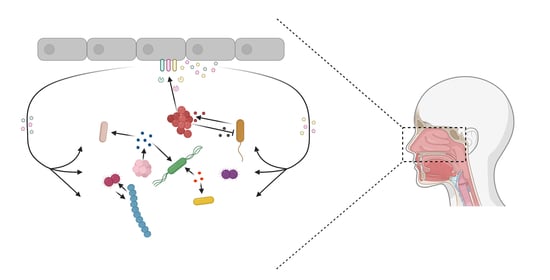
Nutritional Interactions between Bacterial Species Colonising the Human Nasal Cavity: Current Knowledge and Future Prospects
Abstract
1. Introduction
2. Nutritional Interactions amongst Species of the Nasal Microbiome
2.1. Secreted Small Molecules
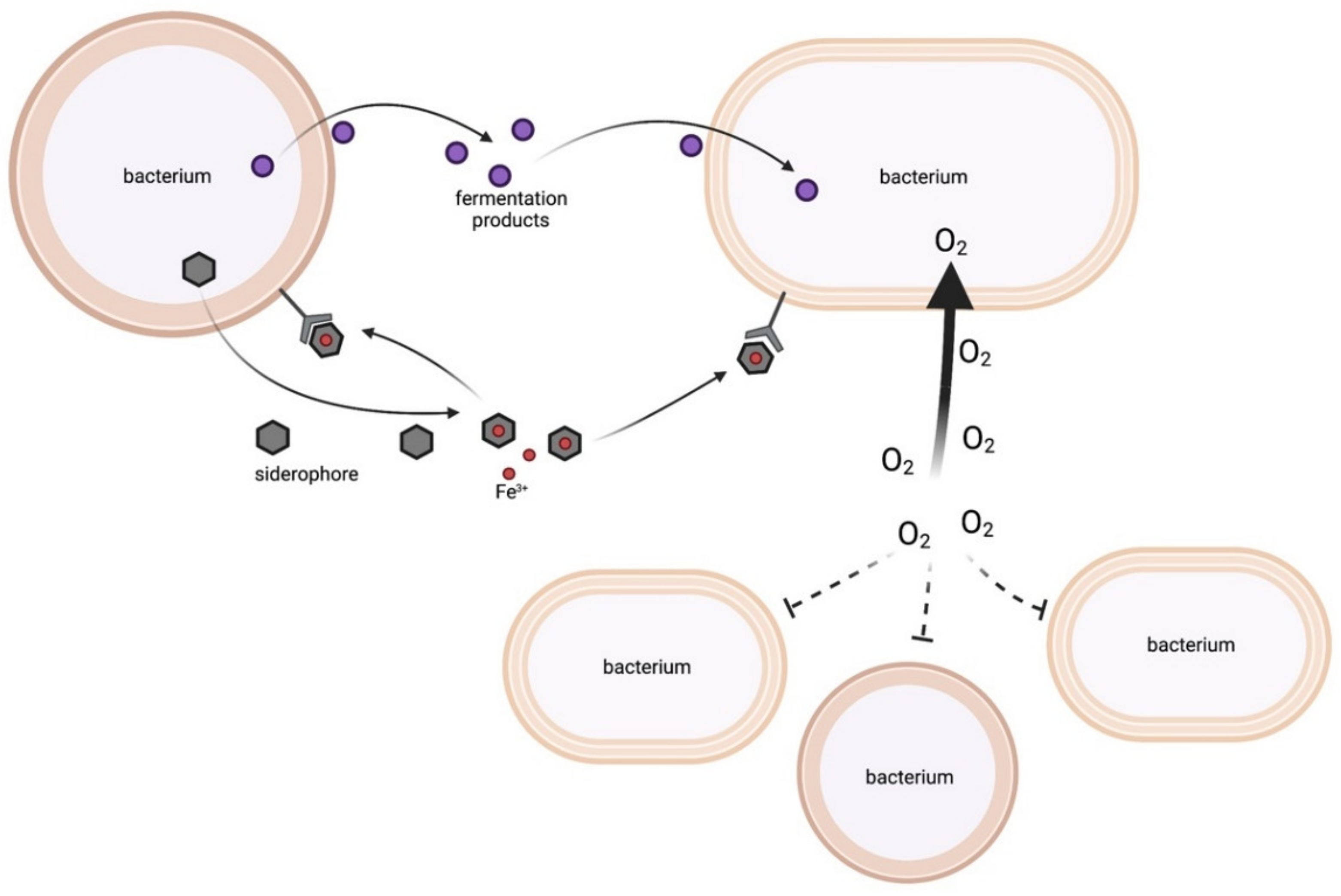
2.1.1. Energy-Rich Fermentation Products
2.1.2. Siderophores
2.1.3. Oxygen Consumption
2.2. Host Cells as a Source of Nutrients
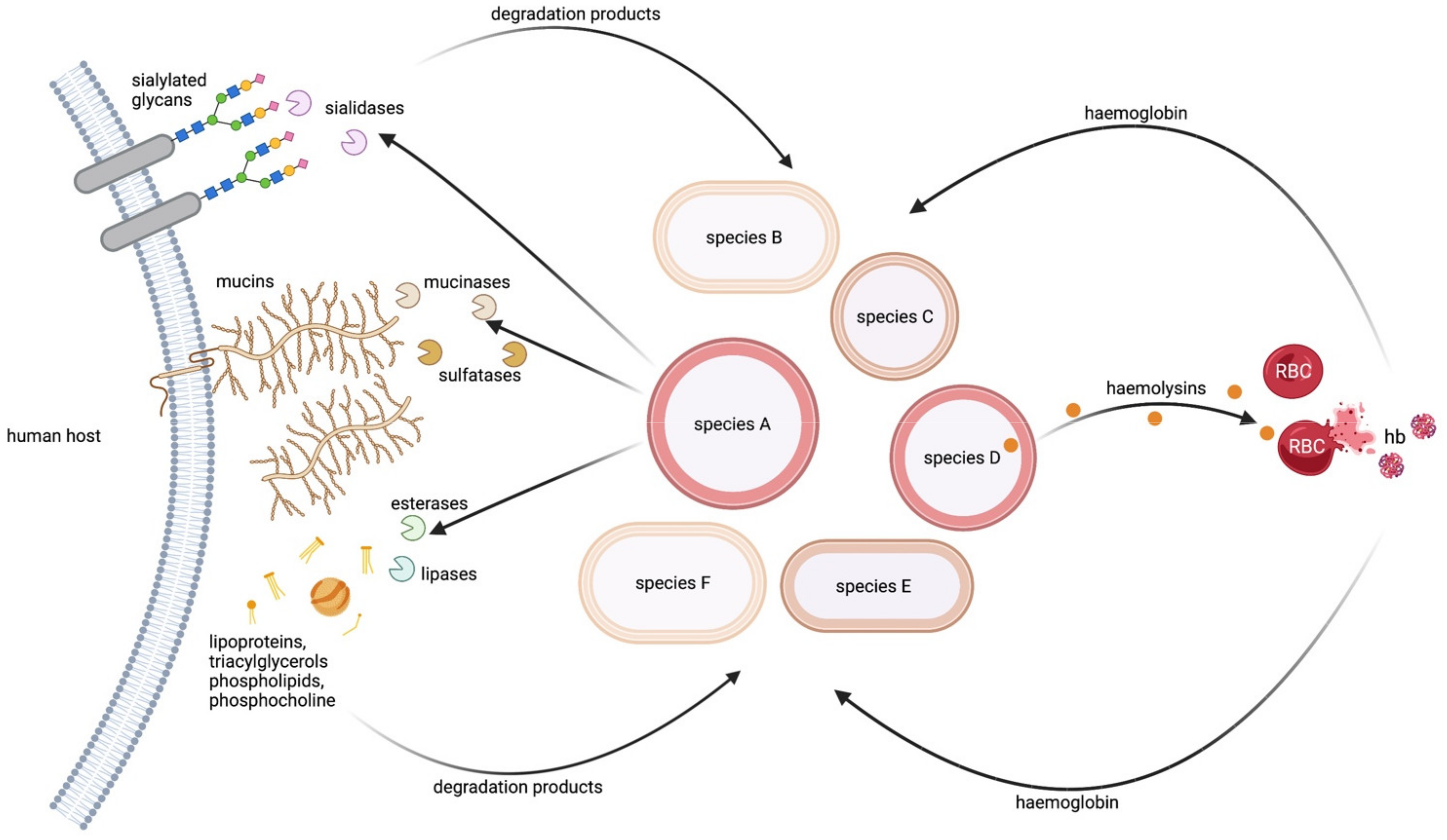
2.2.1. Host Mucins as a Source of Carbon and Sulphate
2.2.2. Host Glycans as a Source of Sialic Acid
2.2.3. Host Fatty Acids and Phospholipids as a Source of Carbon and Phosphorous
2.2.4. Host Erythrocytes as a Source of Haem
2.3. Some Microbiome Members Act as Prey to Obtain Essential Nutrients
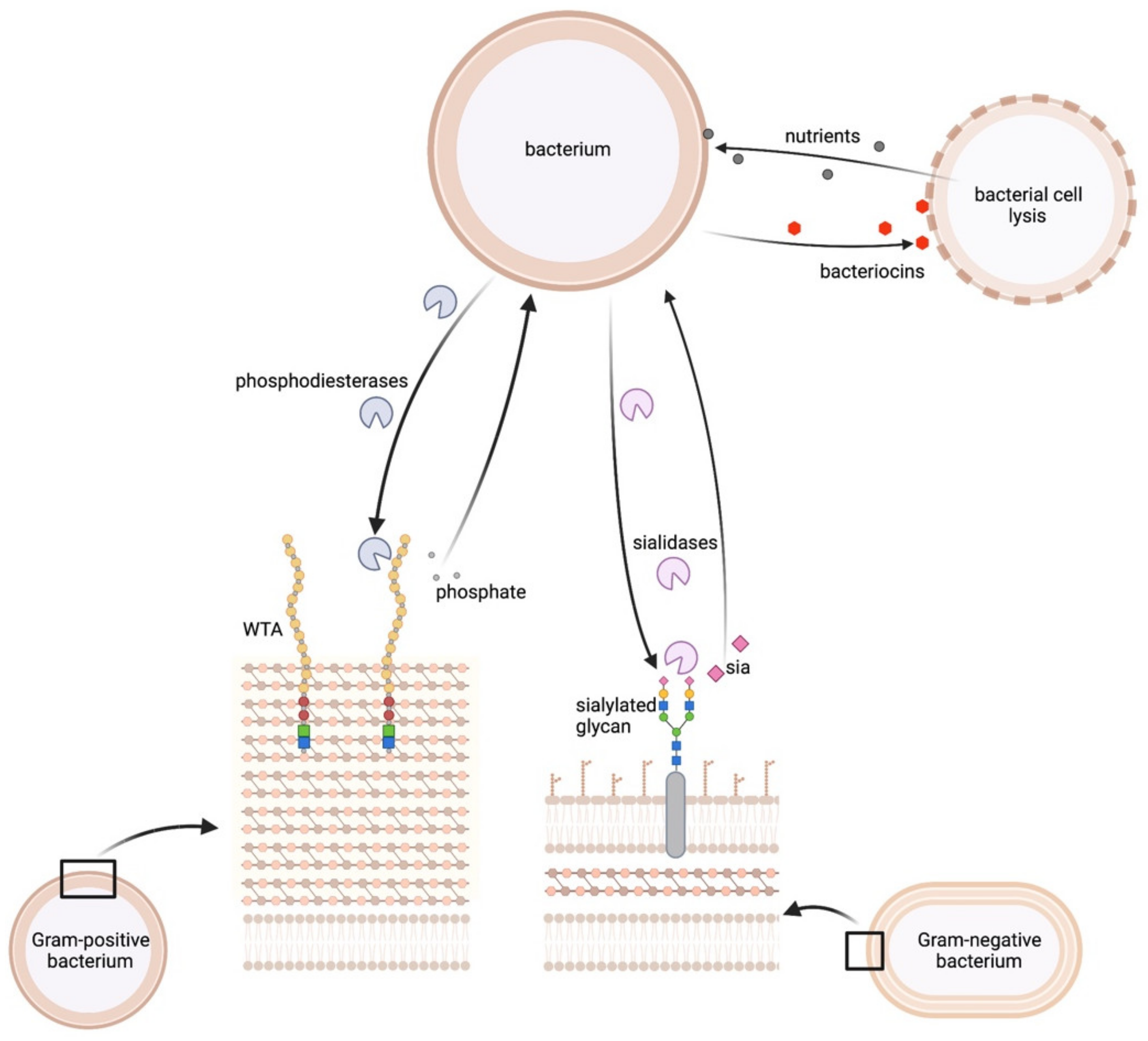
2.3.1. WTA as a Source of Phosphorus
2.3.2. Bacterial Surfaces as a Source for Sialic Acid
2.3.3. Lysis of Bacterial Cells to Release Diverse Nutrients
2.4. Uncharacterised Bacterial Interactions
3. Human Diseases Altering Nutritional Composition in the Upper Respiratory Tract
4. Genome-Based Metabolic Models to Predict Bacterial Interactions
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Ogilvie, L.A.; Jones, B.V. The human gut virome: A multifaceted majority. Front. Microbiol. 2015, 6, 918. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Bomar, L.; Brugger, S.D.; Lemon, K.P. Bacterial microbiota of the nasal passages across the span of human life. Curr. Opin. Microbiol. 2018, 41, 8–14. [Google Scholar] [CrossRef]
- Bode, L.G.; Kluytmans, J.A.; Wertheim, H.F.; Bogaers, D.; Vandenbroucke-Grauls, C.M.; Roosendaal, R.; Troelstra, A.; Box, A.T.; Voss, A.; van der Tweel, I.; et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N. Engl. J. Med. 2010, 362, 9–17. [Google Scholar] [CrossRef]
- Di Stadio, A.; Costantini, C.; Renga, G.; Pariano, M.; Ricci, G.; Romani, L. The Microbiota/Host Immune System Interaction in the Nose to Protect from COVID-19. Life 2020, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Welp, A.L.; Bomberger, J.M. Bacterial Community Interactions During Chronic Respiratory Disease. Front. Cell Infect. Microbiol. 2020, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Rawls, M.; Ellis, A.K. The microbiome of the nose. Ann. Allergy Asthma Immunol. 2019, 122, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Hardy, B.L.; Merrell, D.S. Friend or Foe: Interbacterial Competition in the Nasal Cavity. J. Bacteriol. 2021, 203, e00480-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Price, L.B.; Hungate, B.A.; Abraham, A.G.; Larsen, L.A.; Christensen, K.; Stegger, M.; Skov, R.; Andersen, P.S. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015, 1, e1400216. [Google Scholar] [CrossRef]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R.; et al. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef]
- Krismer, B.; Liebeke, M.; Janek, D.; Nega, M.; Rautenberg, M.; Hornig, G.; Unger, C.; Weidenmaier, C.; Lalk, M.; Peschel, A. Nutrient Limitation Governs Staphylococcus aureus Metabolism and Niche Adaptation in the Human Nose. PLoS Pathog. 2014, 10, e1003862. [Google Scholar] [CrossRef]
- Sassone-Corsi, M.; Raffatellu, M. No vacancy: How beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 2015, 194, 4081–4087. [Google Scholar] [CrossRef]
- Heilbronner, S.; Krismer, B.; Brotz-Oesterhelt, H.; Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 2021, 19, 726–739. [Google Scholar] [CrossRef]
- Coulthurst, S. The Type VI secretion system: A versatile bacterial weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffman, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal Expression of Adhesion Factors and Activity of Global Regulators during Establishment of Staphylococcus aureus Nasal Colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef]
- Burian, M.; Wolz, C.; Goerke, C. Regulatory Adaptation of Staphylococcus aureus during Nasal Colonization of Humans. PLoS ONE 2010, 5, e10040. [Google Scholar] [CrossRef]
- Martens, E.C.; Chiang, H.C.; Gordon, J.I. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe 2008, 4, 447–457. [Google Scholar] [CrossRef]
- Momose, Y.; Hirayama, K.; Itoh, K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek 2008, 94, 165–171. [Google Scholar] [CrossRef]
- Maltby, R.; Leatham-Jensen, M.P.; Gibson, T.; Cohen, P.S.; Conway, T. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. PLoS ONE 2013, 8, e53957. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan Sylvia, H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley Gerald, E.; Flint Harry, J. Two Routes of Metabolic Cross-Feeding between Bifidobacterium adolescentis and Butyrate-Producing Anaerobes from the Human Gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef]
- Moens, F.; Verce, M.; De Vuyst, L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int. J. Food Microbiol. 2017, 241, 225–236. [Google Scholar] [CrossRef]
- Magalhães, A.P.; Grainha, T.; Sousa, A.M.; França, Â.; Cerca, N.; Pereira, M.O. Viable but non-cultivable state: A strategy for Staphylococcus aureus survivable in dual-species biofilms with Pseudomonas aeruginosa? Environ. Microbiol. 2021, 23, 5639–5649. [Google Scholar] [CrossRef]
- Khanolkar Rutvij, A.; Clark Shawn, T.; Wang Pauline, W.; Hwang David, M.; Yau Yvonne, C.W.; Waters Valerie, J.; Guttman David, S.; Cleary David, W. Ecological Succession of Polymicrobial Communities in the Cystic Fibrosis Airways. mSystems 2020, 5, e00809-20. [Google Scholar] [CrossRef]
- Limoli, D.H.; Hoffman, L.R. Help, hinder, hide and harm: What can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 2019, 74, 684. [Google Scholar] [CrossRef]
- Camus, L.; Briaud, P.; Bastien, S.; Elsen, S.; Doléans-Jordheim, A.; Vandenesch, F.; Moreau, K. Trophic cooperation promotes bacterial survival of Staphylococcus aureus and Pseudomonas aeruginosa. ISME J. 2020, 14, 3093–3105. [Google Scholar] [CrossRef]
- Venkataraman, A.; Rosenbaum, M.A.; Werner, J.J.; Winans, S.C.; Angenent, L.T. Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa. ISME J. 2014, 8, 1210–1220. [Google Scholar] [CrossRef]
- Nguyen, M.; Sharma, A.; Wu, W.; Gomi, R.; Sung, B.; Hospodsky, D.; Angenent, L.T.; Worgall, S. The fermentation product 2,3-butanediol alters P. aeruginosa clearance, cytokine response and the lung microbiome. ISME J. 2016, 10, 2978–2983. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Kang, Z.; Xiao, D.; Gao, C.; Xu, P.; Ma, C. 2,3-Butanediol catabolism in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 2018, 20, 3927–3940. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Marahiel Mohamed, A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Cordero, O.X.; Ventouras, L.-A.; DeLong, E.F.; Polz, M.F. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. USA 2012, 109, 20059–20064. [Google Scholar] [CrossRef] [PubMed]
- Griffin, A.S.; West, S.A.; Buckling, A. Cooperation and competition in pathogenic bacteria. Nature 2004, 430, 1024–1027. [Google Scholar] [CrossRef] [PubMed]
- Sebulsky, M.T.; Heinrichs, D.E. Identification and Characterization of fhuD1 and fhuD2, Two Genes Involved in Iron-Hydroxamate Uptake in Staphylococcus aureus. J. Bacteriol. 2001, 183, 4994–5000. [Google Scholar] [CrossRef] [PubMed]
- Tuomanen, E.I.; Morrissey, J.A.; Cockayne, A.; Hill, P.J.; Williams, P. Molecular Cloning and Analysis of a Putative Siderophore ABC Transporter from Staphylococcus aureus. Infect. Immun. 2000, 68, 6281–6288. [Google Scholar] [CrossRef]
- Brozyna, J.R.; Sheldon, J.R.; Heinrichs, D.E. Growth promotion of the opportunistic human pathogen, Staphylococcus lugdunensis, by heme, hemoglobin, and coculture with Staphylococcus aureus. Microbiologyopen 2014, 3, 182–195. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; May, D.S.; Chevrette, M.G.; Temkin, M.I.; Wendt-Pienkowski, E.; Cagnazzo, J.; Carlson, C.M.; Gern, J.E.; Currie, C.R. Competition among Nasal Bacteria Suggests a Role for Siderophore-Mediated Interactions in Shaping the Human Nasal Microbiota. Appl. Environ. Microbiol. 2019, 85, e02406-18. [Google Scholar] [CrossRef]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef]
- Ali, M.Y. Histology of the human nasopharyngeal mucosa. J. Anat. 1965, 99, 657–672. [Google Scholar]
- Brook, I. The role of anaerobic bacteria in sinusitis. Anaerobe 2006, 12, 5–12. [Google Scholar] [CrossRef]
- Varki, N.M.; Varki, A. Diversity in cell surface sialic acid presentations: Implications for biology and disease. Lab. Investig. 2007, 87, 851–857. [Google Scholar] [CrossRef]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef]
- McShane, A.; Bath, J.; Jaramillo, A.M.; Ridley, C.; Walsh, A.A.; Evans, C.M.; Thornton, D.J.; Ribbeck, K. Mucus. Curr. Biol. 2021, 31, R938–R945. [Google Scholar] [CrossRef]
- Lucas, S.K.; Villarreal, A.R.; Ahmad, M.M.; Itabiyi, A.; Feddema, E.; Boyer, H.C.; Hunter, R.C.; Raffatellu, M. Anaerobic Microbiota Derived from the Upper Airways Impact Staphylococcus aureus Physiology. Infect. Immun. 2021, 89, e0015321. [Google Scholar] [CrossRef]
- Bradshaw, D.J.; Homer, K.A.; Marsh, P.D.; Beighton, D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology 1994, 140, 3407–3412. [Google Scholar] [CrossRef]
- Audry, M.; Robbe-Masselot, C.; Barnier, J.-P.; Gachet, B.; Saubaméa, B.; Schmitt, A.; Schönherr-Hellec, S.; Léonard, R.; Nassif, X.; Coureuil, M.; et al. Airway Mucus Restricts Neisseria meningitidis Away from Nasopharyngeal Epithelial Cells and Protects the Mucosa from Inflammation. mSphere 2019, 4, e00494-19. [Google Scholar] [CrossRef]
- Flynn, J.M.; Niccum, D.; Dunitz, J.M.; Hunter, R.C. Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease. PLoS Pathog. 2016, 12, e1005846. [Google Scholar] [CrossRef]
- Cho, D.-Y.; Skinner, D.; Hunter, R.C.; Weeks, C.; Lim, D.J.; Thompson, H.; Walz, C.R.; Zhang, S.; Grayson, J.W.; Swords, W.E.; et al. Contribution of Short Chain Fatty Acids to the Growth of Pseudomonas aeruginosa in Rhinosinusitis. Front. Cell Infect. Microbiol. 2020, 10, 412. [Google Scholar] [CrossRef]
- Roberton, A.M.; Wright, D.P. Bacterial Glycosulphatases and Sulphomucin Degradation. Can. J. Gastroenterol. 1997, 11, 642360. [Google Scholar] [CrossRef]
- Luis, A.S.; Jin, C.; Pereira, G.V.; Glowacki, R.W.P.; Gugel, S.R.; Singh, S.; Byrne, D.P.; Pudlo, N.A.; London, J.A.; Baslé, A.; et al. A single sulfatase is required to access colonic mucin by a gut bacterium. Nature 2021, 598, 332–337. [Google Scholar] [CrossRef]
- Kida, Y.; Yamamoto, T.; Kuwano, K. SdsA1, a secreted sulfatase, contributes to the in vivo virulence of Pseudomonas aeruginosa in mice. Microbiol. Immunol. 2020, 64, 280–295. [Google Scholar] [CrossRef]
- King, S.J.; Hippe, K.R.; Gould, J.M.; Bae, D.; Peterson, S.; Cline, R.T.; Fasching, C.; Janoff, E.N.; Weiser, J.N. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 2004, 54, 159–171. [Google Scholar] [CrossRef]
- Spik, G.; Strecker, G.; Fournet, B.; Bouquelet, S.; Montreuil, J.; Dorland, L.; van Halbeek, H.; Vliegenthart, J.F. Primary structure of the glycans from human lactotransferrin. Eur. J. Biochem. 1982, 121, 413–419. [Google Scholar] [CrossRef]
- Mattu, T.S.; Pleass, R.J.; Willis, A.C.; Kilian, M.; Wormald, M.R.; Lellouch, A.C.; Rudd, P.M.; Woof, J.M.; Dwek, R.A. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J. Biol. Chem. 1998, 273, 2260–2272. [Google Scholar] [CrossRef]
- Vimr Eric, R.; Kalivoda Kathryn, A.; Deszo Eric, L.; Steenbergen Susan, M. Diversity of Microbial Sialic Acid Metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132–153. [Google Scholar] [CrossRef]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology 2007, 153, 2817–2822. [Google Scholar] [CrossRef]
- Jennings, M.P.; Day, C.J.; Atack, J.M. How bacteria utilize sialic acid during interactions with the host: Snip, snatch, dispatch, match and attach. Microbiology 2022, 168, 001157. [Google Scholar] [CrossRef]
- Burnaugh, A.M.; Frantz, L.J.; King, S.J. Growth of Streptococcus pneumoniae on Human Glycoconjugates Is Dependent upon the Sequential Activity of Bacterial Exoglycosidases. J. Bacteriol. 2008, 190, 221–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brüggemann, H.; Henne, A.; Hoster, F.; Liesegang, H.; Wiezer, A.; Strittmatter, A.; Hujer, S.; Dürre, P.; Gottschalk, G. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 2004, 305, 671–673. [Google Scholar] [CrossRef] [PubMed]
- Von Nicolai, H.; Höffler, U.; Zilliken, F. Isolation, Purification, and Properties of Neuraminidase from Propionibacterium acnes. Zent. Bakteriol. 1 Abt. Orig. A Med. Mikrobiol. Infekt. Parasitol. 1980, 247, 84–94. [Google Scholar] [CrossRef]
- Derrien, M.; van Passel, M.W.J.; van de Bovenkamp, J.H.B.; Schipper, R.; de Vos, W.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef]
- Olson, M.E.; King, J.M.; Yahr, T.L.; Horswill, A.R. Sialic Acid Catabolism in Staphylococcus aureus. J. Bacteriol. 2013, 195, 1779–1788. [Google Scholar] [CrossRef]
- Vromman, F.; Subtil, A. Exploitation of host lipids by bacteria. Curr. Opin. Microbiol. 2014, 17, 38–45. [Google Scholar] [CrossRef]
- Fozo, E.M.; Rucks, E.A. Chapter Two—The Making and Taking of Lipids: The Role of Bacterial Lipid Synthesis and the Harnessing of Host Lipids in Bacterial Pathogenesis. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: New York, NY, USA, 2016; Volume 69, pp. 51–155. [Google Scholar]
- Wargo, M.J. Homeostasis and Catabolism of Choline and Glycine Betaine: Lessons from Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2112–2120. [Google Scholar] [CrossRef]
- Kengmo Tchoupa, A.; Eijkelkamp, B.A.; Peschel, A. Bacterial adaptation strategies to host-derived fatty acids. Trends Microbiol. 2022, 30, 241–253. [Google Scholar] [CrossRef]
- Do, T.Q.; Moshkani, S.; Castillo, P.; Anunta, S.; Pogosyan, A.; Cheung, A.; Marbois, B.; Faull, K.F.; Ernst, W.; Chiang, S.M.; et al. Lipids Including Cholesteryl Linoleate and Cholesteryl Arachidonate Contribute to the Inherent Antibacterial Activity of Human Nasal Fluid. J. Immunol. 2008, 181, 4177–4187. [Google Scholar] [CrossRef]
- Corda, D.; Mosca, M.G.; Ohshima, N.; Grauso, L.; Yanaka, N.; Mariggiò, S. The emerging physiological roles of the glycerophosphodiesterase family. FEBS J. 2014, 281, 998–1016. [Google Scholar] [CrossRef]
- Jaeger, K.-E.; Ransac, S.; Dijkstra, B.W.; Colson, C.; van Heuvel, M.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. [Google Scholar] [CrossRef]
- Jorge, A.M.; Schneider, J.; Unsleber, S.; Xia, G.; Mayer, C.; Peschel, A. Staphylococcus aureus counters phosphate limitation by scavenging wall teichoic acids from other staphylococci via the teichoicase GlpQ. J. Biol. Chem. 2018, 293, 14916–14924. [Google Scholar] [CrossRef]
- Jorge, A.M.; Schneider, J.; Unsleber, S.; Göhring, N.; Mayer, C.; Peschel, A. Utilization of glycerophosphodiesters by Staphylococcus aureus. Mol. Microbiol. 2017, 103, 229–241. [Google Scholar] [CrossRef]
- Delekta, P.C.; Shook, J.C.; Lydic, T.A.; Mulks, M.H.; Hammer, N.D.; O’Toole, G. Staphylococcus aureus Utilizes Host-Derived Lipoprotein Particles as Sources of Fatty Acids. J. Bacteriol. 2018, 200, e00728-17. [Google Scholar] [CrossRef]
- Bomar, L.; Brugger, S.D.; Yost, B.H.; Davies, S.S.; Lemon, K.P. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio 2016, 7, e01725. [Google Scholar] [CrossRef]
- Zheng, Y.; Hunt, R.L.; Villaruz, A.E.; Fisher, E.L.; Liu, R.; Liu, Q.; Cheung, G.Y.C.; Li, M.; Otto, M. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe 2022, 30, 301–313. [Google Scholar] [CrossRef]
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Moeller Christensen, G.J.; Nakatsuji, T.; et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef]
- Kang, Y.; Zarzycki-Siek, J.; Walton, C.B.; Norris, M.H.; Hoang, T.T. Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa. PLoS ONE 2010, 5, e13557. [Google Scholar] [CrossRef]
- Choby, J.E.; Skaar, E.P. Heme Synthesis and Acquisition in Bacterial Pathogens. J. Mol. Biol. 2016, 428, 3408–3428. [Google Scholar] [CrossRef]
- Pynnonen, M.; Stephenson, R.E.; Schwartz, K.; Hernandez, M.; Boles, B.R. Hemoglobin Promotes Staphylococcus aureus Nasal Colonization. PLoS Pathog. 2011, 7, e1002104. [Google Scholar] [CrossRef]
- Heilbronner, S.; Holden, M.T.; van Tonder, A.; Geoghegan, J.A.; Foster, T.J.; Parkhill, J.; Bentley, S.D. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 2011, 322, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Donvito, B.; Etienne, J.; Denoroy, L.; Greenland, T.; Benito, Y.; Vandenesch, F. Synergistic hemolytic activity of Staphylococcus lugdunensis is mediated by three peptides encoded by a non-agr genetic locus. Infect. Immun. 1997, 65, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Donvito, B.; Etienne, J.; Greenland, T.; Mouren, C.; Delorme, V.; Vandenesch, F. Distribution of the synergistic haemolysin genes hld and slush with respect to agr in human staphylococci. FEMS Microbiol. Lett. 1997, 151, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Dekio, I.; McDowell, A.; Sakamoto, M.; Tomida, S.; Ohkuma, M. Proposal of new combination, Cutibacterium acnes subsp. elongatum comb. nov., and emended descriptions of the genus Cutibacterium, Cutibacterium acnes subsp. acnes and Cutibacterium acnes subsp. defendens. Int. J. Syst. Evol. Microbiol. 2019, 69, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.A.; Gray, E.; Wang, Y.-P.; Roe, B.A.; Dyer, D.W. Molecular characterization of hpuAB, the haemoglobin–haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 1997, 23, 737–749. [Google Scholar] [CrossRef]
- Morton, D.J.; Seale, T.W.; Madore, L.L.; VanWagoner, T.M.; Whitby, P.W.; Stull, T.L. The haem–haemopexin utilization gene cluster (hxuCBA) as a virulence factor of Haemophilus influenzae. Microbiology 2007, 153, 215–224. [Google Scholar] [CrossRef]
- Marvig, R.L.; Damkiær, S.; Khademi, S.M.H.; Markussen, T.M.; Molin, S.; Jelsbak, L.; Keim, P.S. Within-Host Evolution of Pseudomonas aeruginosa Reveals Adaptation toward Iron Acquisition from Hemoglobin. MBio 2014, 5, e00966-14. [Google Scholar] [CrossRef]
- Schmitt, M.P. Utilization of host iron sources by Corynebacterium diphtheriae: Identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 1997, 179, 838–845. [Google Scholar] [CrossRef]
- Artman, M.; Domenech, E.; Weiner, M. Growth of Haemophilus influenzae in simulated blood cultures supplemented with hemin and NAD. J. Clin. Microbiol. 1983, 18, 376–379. [Google Scholar] [CrossRef]
- Hajishengallis, G. The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral Microbiol. 2014, 29, 248–257. [Google Scholar] [CrossRef]
- Van Dalen, R.; Peschel, A.; van Sorge, N.M. Wall Teichoic Acid in Staphylococcus aureus Host Interaction. Trends Microbiol. 2020, 28, 985–998. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Shakhnovich, E.A.; King, S.J.; Weiser, J.N. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: A paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect. Immun. 2002, 70, 7161–7164. [Google Scholar] [CrossRef]
- González-Pastor José, E.; Hobbs Errett, C.; Losick, R. Cannibalism by Sporulating Bacteria. Science 2003, 301, 510–513. [Google Scholar] [CrossRef]
- Brugger, S.D.; Eslami, S.M.; Pettigrew, M.M.; Escapa, I.F.; Henke, M.T.; Kong, Y.; Lemon, K.P. Dolosigranulum pigrum Cooperation and Competition in Human Nasal Microbiota. mSphere 2020, 5, e00852-20. [Google Scholar] [CrossRef]
- Yan, M.; Sünje, J.P.; Fukuyama, J.; Hwang, P.H.; Cho, D.-Y.; Holmes, S.; Relman, D.A. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 2013, 14, 631–640. [Google Scholar] [CrossRef]
- Kanmani, P.; Clua, P.; Vizoso-Pinto, M.G.; Rodriguez, C.; Alvarez, S.; Melnikov, V.; Takahashi, H.; Kitazawa, H.; Villena, J. Respiratory Commensal Bacteria Corynebacterium pseudodiphtheriticum Improves Resistance of Infant Mice to Respiratory Syncytial Virus and Streptococcus pneumoniae Superinfection. Front. Microbiol. 2017, 8, 1613. [Google Scholar] [CrossRef]
- Raphael, G.D.; Jeney, E.V.; Baraniuk, J.N.; Kim, I.; Meredith, S.D.; Kaliner, M.A. Pathophysiology of rhinitis. Lactoferrin and lysozyme in nasal secretions. J. Clin. Investig. 1989, 84, 1528–1535. [Google Scholar] [CrossRef]
- Wood, D.M.; Brennan, A.L.; Philips, B.J.; Baker, E.H. Effect of hyperglycaemia on glucose concentration of human nasal secretions. Clin. Sci. 2004, 106, 527–533. [Google Scholar] [CrossRef]
- Zhou, W.; Sailani, M.R.; Contrepois, K.; Zhou, Y.; Ahadi, S.; Leopold, S.R.; Zhang, M.J.; Rao, V.; Avina, M.; Mishra, T.; et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 2019, 569, 663–671. [Google Scholar] [CrossRef]
- Ahluwalia, A.; Sood, A.; Sood, A.; Lakshmy, R.; Kapil, A.; Pandey, R.M. Nasal colonization with Staphylococcus aureus in patients with diabetes mellitus. Diabet. Med. 2000, 17, 487–488. [Google Scholar] [CrossRef]
- Gill, S.K.; Hui, K.; Farne, H.; Garnett, J.P.; Baines, D.L.; Moore, L.S.P.; Holmes, A.H.; Filloux, A.; Tregoning, J.S. Increased airway glucose increases airway bacterial load in hyperglycaemia. Sci. Rep. 2016, 6, 27636. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Xu, H.; Yi, H.; Guan, J.; Yin, S. Metabolomics and microbiome profiling as biomarkers in obstructive sleep apnoea: A comprehensive review. Eur. Respir. Rev. 2021, 30, 200220. [Google Scholar] [CrossRef]
- Cai, Y.; Juszczak, H.M.; Cope, E.K.; Goldberg, A.N. The microbiome in obstructive sleep apnea. Sleep 2021, 44, zsab061. [Google Scholar] [CrossRef]
- Yang, W.; Shao, L.; Heizhati, M.; Wu, T.; Yao, X.; Wang, Y.; Wang, L.; Li, N. Oropharyngeal Microbiome in Obstructive Sleep Apnea: Decreased Diversity and Abundance. J. Clin. Sleep Med. 2019, 15, 1777–1788. [Google Scholar] [CrossRef]
- Wu, B.G.; Sulaiman, I.; Wang, J.; Shen, N.; Clemente, J.C.; Li, Y.; Laumbach, R.J.; Lu, S.E.; Udasin, I.; Le-Hoang, O.; et al. Severe Obstructive Sleep Apnea Is Associated with Alterations in the Nasal Microbiome and an Increase in Inflammation. Am. J. Respir. Crit. Care Med. 2019, 199, 99–109. [Google Scholar] [CrossRef]
- Schilling, C.H.; Palsson, B.O. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J. Theor. Biol. 2000, 203, 249–283. [Google Scholar] [CrossRef]
- Papin, J.A.; Price, N.D.; Edwards, J.S.; Palsson, B.Ø. The Genome-Scale Metabolic Extreme Pathway Structure in Haemophilus influenzae Shows Significant Network Redundancy. J. Theor. Biol. 2002, 215, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-C.; Huang, T.-W.; Chen, F.-C.; Charusanti, P.; Hong, J.S.J.; Chang, H.-Y.; Tsai, S.-F.; Palsson, B.O.; Hsiung, C.A. An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, i YL1228. J. Bacteriol. 2011, 193, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Rokem, J.S.; Vongsangnak, W.; Nielsen, J. Comparative metabolic capabilities for Micrococcus luteus NCTC 2665, the “Fleming” strain, and actinobacteria. Biotechnol. Bioeng. 2011, 108, 2770–2775. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.D.; Renz, A.; Dunphy, L.J.; Lewis, T.; Dräger, A.; Papin, J.A. An updated genome-scale metabolic network reconstruction of Pseudomonas aeruginosa PA14 to characterize mucin-driven shifts in bacterial metabolism. NPJ Syst. Biol. Appl. 2021, 7, 37. [Google Scholar] [CrossRef]
- Renz, A.; Dräger, A. Curating and comparing 114 strain-specific genome-scale metabolic models of Staphylococcus aureus. NPJ Syst. Biol. Appl. 2021, 7, 30. [Google Scholar] [CrossRef]
- Renz, A.; Widerspick, L.; Dräger, A. First Genome-Scale Metabolic Model of Dolosigranulum pigrum Confirms Multiple Auxotrophies. Metabolites 2021, 11, 232. [Google Scholar] [CrossRef]
- Shoaie, S.; Karlsson, F.; Mardinoglu, A.; Nookaew, I.; Bordel, S.; Nielsen, J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci. Rep. 2013, 3, 2532. [Google Scholar] [CrossRef]
- Heinken, A.; Thiele, I. Systematic prediction of health-relevant humanmicrobial co-metabolism through a computational framework. Gut Microbes 2015, 6, 120–130. [Google Scholar] [CrossRef]
- Bauer, E.; Zimmermann, J.; Baldini, F.; Thiele, I.; Kaleta, C. BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput. Biol. 2017, 13, e1005544. [Google Scholar] [CrossRef]
- Diener, C.; Gibbons, S.M.; Resendis-Antonio, O. MICOM: Metagenome-Scale Modeling to Infer Metabolic Interactions in the Gut Microbiota. mSystems 2020, 5, e00606-19. [Google Scholar] [CrossRef]
- Heinken, A.; Thiele, I. Anoxic conditions promote species-specific mutualism between gut microbes In Silico. Appl. Environ. Microbiol. 2015, 81, 4049–4061. [Google Scholar] [CrossRef]
- El-Semman, I.E.; Karlsson, F.H.; Shoaie, S.; Nookaew, I.; Soliman, T.H.; Nielsen, J. Genome-scale metabolic reconstructions of Bifidobacterium adolescentis L2-32 and Faecalibacterium prausnitzii A2-165 and their interaction. BMC Syst. Biol. 2014, 8, 41. [Google Scholar] [CrossRef]
- Tzamali, E.; Poirazi, P.; Tollis, I.G.; Reczko, M. A computational exploration of bacterial metabolic diversity identifying metabolic interactions and growth-efficient strain communities. BMC Syst. Biol. 2011, 5, 167. [Google Scholar] [CrossRef]
- Hoek, M.J.A.v.; Merks, R.M.H. Emergence of microbial diversity due to cross-feeding interactions in a spatial model of gut microbial metabolism. BMC Syst. Biol. 2017, 11, 56. [Google Scholar] [CrossRef]
- Glöckler, M.; Dräger, A.; Mostolizadeh, R. NCMW: A Python Package to Analyze Metabolic Interactions in the Nasal Microbiome. Front. Bioinform. 2022, 2, 827024. [Google Scholar] [CrossRef]

| Beneficial Interactions | ||||
|---|---|---|---|---|
| Nutrient Source | Metabolite/Public Good | Producer of Metabolite/Macromolecule Degrading Strain | Beneficiary | Ref. |
| bacterial metabolism | acetoin | Staph. aureus | P. aeruginosa | [28] |
| bacterial metabolism | 2,3-butanediol | fermenting bacteria | P. aeruginosa environmental microbes | [29,30] |
| bacterial metabolism | siderophores staphyloferrin A and B | Staph. aureus | Staph. lugdunensis | [38] |
| bacterial oxygen consumption | oxygen | oxygen-consuming aerobic bacteria | anaerobic bacteria | [42] |
| human mucins | mucin degradation products | mucin-degrading bacteria | Staph. aureus oral microbiome members | [47] |
| human mucins | mucin degradation products | Strep. mitis | N. meningitidis | [49] |
| human mucins | mucin degradation products/SCFAs | anaerobic communities | P. aeruginosa | [50,51] |
| human sialylated molecules | sialic acid | Strep. pneumoniae | Strep. pneumoniae | [61] |
| bacterial sialylated molecules | sialic acid | H. influenzae, N. meningitidis | Strep. pneumoniae | [94] |
| human phospholipids | glycerol-3-phosphate | Staph. aureus | Staph. aureus | [74] |
| WTA from CoNS | glycerol-3-phosphate | Staph. aureus | Staph. aureus | [73] |
| human low-density lipoproteins | fatty acids | Staph. aureus | Staph. aureus | [75] |
| human triacylglycerols | fatty acids | C. accolens | C. accolens | [76] |
| human sphingomyelin | phosphocholine | Staph. epidermidis | Staph. epidermidis | [77] |
| human sphingomyelin | ceramide | Staph. epidermidis | human host | [77] |
| human fatty acids | fatty acids | P. aeruginosa | P. aeruginosa | [79] |
| human erythrocytes | haemin & NAD+ | Staph. aureus | H. influenzae | [90] |
| human tissue destruction | amino acids, haem | P. gingivalis | bacterial community | [91] |
| bacterial metabolism | unknown | Corynebacterium spp. | D. pigrum | [96] |
| Inhibiting Interactions | ||||
| Nutrient Source | Metabolite/Public Good | Producer of Metabolite/Macromolecule Degrading Strain | Inhibited Species | Ref. |
| bacterial metabolism | acetoin | Staph. aureus | Staph. aureus | [28] |
| bacterial metabolism | siderophore dehydroxynocarda-mine | C. propinquum | CoNS | [39] |
| human triacylglycerols | oleic acid | C. accolens | Strep. pneumoniae | [76] |
| bacterial metabolism | bacteriocins | D. pigrum | unknown | [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adolf, L.A.; Heilbronner, S. Nutritional Interactions between Bacterial Species Colonising the Human Nasal Cavity: Current Knowledge and Future Prospects. Metabolites 2022, 12, 489. https://doi.org/10.3390/metabo12060489
Adolf LA, Heilbronner S. Nutritional Interactions between Bacterial Species Colonising the Human Nasal Cavity: Current Knowledge and Future Prospects. Metabolites. 2022; 12(6):489. https://doi.org/10.3390/metabo12060489
Chicago/Turabian StyleAdolf, Lea A., and Simon Heilbronner. 2022. "Nutritional Interactions between Bacterial Species Colonising the Human Nasal Cavity: Current Knowledge and Future Prospects" Metabolites 12, no. 6: 489. https://doi.org/10.3390/metabo12060489
APA StyleAdolf, L. A., & Heilbronner, S. (2022). Nutritional Interactions between Bacterial Species Colonising the Human Nasal Cavity: Current Knowledge and Future Prospects. Metabolites, 12(6), 489. https://doi.org/10.3390/metabo12060489