative stress. Antioxidants 2020, 9, 694. [Google Scholar] [CrossRef]
- Popovici, V.; Matei, E.; Cozaru, G.C.; Aschie, M.; Bucur, L.; Rambu, D.; Costache, T.; Cucolea, I.E.; Vochita, G.; Gherghel, D.; et al. Usnic acid and Usnea barbata (L.) F.H. wigg. dry extracts promote apoptosis and DNA damage in human blood cells through enhancing ROS levels. Antioxidants 2021, 10, 1171. [Google Scholar] [CrossRef]
- Pathak, A.; Upreti, D.K.; Dikshit, A. Antidermatophytic Activity of the Fruticose Lichen Usnea orientalis. Medicines 2016, 3, 24. [Google Scholar] [CrossRef]
- Reddy, S.D.; Siva, B.; Kumar, K.; Babu, V.S.P.; Sravanthi, V.; Boustie, J.; Nayak, V.L.; Tiwari, A.K.; Rao, C.V.; Sridhar, B.; et al. Comprehensive Analysis of Secondary Metabolites in Usnea longissima (Lichenized Ascomycetes, Parmeliaceae) Using UPLC-ESI-QTOF-MS/MS and Pro-Apoptotic Activity of Barbatic Acid. Molecules 2019, 24, 2270. [Google Scholar] [CrossRef]
- Do, T.-H.; Duong, T.-H.; Nguyen, H.T.; Nguyen, T.-H.; Sichaem, J.; Nguyen, C.H.; Nguyen, H.-H.; Long, N.P. Biological Activities of Lichen-Derived Monoaromatic Compounds. Molecules 2022, 27, 2871. [Google Scholar] [CrossRef]
- Padrón, C.E.H.; Pérez, E.M.R.; Padrón, C.E.; Barrera, J.B. Chemical constituents of the lichen stereocaulon azoreum. Z. Naturforsch.-Sect. C J. Biosci. 1992, 47, 503–507. [Google Scholar] [CrossRef]
- Caccamese, S.; Compagnini, A.; Maria Toscano, R.; Cascio, O. Methyl β-orcinolcarboxylate and atranol from the lichen stereocaulon vesuvianum. J. Nat. Prod. 1986, 49, 1159–1160. [Google Scholar] [CrossRef]
- Salgado, F.; Caballero, J.; Vargas, R.; Cornejo, A.; Areche, C. Continental and Antarctic Lichens: Isolation, identification and molecular modeling of the depside tenuiorin from the Antarctic lichen Umbilicaria antarctica as tau protein inhibitor. Nat. Prod. Res. 2020, 34, 646–650. [Google Scholar] [CrossRef]
- Myllys, L.; Lindgren, H.; Aikio, S.; Häkkinen, L.; Högnabba, F. Chemical diversity and ecology of the genus Bryoria section Implexae (Parmeliaceae) in Finland. Bryologist 2016, 119, 29. [Google Scholar] [CrossRef]
- Varol, M. Anti-breast cancer and anti-angiogenic potential of a lichen-derived small-molecule: Barbatolic acid. Cytotechnology 2018, 70, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Ulus, G. Antiangiogenic properties of lichen secondary metabolites. Phyther. Res. 2021, 35, 3046–3058. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; Conde, E.; Miranda, T.; Vicente, C. Regulation mechanisms of phenolic production in the lichen Himantormia lugubris, as deduced from the analysis of metabolite accumulation. Plant Sci. 1991, 77, 1–9. [Google Scholar] [CrossRef]
- Dieu, A.; Mambu, L.; Champavier, Y.; Chaleix, V.; Sol, V.; Gloaguen, V.; Millot, M. Antibacterial activity of the lichens Usnea Florida and Flavoparmelia caperata (Parmeliaceae). Nat. Prod. Res. 2020, 34, 3358–3362. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Bórquez, J.; Schmeda-Hirschmann, G. Antioxidant capacity, polyphenolic content and tandem HPLC-DAD-ESI/MS profiling of phenolic compounds from the South American berries Luma apiculata and L. chequén. Food Chem. 2013, 139, 289–299. [Google Scholar] [CrossRef]
- Odabasoglu, F.; Aslan, A.; Cakir, A.; Suleyman, H.; Karagoz, Y.; Halici, M.; Bayir, Y. Comparison of antioxidant activity and phenolic content of three lichen species. Phyther. Res. 2004, 18, 938–941. [Google Scholar] [CrossRef]
- Fernández-Moriano, C.; Gómez-Serranillos, M.P.; Crespo, A. Antioxidant potential of lichen species and their secondary metabolites. A systematic review. Pharm. Biol. 2016, 54, 1–17. [Google Scholar] [CrossRef]
- Kosanić, M.; Ranković, B.; Stanojković, T. Antioxidant, antimicrobial, and anticancer activity of 3 Umbilicaria species. J. Food Sci. 2012, 77. [Google Scholar] [CrossRef]
- Galanty, A.; Zagrodzki, P.; Gdula-Argasińska, J.; Grabowska, K.; Koczurkiewicz-Adamczyk, P.; Wróbel-Biedrawa, D.; Podolak, I.; Pękala, E.; Paśko, P. A Comparative Survey of Anti-Melanoma and Anti-Inflammatory Potential of Usnic Acid Enantiomers—A Comprehensive In Vitro Approach. Pharmaceuticals 2021, 14, 945. [Google Scholar] [CrossRef]
- Brandão, L.F.G.; Da Silva Santos, N.P.; Pereira, E.C.G.; Da Silva, N.H.; Cepa Matos, M.D.F.; Bogo, D.; Honda, N.K. Effects of Fumarprotocetraric Acid, A Depsidone from the Lichen Cladonia verticillaris, On tyrosinase activity. Orbital 2017, 9, 256–260. [Google Scholar] [CrossRef]
- Mocan, A.; Zengin, G.; Simirgiotis, M.; Schafberg, M.; Mollica, A.; Vodnar, D.C.; Crişan, G.; Rohn, S. Functional constituents of wild and cultivated Goji (L. barbarum L.) leaves: Phytochemical characterization, biological profile, and computational studies. J. Enzyme Inhib. Med. Chem. 2017, 32, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Larrazábal-Fuentes, M.J.; Fernández-Galleguillos, C.; Palma-Ramírez, J.; Romero-Parra, J.; Sepúlveda, K.; Galetovic, A.; González, J.; Paredes, A.; Bórquez, J.; Simirgiotis, M.J.; et al. Chemical Profiling, Antioxidant, Anticholinesterase, and Antiprotozoal Potentials of Artemisia copa Phil. (Asteraceae). Front. Pharmacol. 2020, 11, 1. [Google Scholar] [CrossRef]
- Maryono; Muharram; Suryani, A.I.; Dini, I. Usnic acid derivate from Usnea sp. And bioactivity against Arthemia salina leach. In Materials Science Forum; Trans Tech Publications Ltd.: Aedermannsdorf, Switzerland, 2019; Volume 967, pp. 45–50. [Google Scholar]
- Habib, E.; León, F.; Bauer, J.D.; Hill, R.A.; Carvalho, P.; Cutler, H.G.; Cutler, S.J. Mycophenolic derivatives from Eupenicillium parvum. J. Nat. Prod. 2008, 71, 1915–1918. [Google Scholar] [CrossRef]
- Tram, N.T.T.; Huyen, V.T.; Pascal, R. Study on chemical constituents of the lichen Parmotrema tinctorum (Nyl.) Hale. Vietnam J. Sci. Technol. 2018, 56, 434. [Google Scholar] [CrossRef]
- Simirgiotis, J.M.; Quispe, C.; Bórquez, J.; Areche, C.; Sepúlveda, B. Fast Detection of Phenolic Compounds in Extracts of Easter Pears (Pyrus communis) from the Atacama Desert by Ultrahigh-Performance Liquid Chromatography and Mass Spectrometry (UHPLC–Q/Orbitrap/MS/MS). Molecules 2016, 21, 92. [Google Scholar] [CrossRef]
- Jiménez-Aspee, F.; Quispe, C.; del Soriano, M.P.C.; Fuentes Gonzalez, J.; Hüneke, E.; Theoduloz, C.; Schmeda-Hirschmann, G. Antioxidant activity and characterization of constituents in copao fruits (Eulychnia acida Phil., Cactaceae) by HPLC-DAD-MS/MSn. Food Res. Int. 2014, 62, 286–298. [Google Scholar] [CrossRef]
- Huang, D.; Ou, B.; Hampsch-Woodill, M.; Flanagan, J.A.; Prior, R.L. High-throughput assay of oxygen radical absorbance capacity (ORAC) using a multichannel liquid handling system coupled with a microplate fluorescence reader in 96-well format. J. Agric. Food Chem. 2002, 50, 4437–4444. [Google Scholar] [CrossRef]
- Zengin, G.; Uysal, S.; Ceylan, R.; Aktumsek, A. Phenolic constituent, antioxidative and tyrosinase inhibitory activity of Ornithogalum narbonense L. from Turkey: A phytochemical study. Ind. Crops Prod. 2015, 70, 1–6. [Google Scholar] [CrossRef]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model Seeking for parameter-free double-hybrid functionals: The PBE0-DH model Accurate excitation energies from time-dependent density functional theory: Assessing the PBE0 model Toward reliable density functional methods without adjustable parameters: The PBE0 model. Cit. J. Chem. Phys. 1999, 110, 2889. [Google Scholar] [CrossRef]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Frisch, A. Gaussian 09W Reference; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Release, S. Maestro, Version 11.8. Schrodinger, LLC.: New York, NY, USA. References—Scientific Research Publishing. Available online: https://scirp.org/reference/referencespapers.aspx?referenceid=2581072 (accessed on 4 December 2021).
- Greenblatt, H.M.; Kryger, G.; Lewis, T.; Silman, I.; Sussman, J.L. Structure of acetylcholinesterase complexed with (−)-galanthamine at 2.3 Å resolution. FEBS Lett. 1999, 463, 321–326. [Google Scholar] [CrossRef]
- Nachon, F.; Carletti, E.; Ronco, C.; Trovaslet, M.; Nicolet, Y.; Jean, L.; Renard, P.Y. Crystal structures of human cholinesterases in complex with huprine W and tacrine: Elements of specificity for anti-Alzheimer’s drugs targeting acetyl- and butyryl-cholinesterase. Biochem. J. 2013, 453, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Sussman, J.L.; Harel, M.; Frolow, F.; Oefner, C.; Goldman, A.; Toker, L.; Silman, I. Atomic structure of acetylcholinesterase from Torpedo californica: A prototypic acetylcholine-binding protein. Science 1991, 253, 872–879. [Google Scholar] [CrossRef]
- Silman, I.; Harel, M.; Axelsen, P.; Raves, M.; Sussman, J.L. Three-dimensional structures of acetylcholinesterase and of its complexes with anticholinesterase agents. In Biochemical Society Transactions; Portland Press Ltd.: London, UK, 1994; Volume 22, pp. 745–749. [Google Scholar]
- Nicolet, Y.; Lockridge, O.; Masson, P.; Fontecilla-Camps, J.C.; Nachon, F. Crystal Structure of Human Butyrylcholinesterase and of Its Complexes with Substrate and Products. J. Biol. Chem. 2003, 278, 41141–41147. [Google Scholar] [CrossRef]
- Tallini, L.R.; Bastida, J.; Cortes, N.; Osorio, E.H.; Theoduloz, C.; Schmeda-Hirschmann, G. Cholinesterase inhibition activity, alkaloid profiling and molecular docking of chilean rhodophiala (Amaryllidaceae). Molecules 2018, 23, 1532. [Google Scholar] [CrossRef]
- Macedo, S.K.S.; dos Almeida, T.S.; Ferraz, C.A.A.; Oliveira, A.P.; Hugo Almeida, A.V.; da Almeida, J.R.G.S.; Silva, N.D.S.; Nunes, X.P. Identification of flavonol glycosides and in vitro photoprotective and antioxidant activities of Triplaris gardneriana Wedd. J. Med. Plants Res. 2015, 9, 207–215. [Google Scholar] [CrossRef]
- Chen, J.; Ye, Y.; Ran, M.; Li, Q.; Ruan, Z.; Jin, N. Inhibition of Tyrosinase by Mercury Chloride: Spectroscopic and Docking Studies. Front. Pharmacol. 2020, 11, 81. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System. Available online: http://www.pymol.org (accessed on 27 May 2022).




| Peak | Retention Time (min.) | Tentative Identification | [M-H]− | Theoretical Mass (m/z) | Measured Mass (m/z) | Accuracy (ppm) | Metabolite Type | MS Ions (ppm) |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.34 | Mannitol | C6H13O6 | 181.0712 | 181.0705 | 3.9 | CH | 151.0598 |
| 2 | 1.78 | Citric acid | C6H7O7 | 191.0192 | 191.0184 | 4.2 | CH | 111.0074 |
| 3 | 11.43 | Atranol * | C8H7O3 | 151.0395 | 151.0387 | 5.3 | A | 135.0438; 123.0438; 107.0488 |
| 4 | 12.91 | 5,7-Dihydroxy-4-methylphthalide | C9H7O4 | 179.0344 | 179.0336 | 4.5 | A | 107.0488; 135.0437; 151.0386 |
| 5 | 13.18 | Haematommic acid (3-formyl-2,4-dihydroxy-6-methylbenzoic acid) | C9H7O5 | 195.0293 | 195.0286 | 3.6 | A | 179.0335; 151.0387; 123. 0438; 149.0230 |
| 6 | 14.93 | 5,7-Dihydroxy-6-methylphthalide * | C9H7O4 | 179.0344 | 179.0337 | 3.9 | A | 135.0438; 107.0488 |
| 7 | 16.28 | 9,10,12,13-Tetrahydroxyheneicosanoic acid | C21H41O6 | 389.2903 | 389.2892 | 2.8 | L | 371.2784 |
| 8 | 17.35 | 9,10,12,13,14-Pentahydroxytetracosanoic acid | C24H47O7 | 447.3322 | 447.3306 | 3.6 | L | 389.2891; 429.3199; 361.2581 |
| 9 | 18.71 | Evernic acid isomer (3-hydroxy-4-(2-hydroxy-4-methoxy-6-methylbenzoyl)oxy-6-methylbenzoic acid) | C17H15O7 | 331.0818 | 331.0809 | 2.7 | d | 135.0438; 123.0439; 181.0494 151.0386; 167.0336; 313.0703 |
| 10 | 19.75 | Methyl orsellinate | C9H9O4 | 181.0501 | 181.0494 | 3.9 | A | 151.0387; 123,0439; 135.0438 |
| 11 | 19.83 | 9,10,12,13-Tetrahydroxydocosanoic acid | C22H43O6 | 403.3060 | 403.3047 | 3.2 | L | 385.2939; 215.1273 |
| 12 | 20.16 | Evernic acid II isomer (3-hydroxy-4-(3-hydroxy-4-methoxy-6-methylbenzoyl)oxy-6-methylbenzoic acid) | C17H15O7 | 331.0818 | 331.0809 | 2.7 | d | 195.0284; 151.0386; 123.0438; 135.0436; 167.0336 |
| 13 | 20.25 | Pentahydroxyhexacosanoic acid | C26H51O7 | 475.3635 | 475.3618 | 3.6 | L | - |
| 14 | 20.33 | 9,10,12,13-Tetrahidroxytricosanoic acid | C23H45O6 | 417.3236 | 417.3204 | 7.7 | L | 399.3095 |
| 15 | 20.42 | Barbatolic acid * | C18H13O10 | 389.05080 | 389.05086 | 2.5 | A | 211.0246, 195.02122 |
| 16 | 20.49 | Isomer haematommic acid (3-formyl-2,5-dihydroxy-6-methylbenzoic acid) | C9H7O5 | 195.0293 | 195.0285 | 4.1 | A | 179.0335; 151.0387; 123.0438 |
| 17 | 20.81 | 2,4-Diformyl-3,5-dihydroxytoluene o 2,6-Diformyl-3,5-dihydroxytoluene | C9H7O4 | 179.0344 | 179.0338 | 3.4 | A | 151.0386; 107.0488; 135.0437 |
| 18 | 21.74 | Evernic acid (2-hydroxy-4-(2-hydroxy-4-methoxy-6-methylbenzoyl)oxy-6-methylbenzoic acid) | C17H15O7 | 331.0818 | 331.0808 | 3.0 | d | 167.0334; 151.0386; 135.0437 313.0703; 123.0439; 181.0494 |
| 19 | 21.85 | 9,10,12,13,14,15-Hexahydroxyheptacosenoic acid | C27H51O8 | 503.3584 | 503.3564 | 4.0 | L | 475.3615; 443.3355 |
| 20 | 22.30 | Methyl 9,10,11,12,13-pentahydroxy-14-oxoheptacosanoate | C28H53O8 | 517.3740 | 517.3719 | 4.1 | L | 457.3510; 439.3404 |
| 21 | 23.08 | Lichesterinic acid o Protolichesterinic acid | C19H31O4 | 323.2222 | 323.2213 | 2.8 | L | 279.2315; 267.2314 |
| 22 | 23.49 | Tetrahydroxydioxoheneicosanoic acid | C21H37O8 | 417.2494 | 417.2496 | 0.9 | L | 399.3091 |
| 23 | 23.85 | Tetrahydroxydocosanoic acid | C22H43O6 | 403.3065 | 403.3067 | 0.6 | L | - |
| 24 | 24.30 | Sphaerophorin | C23H27O7 | 415.1757 | 415.1744 | 3.1 | d | 233.1166; 207.1376; 251.1275 |
| 25 | 24.67 | Pseudoplacodiolic acid or Placodiolic acid | C19H19O8 | 375.1080 | 375.1070 | 2.7 | DBF | 343.0807; 259.0598; 231.0648 |
| 26 | 25.63 | Isomer barbatic acid (3-hydroxy-4-(2-hydroxy-4-methoxy-3,6-dimethylbenzoyl)oxy-3,6-dimethylbenzoic acid) | C19H19O7 | 359.1131 | 359.1121 | 2.8 | d | 181.0493; 163.0387; 137.0594 |
| 27 | 25.78 | Barbatic acid (2-hydroxy-4-(2-hydroxy-4-methoxy-3,6-dimethylbenzoyl)oxy-3,6-dimethylbenzoic acid) | C19H19O7 | 359.1131 | 359.1120 | 3.1 | d | 181.0493; 163.0387; 137.0594 |
| 28 | 26.16 | Usnic acid * | C18H15O7 | 343.0818 | 343.0808 | 2.9 | DBF | 259.0598; 231.0647; 328.0570 |
| Usnic Acid * | Barbatolic Acid * | Atranol * | 5,7-Dihydroxy-6-methylphthalide * | |
|---|---|---|---|---|
| H. lugubris lichen | 8.921 ± 0.372 | 85.833 ± 0.325 | 32.345 ± 0.071 | 49.374 ± 0.095 |
| Assay | TPC a | FRAP b | ORAC b | DPPH c | AChE d | BChE d | Tyr d |
|---|---|---|---|---|---|---|---|
| H. lugubris Ethanol extract | 47.4 ± 0.05 | 27.8 ± 0.0 | 32.7 ± 0.70 | 75.3 ± 0.02 | 12.38 ± 0.09 b | 31.54 ± 0.20 | 22.32 ± 0.21 |
| Usnic acid | 22.4 ± 0.00 | 122.73 ± 1.0 | 55.25 ± 0.04 | 2.21± 0.03 | 4.36 ± 0.03 | 132.23 ± 0.12 | |
| Barbatolic acid | - | 28.10 ± 0.0 a | 101.11 ± 0.71 | 62.55 ± 0.01 | 17.42 ± 0.03 | 23.95 ± 0.02 | 35.23 ± 0.11 |
| Atranol | - | 29.32 ± 0.0 a | 176.28 ± 0.84 | 21.04 ± 0.02 | 28.82 ± 0.10 | 36.43 ± 0.08 | 7.25 ± 0.18 |
| 5,7-dihydroxy-6-methylphthalide | - | 36.91 ± 0.0 | 373.65 ± 1.05 | 12.52 ± 0.02 | 12.71 ± 0.12 b | 19.47 ± 0.10 | 12.13 ± 0.15 |
| Gallic acid | - | 45.5 ± 0.00 | - | 2.24 ± 0.04 | - | - | |
| Galantamine | - | - | - | 0.27 ± 0.03 | 3.82 ± 0.02 | - | |
| Kojic acid | - | - | - | - | - | - | 0.76 ± 0.05 |
| Compound | Binding Energy (kcal/mol) Acetylcholinesterase (TcAChE) | Binding Energy (kcal/mol) Butyrylcholinesterase (hBuChE) | Binding Energy (kcal/mol) Tyrosinase |
|---|---|---|---|
| Usnic acid | −10.779 | −8.844 | −5.744 |
| Barbatolic acid | −8.027 | −8.165 | −6.490 |
| 7-dihydroxy-6-methylphthalide | −7.913 | −6.855 | −4.639 |
| Atranol | −6.197 | −6.343 | −4.964 |
| Galantamine | −12.989 | −7.125 | - |
| Kojic acid | - | - | −6.050 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Areche, C.; Parra, J.R.; Sepulveda, B.; García-Beltrán, O.; Simirgiotis, M.J. UHPLC-MS Metabolomic Fingerprinting, Antioxidant, and Enzyme Inhibition Activities of Himantormia lugubris from Antarctica. Metabolites 2022, 12, 560. https://doi.org/10.3390/metabo12060560
Areche C, Parra JR, Sepulveda B, García-Beltrán O, Simirgiotis MJ. UHPLC-MS Metabolomic Fingerprinting, Antioxidant, and Enzyme Inhibition Activities of Himantormia lugubris from Antarctica. Metabolites. 2022; 12(6):560. https://doi.org/10.3390/metabo12060560
Chicago/Turabian StyleAreche, Carlos, Javier Romero Parra, Beatriz Sepulveda, Olimpo García-Beltrán, and Mario J. Simirgiotis. 2022. "UHPLC-MS Metabolomic Fingerprinting, Antioxidant, and Enzyme Inhibition Activities of Himantormia lugubris from Antarctica" Metabolites 12, no. 6: 560. https://doi.org/10.3390/metabo12060560
APA StyleAreche, C., Parra, J. R., Sepulveda, B., García-Beltrán, O., & Simirgiotis, M. J. (2022). UHPLC-MS Metabolomic Fingerprinting, Antioxidant, and Enzyme Inhibition Activities of Himantormia lugubris from Antarctica. Metabolites, 12(6), 560. https://doi.org/10.3390/metabo12060560









