NMR-Based Metabolomics in Differential Diagnosis of Chronic Kidney Disease (CKD) Subtypes
Abstract
:1. Introduction
2. Results
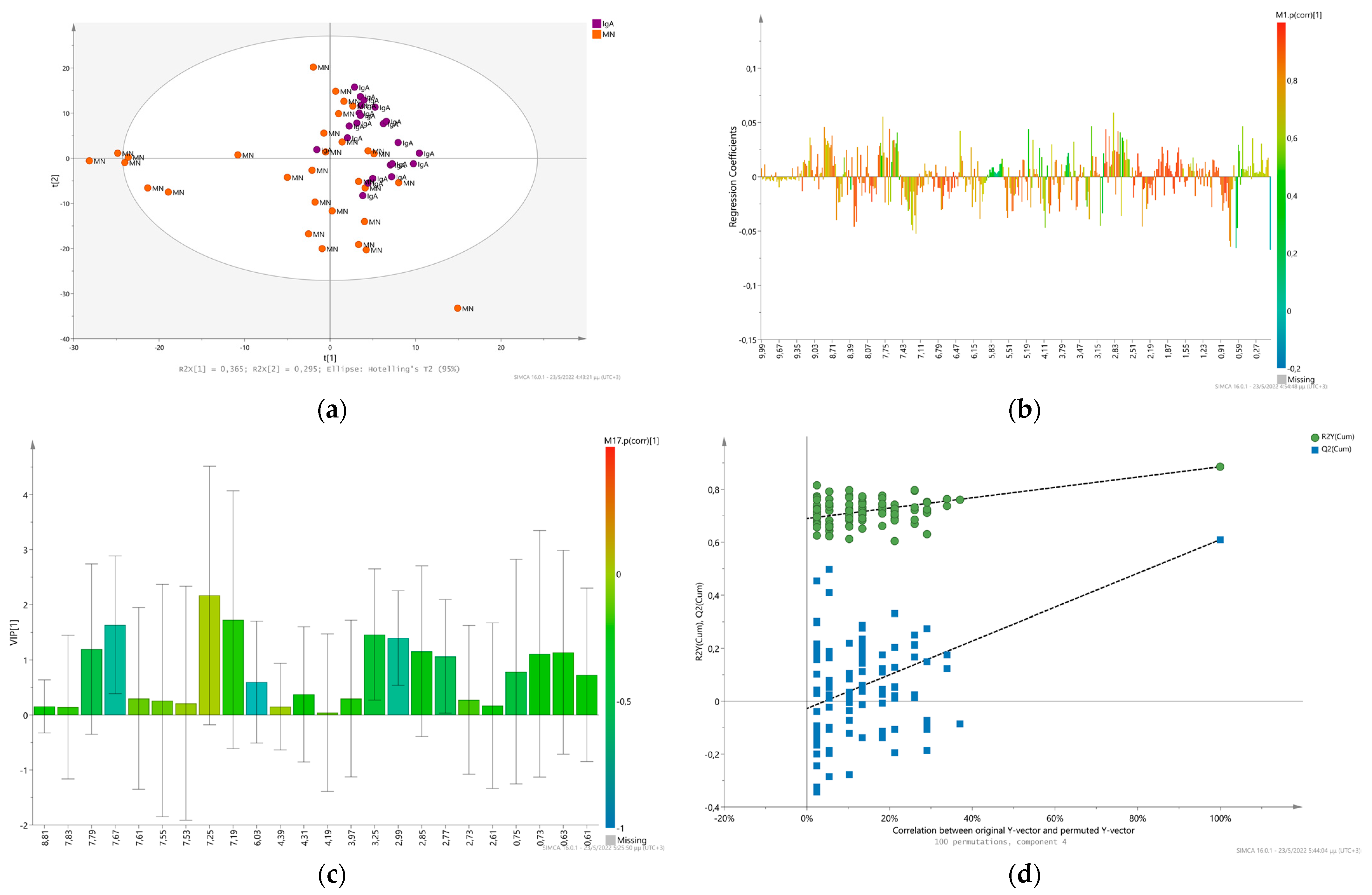
2.1. Glomerulonephritis. Membranous versus IgA Nephropathy
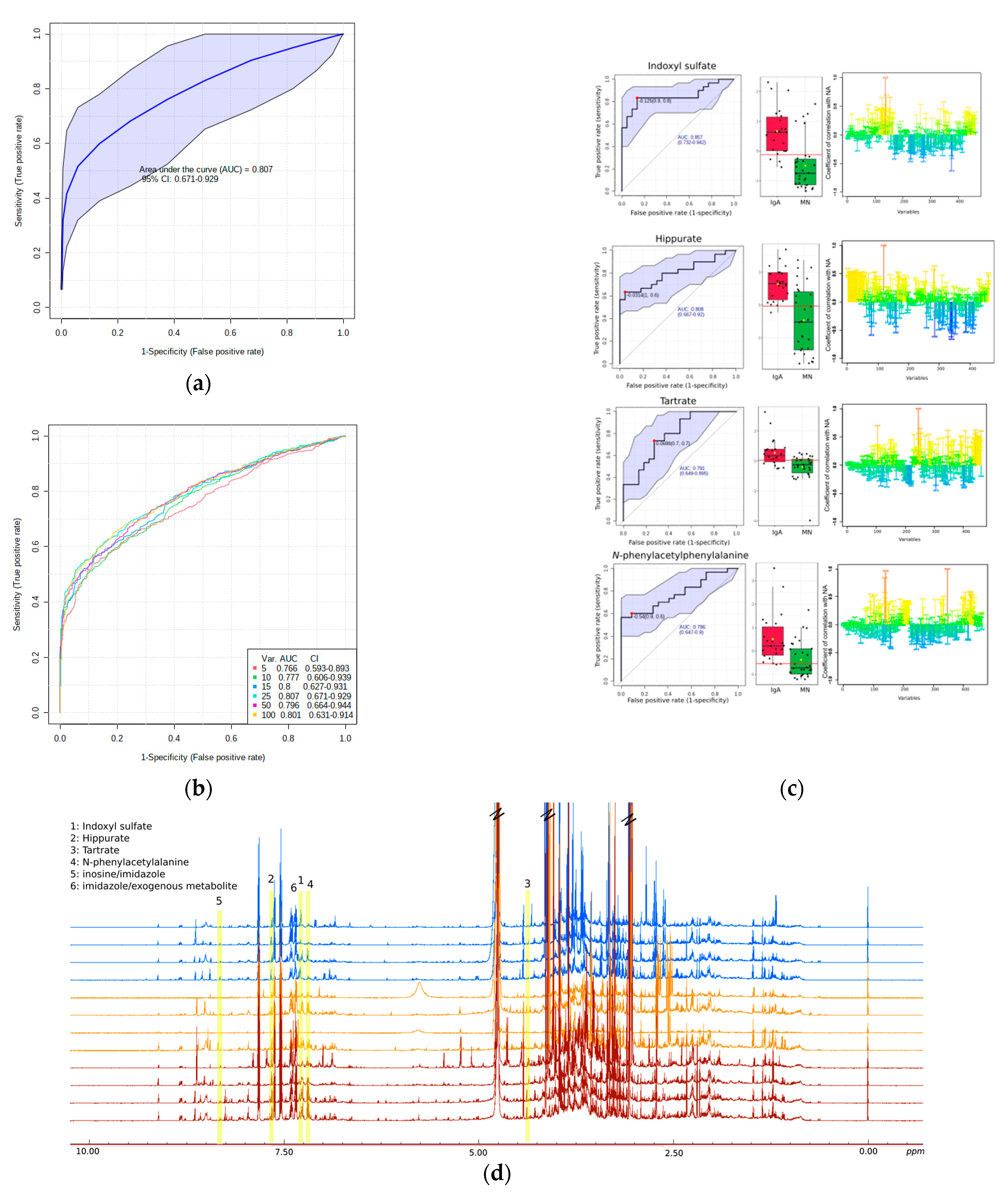
Univariate Analysis-Biomarker Evaluation on IgAN and MN Discrimination
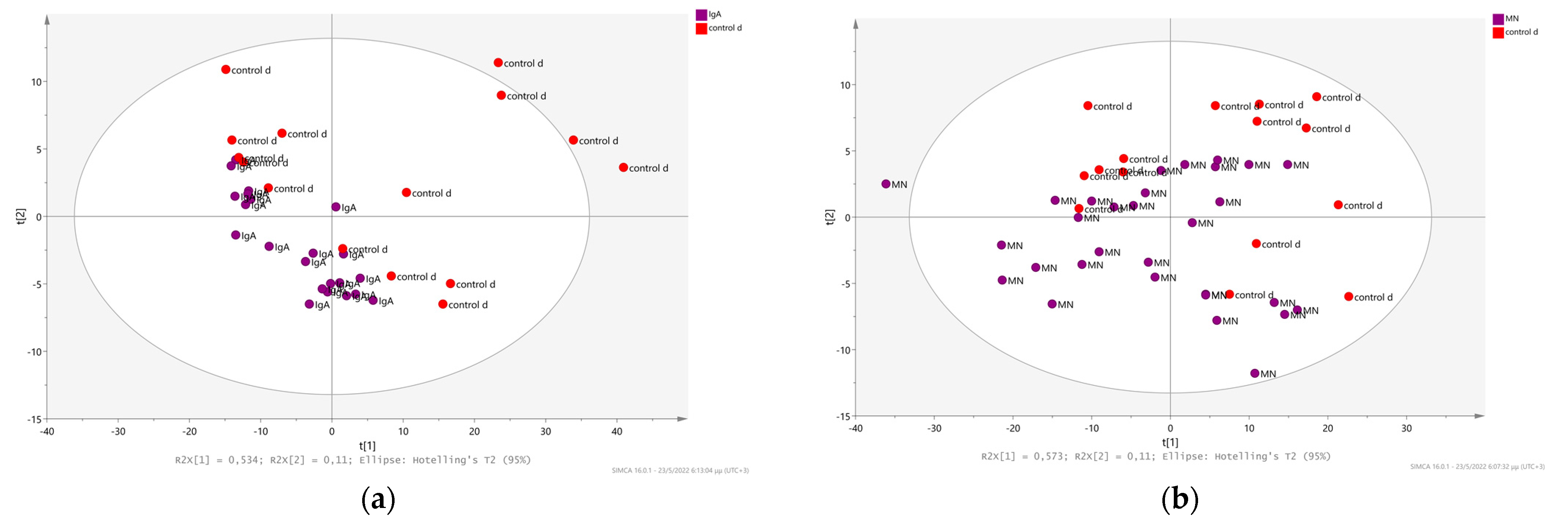
2.2. Chronic Kidney Disease Diagnosis as a Result of Other Systemic Diseases and its Relation with Membranous and IgA Nephritis
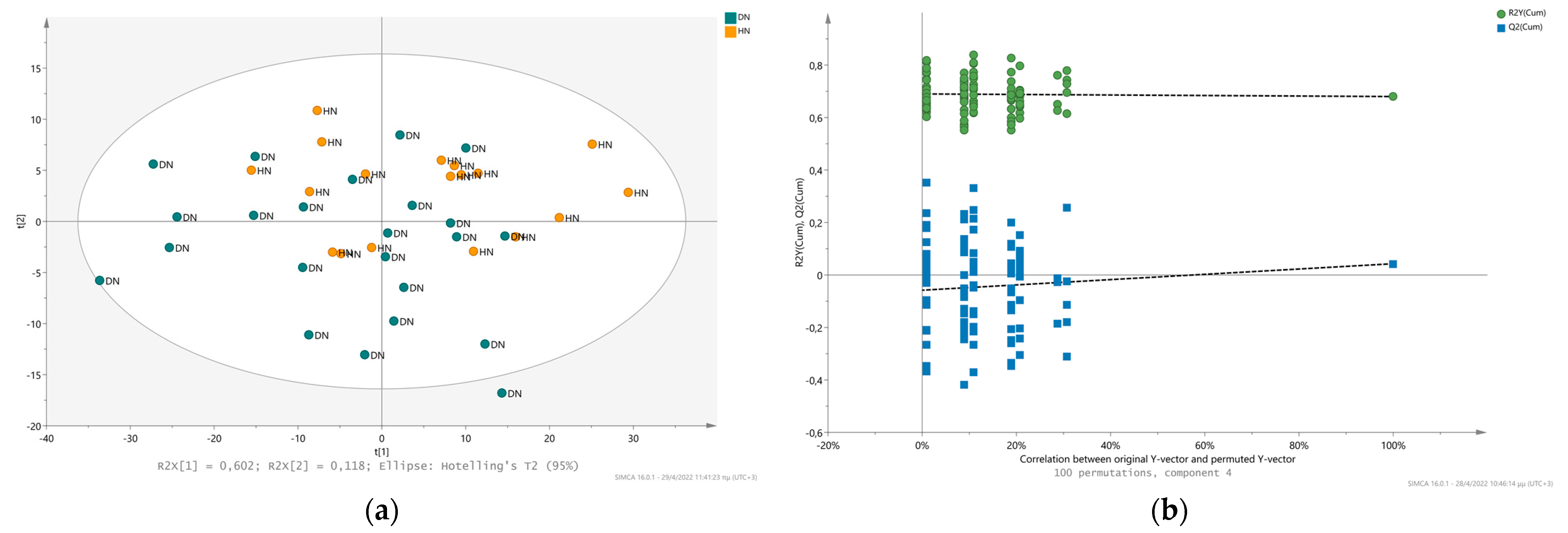
2.3. Comparison of the Metabolic Fingerprint of Hypertensive Nephrosclerosis (HN) and Diabetic Nephropathy (DN)
3. Materials and Methods
3.1. Population Samples
3.2. Sample Collection
3.3. NMR Analysis
3.4. NMR Data Processing
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolf, G. Renal injury due to renin–angiotensin–aldosterone system activation of the transforming growth factor-β pathway. Kidney Int. 2006, 70, 1914–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levey, A.S.; Coresh, J.; Balk, E.; Kausz, A.T.; Levin, A.; Steffes, M.W.; Hogg, R.J.; Perrone, R.D.; Lau, J.; Eknoyan, G. National Kidney Foundation practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Ann. Intern. Med. 2003, 139, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Warnock, D.G.; Muntner, P.; McCullough, P.A.; Zhang, X.; McClure, L.A.; Zakai, N.; Cushman, M.; Newsome, B.B.; Kewalramani, R.; Steffes, M.W. Kidney function, albuminuria, and all-cause mortality in the REGARDS (Reasons for Geographic and Racial Differences in Stroke) study. Am. J. Kidney Dis. 2010, 56, 861–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levey, A.S.; Eckardt, K.-U.; Tsukamoto, Y.; Levin, A.; Coresh, J.; Rossert, J.; Zeeuw, D.D.; Hostetter, T.H.; Lameire, N.; Eknoyan, G. Definition and classification of chronic kidney disease: A position statement from Kidney Disease: Improving Global Outcomes (KDIGO). Kidney Int. 2005, 67, 2089–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, A.; Stevens, P.; Bilous, R.; Coresh, J.; De Francisco, A.; De Jong, P.; Griffith, K.; Hemmelgarn, B.; Iseki, K.; Lamb, E. Kidney. Int. Suppl. 2013, 158, 825–830. [Google Scholar]
- Chen, X.; Chen, Y.; Shi, K.; Lv, Y.; Tong, H.; Zhao, G.; Chen, C.; Chen, B.; Li, D.; Lu, Z. Comparison of prognostic, clinical, and renal histopathological characteristics of overlapping idiopathic membranous nephropathy and IgA nephropathy versus idiopathic membranous nephropathy. Sci. Rep. 2017, 7, 11468. [Google Scholar] [CrossRef] [Green Version]
- Kazi, A.M.; Hashmi, M.F. Glomerulonephritis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Vinen, C.; Oliveira, D. Acute glomerulonephritis. Postgrad. Med. J. 2003, 79, 206–213. [Google Scholar] [CrossRef]
- Assfalg, M.; Bertini, I.; Colangiuli, D.; Luchinat, C.; Schäfer, H.; Schütz, B.; Spraul, M. Evidence of different metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 1420–1424. [Google Scholar] [CrossRef] [Green Version]
- Emwas, A.-H.M.; Salek, R.M.; Griffin, J.L.; Merzaban, J. NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics 2013, 9, 1048–1072. [Google Scholar] [CrossRef]
- Bruzzone, C.; Loizaga-Iriarte, A.; Sánchez-Mosquera, P.; Gil-Redondo, R.n.; Astobiza, I.; Diercks, T.; Cortazar, A.R.; Ugalde-Olano, A.; Schäfer, H.; Blanco, F.J. 1H NMR-Based Urine Metabolomics Reveals Signs of Enhanced Carbon and Nitrogen Recycling in Prostate Cancer. J. Proteome Res. 2020, 19, 2419–2428. [Google Scholar] [CrossRef]
- Park, S.; Lee, J.; Yang, S.H.; Lee, H.; Kim, J.Y.; Park, M.; Kim, K.H.; Moon, J.J.; Cho, S.; Lee, S. Comprehensive metabolomic profiling in early IgA nephropathy patients reveals urine glycine as a prognostic biomarker. J. Cell. Mol. Med. 2021, 25, 5177–5190. [Google Scholar] [CrossRef] [PubMed]
- Taherkhani, A.; Farrokhi Yekta, R.; Mohseni, M.; Saidijam, M.; Arefi Oskouie, A. Chronic kidney disease: A review of proteomic and metabolomic approaches to membranous glomerulonephritis, focal segmental glomerulosclerosis, and IgA nephropathy biomarkers. Proteome Sci. 2019, 17, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanouchi, M.; Furuichi, K.; Wada, T. Nephrosclerosis and Diabetic Kidney Disease. In Diabetic Kidney Disease; Springer: Berlin/Heidelberg, Germany, 2021; pp. 149–158. [Google Scholar]
- Øvrehus, M.A.; Oldereid, T.S.; Dadfar, A.; Bjørneklett, R.; Aasarød, K.I.; Fogo, A.B.; Ix, J.H.; Hallan, S.I. Clinical phenotypes and long-term prognosis in white patients with biopsy-verified hypertensive nephrosclerosis. Kidney Int. Rep. 2020, 5, 339–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diez Ojea, B.; Marín, R.; Coto, E.; Fernández Vega, F.; Alvarez Navascués, R.; Fernández Fresnedo, G.; Pobes Martínez de Salinas, A.; Suárez Laurés, A.; García Monteavaro, C.; Gorostidi, M. Clinical and genetic basis of hypertensive nephrosclerosis. Nefrol. (Engl. Ed.) 2010, 30, 687–697. [Google Scholar]
- Nakayama, M.; Sato, T.; Sato, H.; Yamaguchi, Y.; Obara, K.; Kurihara, I.; Sato, K.; Hotta, O.; Seino, J.; Miyata, M. Different clinical outcomes for cardiovascular events and mortality in chronic kidney disease according to underlying renal disease: The Gonryo study. Clin. Exp. Nephrol. 2010, 14, 333–339. [Google Scholar] [CrossRef]
- Nakayama, M.; Sato, T.; Miyazaki, M.; Matsushima, M.; Sato, H.; Taguma, Y.; Ito, S. Increased risk of cardiovascular events and mortality among non-diabetic chronic kidney disease patients with hypertensive nephropathy: The Gonryo study. Hypertens. Res. 2011, 34, 1106–1110. [Google Scholar] [CrossRef]
- Seccia, T.M.; Caroccia, B.; Calò, L.A. Hypertensive nephropathy. Moving from classic to emerging pathogenetic mechanisms. J. Hypertens. 2017, 35, 205–212. [Google Scholar] [CrossRef]
- Selby, N.M.; Taal, M.W. An updated overview of diabetic nephropathy: Diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes. Metab. 2020, 22, 3–15. [Google Scholar] [CrossRef]
- Titan, S.M.; Venturini, G.; Padilha, K.; Goulart, A.C.; Lotufo, P.A.; Bensenor, I.J.; Krieger, J.E.; Thadhani, R.I.; Rhee, E.P.; Pereira, A.C. Metabolomics biomarkers and the risk of overall mortality and ESRD in CKD: Results from the Progredir Cohort. PLoS ONE 2019, 14, e0213764. [Google Scholar] [CrossRef] [Green Version]
- Zacharias, H.U.; Schley, G.; Hochrein, J.; Klein, M.S.; Köberle, C.; Eckardt, K.-U.; Willam, C.; Oefner, P.J.; Gronwald, W. Analysis of human urine reveals metabolic changes related to the development of acute kidney injury following cardiac surgery. Metabolomics 2013, 9, 697–707. [Google Scholar] [CrossRef]
- Georgiopoulou, P.D.; Chasapi, S.A.; Christopoulou, I.; Varvarigou, A.; Spyroulias, G.A. Untargeted 1H-NMR Urine Metabolomic Analysis of Preterm Infants with Neonatal Sepsis. Appl. Sci. 2022, 12, 1932. [Google Scholar] [CrossRef]
- Georgakopoulou, I.; Chasapi, S.A.; Bariamis, S.E.; Varvarigou, A.; Spraul, M.; Spyroulias, G.A. Metabolic changes in early neonatal life: NMR analysis of the neonatal metabolic profile to monitor postnatal metabolic adaptations. Metabolomics 2020, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Takis, P.G.; Taddei, A.; Pini, R.; Grifoni, S.; Tarantini, F.; Bechi, P.; Luchinat, C. Fingerprinting acute digestive diseases by untargeted NMR based metabolomics. Int. J. Mol. Sci. 2018, 19, 3288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Rong, G.; Gu, J.; Fan, C.; Guo, T.; Jiang, T.; Deng, W.; Xie, J.; Su, Z.; Yu, Q. Nicotinamide N-methyltransferase ameliorates renal fibrosis by its metabolite 1-methylnicotinamide inhibiting the TGF-β1/Smad3 pathway. FASEB J. 2022, 36, e22084. [Google Scholar] [CrossRef]
- Tan, X.; Cao, X.; Zou, J.; Shen, B.; Zhang, X.; Liu, Z.; Lv, W.; Teng, J.; Ding, X. Indoxyl sulfate, a valuable biomarker in chronic kidney disease and dialysis. Hemodial. Int. 2017, 21, 161–167. [Google Scholar] [CrossRef]
- Deguchi, T.; Takemoto, M.; Uehara, N.; Lindup, W.E.; Suenaga, A.; Otagiri, M. Renal clearance of endogenous hippurate correlates with expression levels of renal organic anion transporters in uremic rats. J. Pharmacol. Exp. Ther. 2005, 314, 932–938. [Google Scholar] [CrossRef] [Green Version]
- Kopple, J.D. Phenylalanine and tyrosine metabolism in chronic kidney failure. J. Nutr. 2007, 137, 1586S–1590S. [Google Scholar] [CrossRef]
- Chang, H.-H.; Chao, H.-N.; Walker, C.S.; Choong, S.-Y.; Phillips, A.; Loomes, K.M. Renal depletion of myo-inositol is associated with its increased degradation in animal models of metabolic disease. Am. J. Physiol.-Ren. Physiol. 2015, 309, F755–F763. [Google Scholar] [CrossRef] [Green Version]
- Niewczas, M.A.; Sirich, T.L.; Mathew, A.V.; Skupien, J.; Mohney, R.P.; Warram, J.H.; Smiles, A.; Huang, X.; Walker, W.; Byun, J. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: Metabolomic study. Kidney Int. 2014, 85, 1214–1224. [Google Scholar] [CrossRef] [Green Version]
- Scialla, J.J.; Asplin, J.; Dobre, M.; Chang, A.R.; Lash, J.; Hsu, C.-y.; Kallem, R.R.; Hamm, L.L.; Feldman, H.I.; Chen, J. Higher net acid excretion is associated with a lower risk of kidney disease progression in patients with diabetes. Kidney Int. 2017, 91, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Connor, S.C.; Hansen, M.K.; Corner, A.; Smith, R.F.; Ryan, T.E. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Mol. BioSystems 2010, 6, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Fresno, R.; Llorach, R.; Alcaro, F.; Rodríguez, M.Á.; Vinaixa, M.; Chiva-Blanch, G.; Estruch, R.; Correig, X.; Andrés-Lacueva, C. 1H-NMR-based metabolomic analysis of the effect of moderate wine consumption on subjects with cardiovascular risk factors. Electrophoresis 2012, 33, 2345–2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bletsa, E.; Filippas-Dekouan, S.; Kostara, C.; Dafopoulos, P.; Dimou, A.; Pappa, E.; Chasapi, S.; Spyroulias, G.; Koutsovasilis, A.; Bairaktari, E. Effect of dapagliflozin on urine metabolome in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2021, 106, 1269–1283. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; Bosch, J.P.; Lewis, J.B.; Greene, T.; Rogers, N.; Roth, D.; Modification of Diet in Renal Disease Study Group*. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Ann. Intern. Med. 1999, 130, 461–470. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Luchinat, C.; Turano, P.; Tenori, L.; Roy, R.; Salek, R.M.; Ryan, D.; Merzaban, J.S.; Kaddurah-Daouk, R.; Zeri, A.C. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: A review. Metabolomics 2015, 11, 872–894. [Google Scholar] [CrossRef] [Green Version]
- Mckay, R.T. How the 1D-NOESY suppresses solvent signal in metabonomics NMR spectroscopy: An examination of the pulse sequence components and evolution. Concepts Magn. Reson. Part A 2011, 38, 197–220. [Google Scholar] [CrossRef]
- Chasapi, S.A.; Karagkouni, E.; Matzarapi, K.; Marousis, K.D.; Varvarigou, A.; Spyroulias, G.A. NMR and Metabolomics. eLS 2019, 1–9. [Google Scholar]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Vignoli, A.; Rodio, D.M.; Bellizzi, A.; Sobolev, A.P.; Anzivino, E.; Mischitelli, M.; Tenori, L.; Marini, F.; Priori, R.; Scrivo, R. NMR-based metabolomic approach to study urine samples of chronic inflammatory rheumatic disease patients. Anal. Bioanal. Chem. 2017, 409, 1405–1413. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14-10. [Google Scholar] [CrossRef]
- Gaude, E.; Chignola, F.; Spiliotopoulos, D.; Spitaleri, A.; Ghitti, M.; Garcìa-Manteiga, J.M.; Mari, S.; Musco, G. muma, an R package for metabolomics univariate and multivariate statistical analysis. Curr. Metab. 2013, 1, 180–189. [Google Scholar] [CrossRef]
| No | Metabolites | 1H NMR Chemical Shifts * |
|---|---|---|
| 1. | 1-methylhistidine | 7.91 (m) |
| 2. | Trigonelline | 9.25...9.11 (s), 8.84…8.80 (m), 8.08…8.06 (m), 4.43…4.42 (s) |
| 3. | 1-methylnicotinamide | 9.26 (s), 8.96…8.93 (d), 8.89…8.87(d), 4.46 (s) |
| 4. | NADH/NADPH | 8.46 (s), 8.17 (s), 6.28 (d), 5.94 (d)/ 2.79, 2.77 (s) |
| 5. | NAD+/NADP+ | 9.35/8.37 (s), 6.04 (d), 6.06 (d)/ 8.43 (s), 6.06 (d), 6.09 (d)/ |
| 6. | ATP, AMP | 8.55 (s), 8.26 (s) |
| 7. | Hippurate | 7.83…7.81 (d), 7.64….7.60 (m), 7.55…7.52 (m) |
| 8. | Indoxyl sulfate | 7.35 (s), 7.28…7.25 (m), 7.20… 7.16(m) |
| 9. | Histidine | 7.07 (m) |
| 10. | 3-Hydroxymandelate | 7.31…7.27 (m/t), 6.97 (m), 6.90 (m), 6.85…6.82 (m) |
| 11. | Anserine (-NH) | 8.1 (s), 7.1 (s), 4.5 (m) |
| 12. | Glycolate | 3.94…3.97 (s) |
| 13. | 2-Hydroxyisobutyrate | 1.3 (s) |
| 14. | Tartrate | 4.3 (s) |
| 15. | creatinine | 4.04 (s), 3.03 (s) |
| 16. | Mannitol | 3.87…3.84 (dd), 3.80…3.78 (m), 3.77…3.73 (m), 3.69…3.66 (m) |
| 17. | Myo-inositol | 4.06 (m), 3.68 (br), 3.55–3.54 (dd) |
| 18. | sn-glycerol- | 3.65 (m) |
| 19. | TMAO | 3.25 (s) |
| 20. | Sarcosine | 2.76 (s) broad- 3.60 (s) overlapped |
| 21. | N-phenylacetylglycine | 7.43…7.40 (m), 7.36…7.33 (m) |
| 22. | N-phenylacetylphenylalanine | 7.75(d), 7.31…7.26 (m), 7.18…7.15 (m), 7.10…7.08 (m) |
| 23. | Gentisate | 7.29…7.27 (overlapped) 6.98…6.96 (dd), 6.86…6.84 (d) |
| 24. | 3, 4-Dihydroxymandelate | 6.91…6.87 (m), 6.84…6.82 (dd) |
| 25. | Salicylate | 7.85 (dd), 7.47 (m), 6.97…6.95 (m) |
| 26. | Trans-Aconitate | 6.59–6.57 (s) |
| 27. | Xanthosine | 7.9 (s), 5.81 (d) |
| 28. | uracil | 7.5 (d), 5.77 (d) |
| 29. | urea | 5.75 (s, br) |
| 30. | cis-Aconitate | 5.71…5.65 (m) |
| 31. | Allantoin | 5.38…5.36 (s), br |
| 32. | 1,3- Dihydroxyacetone | 4.42…4.41 |
| 33. | UDP-glucose/ | 4.39 (m) |
| 34. | Glucose | 3.25…3.24 |
| 35. | Taurine | 3.44…3.41 (t), 3.27…3.24 (t) |
| 36. | Citrate | 2.70...2.67 (d), 2.56…2.52 (d) |
| 37. | N, N-Dimethylglycine | 2.93…2.91 (s) |
| 38. | N-methylhydantoin | 4.1 (s), 2.9 (s) |
| 39. | 2-hydroxybutyrate | 1.70...1.63 (m), 1.63…1.57 (m) |
| 40. | Betaine | 3.9 (s), 3.3 (s) |
| 41. | Lysine | 3.01...2.9 (t), 1.92...1.87 (m) |
| 42. | Gamma-aminobutyrate | 3.00…2.97 (t), 1.94…1.90 (m) |
| 43. | Proline | 1.94…1.92 (m) |
| 44. | Isoleucine | 0.99…0.97 (d) |
| 45. | Leucine | 0.96…0.94 (d), 0.95…0.93 (d) |
| 46. | Valine | 1.04…1.02 (d), 0.98…0.97 (d) |
| 47. | 3-aminoisobutyrate | 1.19…1.17 (d) |
| 48. | 3-methyl-2-oxovalerate | 1.07…1.05 (d), 0.89…0.86 (t) |
| 49. | Isobutyrate | 1.10…1.08 (d) |
| 50. | Alanine | 1.48…1.46 (d) |
| 51. | Succinate | 2.39 (s) |
| 52. | Glucuronate | 5.2 (d), 4.7 (d) |
| 53. | Erythritol | 3.8 (d), 3.7 (d) |
| 54. | Lactose | 5.22 (d) |
| 55. | Uridine | 7.9 (m), 5.9 (d), 4.39 (m), 4.25 (m) |
| 56. | L-Glutamate | 2.37…2.30 (m) |
| 57. | Sialic acid | 2.33 (dd) |
| 58. | Lisinopril (zestril) | 7.40 (t), 7.32 (m), 2.33 (m) |
| 59. | Aspartate | 2.83 (dd) |
| 60. | Pyroglutamate | 4.19…4.15 (m) |
| Metabolite | Raw p-Value | Log2 (FC) 1 | Fold Change in DN/HN |
|---|---|---|---|
| Sarcosine | 0.095 | −1.158 | 0.44801 ▲2 HN |
| 1-Methylnicotinamide | 0.178 | −0.976 | 0.50828 ▲ HN |
| Hippurate | 0.290 | −0.906 | 0.52994 ▲ HN |
| Myo-inositol | 0.691 | 0.856 | 1.8102 ▼ HN |
| Creatinine | 0.056 | −0.720 | 0.60693 ▲ HN |
| Trigonelline | 0.49 | −0.714 | 0.60954 ▲ HN |
| Isoleucine | 0.152 | −0.720 | 0.60696 ▲ HN |
| 1,3- Dihydroxyacetone | 0.427 | −0.717 | 0.60828 ▲ HN |
| NAD+/NADP+ | 0.866 | −0.350 | 0.78433 ▲ HN |
| τ-Μethylhistidine | 0.056 | −0.749 | 0.59479 ▲ HN |
| Allantoin | 0.119 | −0.355 | 0.7817 ▲ HN |
| Citrate | 0.664 | 0.156 | 1.1144 ▼ HN |
| Acetone | 0.071 | −0.353 | 0.78286 ▲ HN |
| Alanine | 0.630 | −0.205 | 0.86759 ▲ HN |
| Lactate | 0.687 | −0.072 | 0.95136 ▲ HN |
| 3-methyl-2-oxovalerate | 0.948 | 0.093 | 1.0665 ▼ HN |
| Valine | 0.524 | −0.698 | 0.61621 ▲ HN |
| Cause | Age | Sex (M/F) | Rate Decline eGFR | Urine Creatinine |
|---|---|---|---|---|
| Membranous Nephropathy | 55.83 | 25/5 | 5.19 ± 61.94 | 78.63 ± 39.49 |
| Immunoglobulin A Nephropathy | 43.46 | 16/6 | −3.94 ± 32.78 | 86.34 ± 49.25 |
| Diabetic Nephropathy | 68 ± 8.1 | 22/1 | −0.64 ± 5.38 | 55.98 ± 44.57 |
| Hypertensive Nephrosclerosis | 59.9 ± 1.38 | 13/5 | −3.92 ± 11.5 | 61.14 ± 48.21 |
| Control Diseased Glomerulonephritis | 57.13 ± 14.46 | 8/7 | −6.17 ± 20.28 | 71.33 ± 48.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chasapi, S.A.; Karagkouni, E.; Kalavrizioti, D.; Vamvakas, S.; Zompra, A.; Takis, P.G.; Goumenos, D.S.; Spyroulias, G.A. NMR-Based Metabolomics in Differential Diagnosis of Chronic Kidney Disease (CKD) Subtypes. Metabolites 2022, 12, 490. https://doi.org/10.3390/metabo12060490
Chasapi SA, Karagkouni E, Kalavrizioti D, Vamvakas S, Zompra A, Takis PG, Goumenos DS, Spyroulias GA. NMR-Based Metabolomics in Differential Diagnosis of Chronic Kidney Disease (CKD) Subtypes. Metabolites. 2022; 12(6):490. https://doi.org/10.3390/metabo12060490
Chicago/Turabian StyleChasapi, Styliani A., Evdokia Karagkouni, Dimitra Kalavrizioti, Sotirios Vamvakas, Aikaterini Zompra, Panteleimon G. Takis, Dimitrios S. Goumenos, and Georgios A. Spyroulias. 2022. "NMR-Based Metabolomics in Differential Diagnosis of Chronic Kidney Disease (CKD) Subtypes" Metabolites 12, no. 6: 490. https://doi.org/10.3390/metabo12060490
APA StyleChasapi, S. A., Karagkouni, E., Kalavrizioti, D., Vamvakas, S., Zompra, A., Takis, P. G., Goumenos, D. S., & Spyroulias, G. A. (2022). NMR-Based Metabolomics in Differential Diagnosis of Chronic Kidney Disease (CKD) Subtypes. Metabolites, 12(6), 490. https://doi.org/10.3390/metabo12060490








