Early Metabolomic Markers of Acute Low-Dose Exposure to Uranium in Rats
Abstract
1. Introduction
2. Results
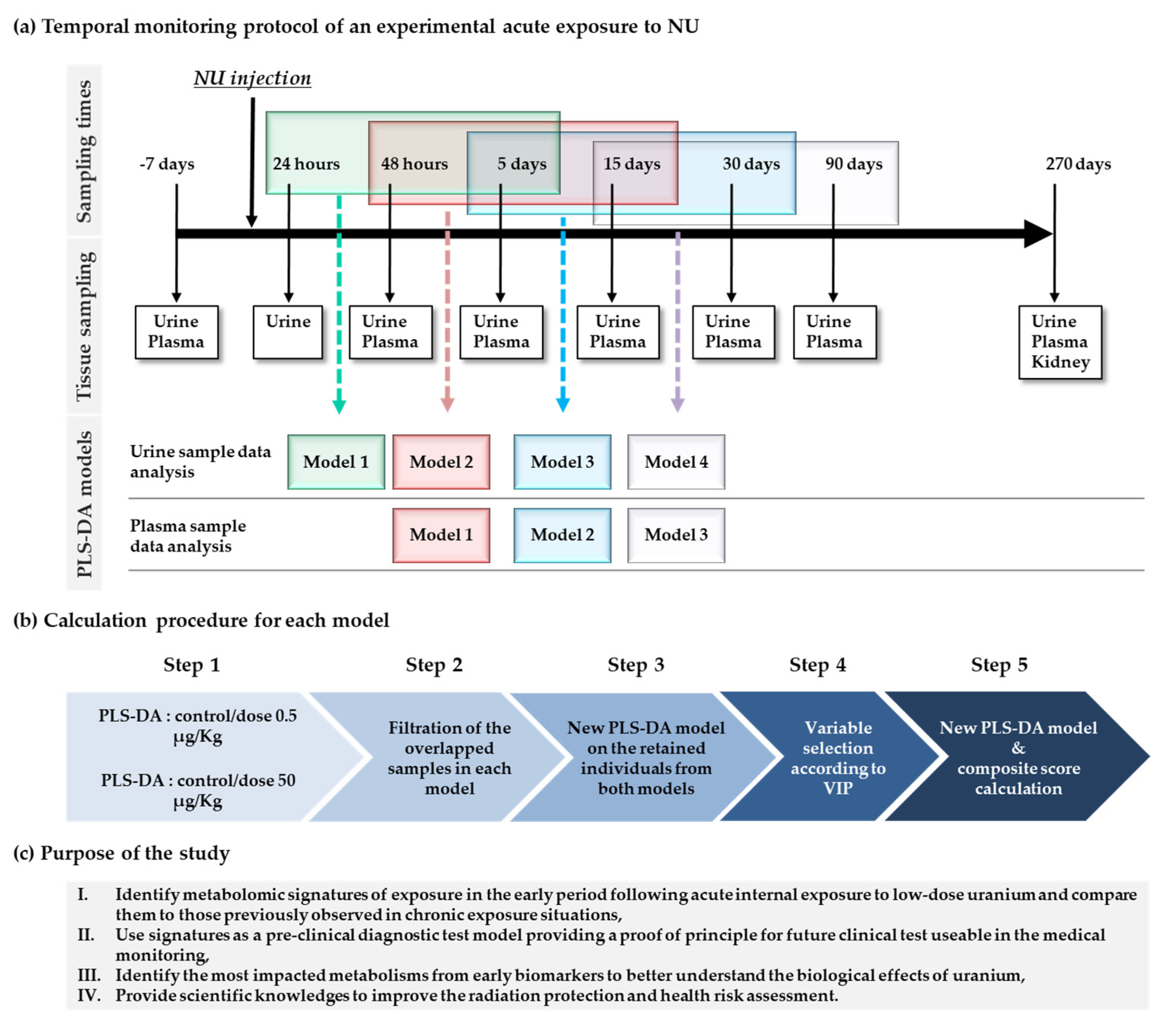
2.1. Clinical Monitoring of Animals
2.2. Metabolic Profile Analysis of Mass Data Features
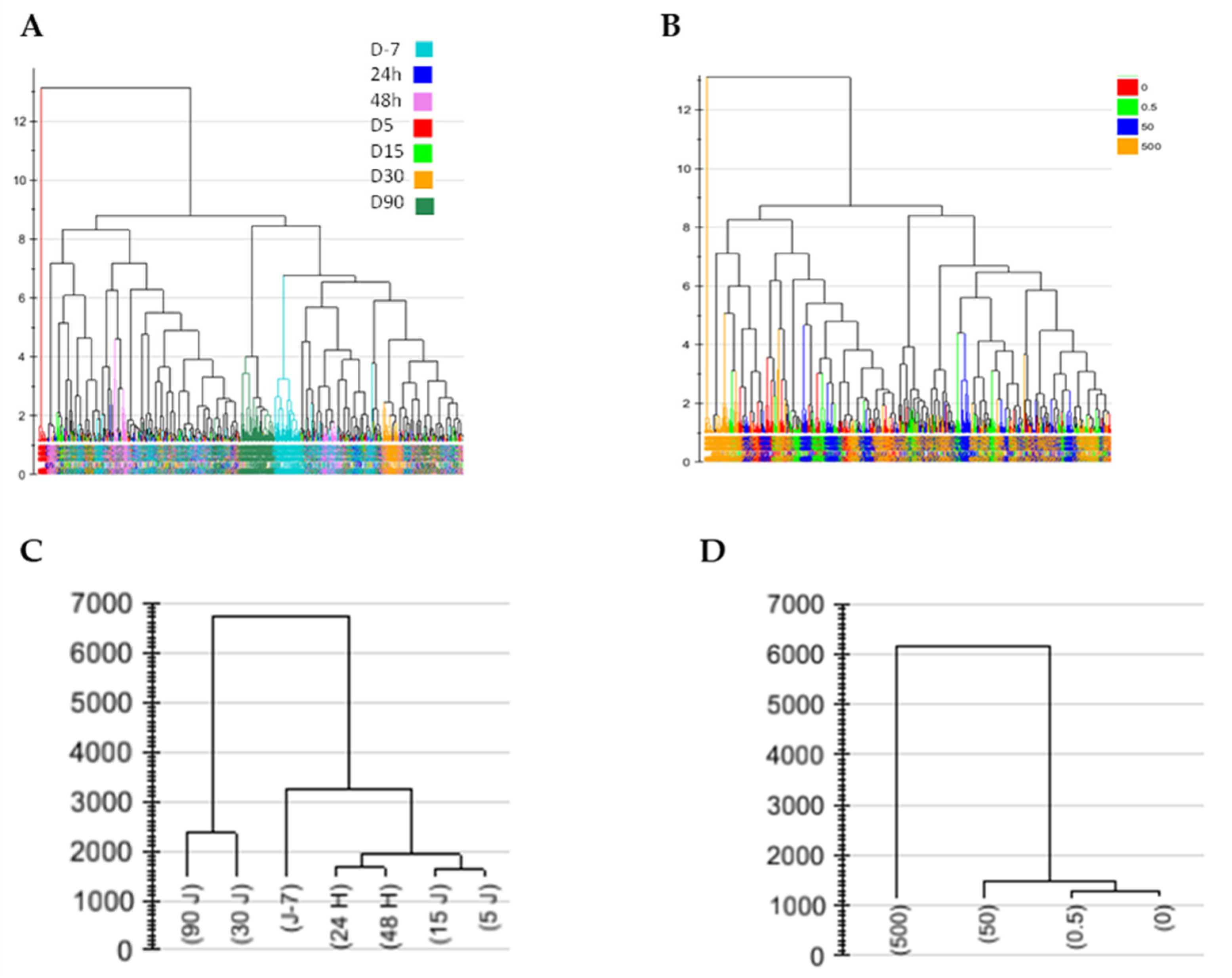
2.2.1. Change in Urinary Metabolic Profiles over Time
2.2.2. Time Effect in Plasma Profiles
2.2.3. Long-Term Kidney Profile after Low-Dose Exposure
2.3. Metabolic Profile Analysis of Annotated Data Matrixes
Discrimination of Rats Contaminated with Low Doses of NU as a Function of Time in the Concatenated Urine and Plasma Annotated Data Tables
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animal Treatment and Sample Collection
4.3. Clinical Monitoring
4.3.1. Animal Monitoring
4.3.2. Chemical Monitoring in Urine and Plasma Samples
4.3.3. Uranium Level in Urine and Kidney Samples
4.4. Metabolomics Analysis
4.4.1. Sample Preparation
- a.
- Urine samples
- b.
- Plasma samples
- c.
- Kidney samples
- d.
- Quality control (QC) and blanks
4.4.2. Liquid Chromatography Mass Spectrometry Analysis
4.4.3. Data Pre-Processing and Statistical Analyses
- a.
- Data pre-processing
- b.
- Data processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO|An Estimated 12.6 Million Deaths Each Year are Attributable to Unhealthy Environments; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar] [CrossRef]
- Jakhu, R.; Mehra, R.; Mittal, H.M. Exposure assessment of natural uranium from drinking water. Environ. Sci. Processes Impacts 2016, 18, 1540–1549. [Google Scholar] [CrossRef] [PubMed]
- Salonen, L. 238U series radionuclides as a source of increased radioactivity in groundwater originating from Finnish bedrock. IAHS Publ. 1994, 222, 71. [Google Scholar]
- Bigalke, M.; Schwab, L.; Rehmus, A.; Tondo, P.; Flisch, M. Uranium in agricultural soils and drinking water wells on the Swiss Plateau. Environ. Pollut 2018, 233, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Papp, Z.; Dezso, Z.; Daroczy, S. Significant radioactive contamination of soil around a coal-fired thermal power plant. J. Environ. Radioact. 2002, 59, 191–205. [Google Scholar] [CrossRef]
- Roper, A.R.; Stabin, M.G.; Delapp, R.C.; Kosson, D.S. Analysis of naturally-occurring radionuclides in coal combustion fly ash, gypsum, and scrubber residue samples. Health Phys. 2013, 104, 264–269. [Google Scholar] [CrossRef]
- Pereira, R.; Barbosa, S.; Carvalho, F.P. Uranium mining in Portugal: A review of the environmental legacies of the largest mines and environmental and human health impacts. Environ. Geochem. Health 2014, 36, 285–301. [Google Scholar] [CrossRef]
- Strand, L.A.; Martinsen, J.I.; Borud, E.K. A 5-Year Continued Follow-up of Cancer Risk and All-Cause Mortality Among Norwegian Military Peacekeepers Deployed to Kosovo During 1999–2016. Mil. Med. 2020, 185, e239–e243. [Google Scholar] [CrossRef]
- IRSN. Natural Uranium and the Environment. Available online: https://www.irsn.fr/EN/Research/publications-documentation/radionuclides-sheets/environment/Pages/Natural-uranium-environment.aspx (accessed on 8 March 2022).
- Craft, E.; Abu-Qare, A.; Flaherty, M.; Garofolo, M.; Rincavage, H.; Abou-Donia, M. Depleted and natural uranium: Chemistry and toxicological effects. J. Toxicol. Environ. Health 2004, 7, 297–317. [Google Scholar] [CrossRef]
- Ménager, M.-T.; Garnier-Laplace, J.; Goyffon, M. Toxicologie Nucléaire Environnementale et Humaine; Éditions Tec & Doc. Lavoisier Éditions Médicales Internationales: Cachan, France, 2009. [Google Scholar]
- Souidi, M.; Tissandie, E.; Racine, R.; Ben Soussan, H.; Rouas, C.; Grignard, E.; Dublineau, I.; Gourmelon, P.; Lestaevel, P.; Gueguen, Y. Uranium: Properties and biological effects after internal contamination. Ann. Biol. Clin. 2009, 67, 23–38. [Google Scholar]
- Lopez-Giacoman, S.; Madero, M. Biomarkers in chronic kidney disease, from kidney function to kidney damage. World J. Nephrol. 2015, 4, 57–73. [Google Scholar] [CrossRef]
- Shaki, F.; Zamani, E.; Arjmand, A.; Pourahmad, J. A Review on Toxicodynamics of Depleted Uranium. Iran. J. Pharm. Res. 2019, 18, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Hubert, P. Effects of low doses: Proof and inferences. Environ. Risques St. 2010, 9, 295–302. [Google Scholar] [CrossRef]
- Vetter, T.R.; Mascha, E.J. Bias, Confounding, and Interaction: Lions and Tigers, and Bears, Oh My! Anesth. Analg. 2017, 125, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Marano, F.; Barouki, R. La toxicologie prédictive: Quel apport pour l’évaluation des risques en santé environnementale ? Environ. Risque St. 2011, 10, 404–411. [Google Scholar] [CrossRef]
- Vlaanderen, J.; Moore, L.E.; Smith, M.T.; Lan, Q.; Zhang, L.; Skibola, C.F.; Rothman, N.; Vermeulen, R. Application of OMICS technologies in occupational and environmental health research; current status and projections. Occup. Environ. Med. 2010, 67, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Grison, S.; Fave, G.; Maillot, M.; Manens, L.; Delissen, O.; Blanchardon, E.; Banzet, N.; Defoort, C.; Bott, R.; Dublineau, I.; et al. Metabolomics identifies a biological response to chronic low-dose natural uranium contamination in urine samples. Metabolomics 2013, 9, 1168–1180. [Google Scholar] [CrossRef]
- Grison, S.; Fave, G.; Maillot, M.; Manens, L.; Delissen, O.; Blanchardon, E.; Dublineau, I.; Aigueperse, J.; Bohand, S.; Martin, J.C.; et al. Metabolomics reveals dose effects of low-dose chronic exposure to uranium in rats: Identification of candidate biomarkers in urine samples. Metabolomics 2016, 12, 154. [Google Scholar] [CrossRef]
- Shim, C.K.; Sawada, Y.; Iga, T.; Hanano, M. Estimation of renal secretory function for organic cations by endogenous N1-methylnicotinamide in rats with experimental renal failure. J. Pharmacokinet. Biopharm. 1984, 12, 23–42. [Google Scholar] [CrossRef]
- Parasuraman, S. Toxicological screening. J. Pharmacol. Pharmacother. 2011, 2, 74–79. [Google Scholar] [CrossRef]
- Masuda, S.; Hisamatsu, T.; Seko, D.; Urata, Y.; Goto, S.; Li, T.-S.; Ono, Y. Time- and dose-dependent effects of total-body ionizing radiation on muscle stem cells. Physiol. Rep. 2015, 3, e12377. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, Y.; Rouas, C.; Leblond, F.A. Kidney injury biomarkers. Nephrol. Ther. 2012, 8, 146–155. [Google Scholar] [CrossRef]
- Gueguen, Y.; Roy, L.; Hornhardt, S.; Badie, C.; Hall, J.; Baatout, S.; Pernot, E.; Tomasek, L.; Laurent, O.; Ebrahimian, T.; et al. Biomarkers for Uranium Risk Assessment for the Development of the CURE (Concerted Uranium Research in Europe) Molecular Epidemiological Protocol. Radiat. Res. 2017, 187, 107–127. [Google Scholar] [CrossRef] [PubMed]
- Grison, S.; Kereselidze, D.; Cohen, D.; Gloaguen, C.; Elie, C.; Lestaevel, P.; Legendre, A.; Manens, L.; Habchi, B.; Benadjaoud, M.A.; et al. Applying a multiscale systems biology approach to study the effect of chronic low-dose exposure to uranium in rat kidneys. Int. J. Radiat. Biol. 2019, 95, 737–752. [Google Scholar] [CrossRef] [PubMed]
- Souidi, M.; Dublineau, I.; Lestaevel, P. Depleted uranium: Metabolic disruptor? Environ. Risques Sante 2011, 10, 469–476. [Google Scholar]
- Dublineau, I.; Souidi, M.; Gueguen, Y.; Lestaevel, P.; Bertho, J.M.; Manens, L.; Delissen, O.; Grison, S.; Paulard, A.; Monin, A.; et al. Unexpected lack of deleterious effects of uranium on physiological systems following a chronic oral intake in adult rat. BioMed Res. Int. 2014, 2014, 181989. [Google Scholar] [CrossRef]
- Ma, M.; Wang, R.; Xu, L.; Xu, M.; Liu, S. Emerging health risks and underlying toxicological mechanisms of uranium contamination: Lessons from the past two decades. Environ. Int. 2020, 145, 106107. [Google Scholar] [CrossRef]
- Paquet, F.; Houpert, P.; Blanchardon, E.; Delissen, O.; Maubert, C.; Dhieux, B.; Moreels, A.M.; Frelon, S.; Gourmelon, P. Accumulation and distribution of uranium in rats after chronic exposure by ingestion. Health Phys. 2006, 90, 139–147. [Google Scholar] [CrossRef]
- Coecke, S.; Pelkonen, O.; Leite, S.B.; Bernauer, U.; Bessems, J.G.; Bois, F.Y.; Gundert-Remy, U.; Loizou, G.; Testai, E.; Zaldivar, J.M. Toxicokinetics as a key to the integrated toxicity risk assessment based primarily on non-animal approaches. Toxicol Vitr. 2013, 27, 1570–1577. [Google Scholar] [CrossRef]
- Gao, Y.; Kang, L.; Zhang, Y.; Feng, J.; Zhu, L. Toxicokinetic and toxicodynamic (TK-TD) modeling to study oxidative stress-dependent toxicity of heavy metals in zebrafish. Chemosphere 2019, 220, 774–782. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica Fate Foreign Compd. Biol. Syst. 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Thevenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- French, B.; Saha-Chaudhuri, P.; Ky, B.; Cappola, T.P.; Heagerty, P.J. Development and evaluation of multi-marker risk scores for clinical prognosis. Stat. Methods Med. Res. 2016, 25, 255–271. [Google Scholar] [CrossRef]
- Hirsch, G.H. Stimulation of renal organic base transport by uranyl nitrate. Can. J. Physiol. Pharmacol. 1972, 50, 533–538. [Google Scholar] [CrossRef]
- Goodson, J.M.; Hardt, M.; Hartman, M.L.; Alqaderi, H.; Green, D.; Tavares, M.; Mutawa, A.S.; Ariga, J.; Soparkar, P.; Behbehani, J.; et al. Salivary N1-Methyl-2-Pyridone-5-Carboxamide, a Biomarker for Uranium Uptake, in Kuwaiti Children Exhibiting Exceptional Weight Gain. Front. Endocrinol. 2019, 10, 382. [Google Scholar] [CrossRef]
- Ito, S.; Kusuhara, H.; Kumagai, Y.; Moriyama, Y.; Inoue, K.; Kondo, T.; Nakayama, H.; Horita, S.; Tanabe, K.; Yuasa, H.; et al. N-methylnicotinamide is an endogenous probe for evaluation of drug-drug interactions involving multidrug and toxin extrusions (MATE1 and MATE2-K). Clin. Pharmacol. Ther. 2012, 92, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.; Pontones, C.; Renner, B.; Mieth, M.; Hoier, E.; Auge, D.; Maas, R.; Zolk, O.; Fromm, M. N(1)-methylnicotinamide as an endogenous probe for drug interactions by renal cation transporters: Studies on the metformin-trimethoprim interaction. Eur. J. Clin. Pharmacol. 2015, 71, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Ribbenstedt, A.; Ziarrusta, H.; Benskin, J.P. Development, characterization and comparisons of targeted and non-targeted metabolomics methods. PLoS ONE 2018, 13, e0207082. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Ake, P.; Strawn, S.J.; Wang, Y.W.; Fornace, A.J., Jr. Effects of Genetic Variation on Urinary Small Molecule Signatures of Mice after Exposure to Ionizing Radiation: A Study of p53 Deficiency. Metabolites 2020, 10, 234. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, D.; Song, P.; Zou, M.H. Tryptophan-kynurenine pathway is dysregulated in inflammation, and immune activation. Front. Biosci. (Landmark Ed.) 2015, 20, 1116–1143. [Google Scholar] [CrossRef]
- Debnath, S.; Velagapudi, C.; Redus, L.; Thameem, F.; Kasinath, B.; Hura, C.E.; Lorenzo, C.; Abboud, H.E.; O’Connor, J.C. Tryptophan Metabolism in Patients With Chronic Kidney Disease Secondary to Type 2 Diabetes: Relationship to Inflammatory Markers. Int. J. Tryptophan Res. 2017, 10, 1178646917694600. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Gong, J.; Liu, Y. Indoleamine 2, 3-dioxygenase regulation of immune response (Review). Mol. Med. Rep. 2018, 17, 4867–4873. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Natividad, J.M.; Sokol, H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 2018, 11, 1024–1038. [Google Scholar] [CrossRef]
- Gershon, M.D. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Bussy, C.; Lestaevel, P.; Dhieux, B.; Amourette, C.; Paquet, F.; Gourmelon, P.; Houpert, P. Chronic ingestion of uranyl nitrate perturbs acetylcholinesterase activity and monoamine metabolism in male rat brain. Neurotoxicology 2006, 27, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in Inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef] [PubMed]
- Olli, K.; Lahtinen, S.; Rautonen, N.; Tiihonen, K. Betaine reduces the expression of inflammatory adipokines caused by hypoxia in human adipocytes. Br. J. Nutr. 2013, 109, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Kar, F.; Hacioglu, C.; Kacar, S.; Sahinturk, V.; Kanbak, G. Betaine suppresses cell proliferation by increasing oxidative stress-mediated apoptosis and inflammation in DU-145 human prostate cancer cell line. Cell Stress Chaperones 2019, 24, 871–881. [Google Scholar] [CrossRef]
- Ueland, P.M.; Holm, P.I.; Hustad, S. Betaine: A key modulator of one-carbon metabolism and homocysteine status. Clin. Chem. Lab. Med. CCLM/FESCC 2005, 43, 1069–1075. [Google Scholar] [CrossRef]
- Shedid, S.M.; Abdel-Magied, N.; Saada, H.N. Role of betaine in liver injury induced by the exposure to ionizing radiation. Environ. Toxicol. 2019, 34, 123–130. [Google Scholar] [CrossRef]
- Monobe, M.; Uzawa, A.; Hino, M.; Ando, K.; Kojima, S. Glycine betaine, a beer component, protects radiation-induced injury. J. Radiat. Res. 2005, 46, 117–121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ali, S.; Abbas, Z.; Seleiman, M.F.; Rizwan, M.; Yava, S.I.; Alhammad, B.A.; Shami, A.; Hasanuzzaman, M.; Kalderis, D. Glycine Betaine Accumulation, Significance and Interests for Heavy Metal Tolerance in Plants. Plants 2020, 9, 896. [Google Scholar] [CrossRef] [PubMed]
- Lever, M.; Sizeland, P.C.; Bason, L.M.; Hayman, C.M.; Robson, R.A.; Chambers, S.T. Abnormal glycine betaine content of the blood and urine of diabetic and renal patients. Clin. Chim. Acta Int. J. Clin. Chem. 1994, 230, 69–79. [Google Scholar] [CrossRef]
- Maleki, V.; Mahdavi, R.; Hajizadeh-Sharafabad, F.; Alizadeh, M. The effects of taurine supplementation on oxidative stress indices and inflammation biomarkers in patients with type 2 diabetes: A randomized, double-blind, placebo-controlled trial. Diabetol. Metab. Syndr. 2020, 12, 9. [Google Scholar] [CrossRef]
- Choi, K.S.; Yoo, I.S.; Shin, K.O.; Chung, K.H. Effects of taurine on cadmium exposure in muscle, gill, and bone tissues of Carassius auratus. Nutr. Res. Pract. 2013, 7, 22–25. [Google Scholar] [CrossRef][Green Version]
- Yeh, Y.-H.; Lee, Y.-T.; Hsieh, H.-S.; Hwang, D.-F. Effect of taurine on toxicity of aluminum in rats. e-SPEN Eur. E J. Clin. Nutr. Metab. 2009, 4, e187–e192. [Google Scholar] [CrossRef]
- Hwang, D.F.; Wang, L.C. Effect of taurine on toxicity of cadmium in rats. Toxicology 2001, 167, 173–180. [Google Scholar] [CrossRef]
- Marcinkiewicz, J.; Kontny, E. Taurine and inflammatory diseases. Amino Acids 2014, 46, 7–20. [Google Scholar] [CrossRef]
- Jan, A.T.; Azam, M.; Siddiqui, K.; Ali, A.; Choi, I.; Haq, Q.M. Heavy Metals and Human Health: Mechanistic Insight into Toxicity and Counter Defense System of Antioxidants. Int. J. Mol. Sci. 2015, 16, 29592–29630. [Google Scholar] [CrossRef]
- Han, X.; Chesney, R.W. The role of taurine in renal disorders. Amino Acids 2012, 43, 2249–2263. [Google Scholar] [CrossRef]
- Lee, H.; Jang, H.B.; Yoo, M.G.; Park, S.I.; Lee, H.J. Amino Acid Metabolites Associated with Chronic Kidney Disease: An Eight-Year Follow-Up Korean Epidemiology Study. Biomedicines 2020, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z. Impaired Citric Acid Cycle in Nondiabetic Chronic Kidney Disease. EBioMedicine 2017, 26, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Kand’ar, R.; Zakova, P. Allantoin as a marker of oxidative stress in human erythrocytes. Clin. Chem. Lab. Med. 2008, 46, 1270–1274. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.S.; Hancock, S.K.; McNally, A.M.; Hinckley, J.; Binder, E.; Zimmerman, K.; Ehrich, M.F.; Jortner, B.S. Neurological effects of acute uranium exposure with and without stress. Neurotoxicology 2007, 28, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yin, Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef]
- Boysen, A.K.; Heal, K.R.; Carlson, L.T.; Ingalls, A.E. Best-Matched Internal Standard Normalization in Liquid Chromatography-Mass Spectrometry Metabolomics Applied to Environmental Samples. Anal. Chem. 2018, 90, 1363–1369. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Giacomoni, F.; Le Corguille, G.; Monsoor, M.; Landi, M.; Pericard, P.; Petera, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform./Ed. Board D. Baxevanis 2019, 68, e86. [Google Scholar] [CrossRef]
- Aidoud, N.; Delplanque, B.; Baudry, C.; Garcia, C.; Moyon, A.; Balasse, L.; Guillet, B.; Antona, C.; Darmaun, D.; Fraser, K.; et al. A combination of lipidomics, MS imaging, and PET scan imaging reveals differences in cerebral activity in rat pups according to the lipid quality of infant formulas. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 4776–4790. [Google Scholar] [CrossRef]
- Martin, J.C.; Berton, A.; Ginies, C.; Bott, R.; Scheercousse, P.; Saddi, A.; Gripois, D.; Landrier, J.F.; Dalemans, D.; Alessi, M.C.; et al. Multilevel systems biology modeling characterized the atheroprotective efficiencies of modified dairy fats in a hamster model. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H935–H945. [Google Scholar] [CrossRef] [PubMed]
- Dickson, L.; Tenon, M.; Svilar, L.; Fanca-Berthon, P.; Lugan, R.; Martin, J.C.; Vaillant, F.; Rogez, H. Main Human Urinary Metabolites after Genipap (Genipa americana L.) Juice Intake. Nutrients 2018, 10, 1155. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform./Ed. Board D. Baxevanis 2016, 55, 14.10.11–14.10.91. [Google Scholar] [CrossRef] [PubMed]

| Experimental Groups (NU Doses) | Control (20) | NU 0.5 µg/kg (20) | NU 50 µg/kg (20) | NU 500 µg/kg (20) | |
|---|---|---|---|---|---|
| Time: Day 5 | |||||
| (a) | Body weight (g) | 352.22 ± 4.94 | 363.63 ± 3.8 | 346.87 ± 4.78 | 345.75 ± 5.25 |
| Urine analysis | |||||
| Urine volume (g/24 h) | 12.76 ± 0.78 | 13.97 ± 0.87 | 13.40 ± 0.73 | 23.34 ± 2.47 *** | |
| Chlorine (mmol) | 4.27 ± 0.35 | 4.69 ± 0.30 | 3.90 ± 0.34 | 3.40 ± 0.36 | |
| Creatinine (µmol) | 97.26 ± 3.38 | 100.87 ± 2.01 | 96.32 ± 3.07 | 96.24 ± 3.81 | |
| Magnesium (mmol) | 0.15 ± 0.01 | 0.16 ± 0.01 | 0.14 ± 0.02 | 0.27 ± 0.02 *** | |
| Phosphorus (mg) | 0.52 ± 0.07 | 0.63 ± 0.06 | 0.57 ± 0.06 | 0.86 ± 0.07 *** | |
| Potassium (mmol) | 2.94 ± 0.22 | 3.05 ± 0.18 | 2.63 ± 0.16 | 2.62 ± 0.18 | |
| Sodium (mmol) | 1.50 ± 0.08 | 1.62 ± 0.06 | 1.44 ± 0.08 | 1.48 ± 0.09 | |
| Total proteins (mg) | 0.007 ± 0.001 | 0.006 ± 0.001 | 0.006 ± 0.001 | 0.05 ± 0.01 *** | |
| Urea (mmol) | 13.49 ± 0.61 | 14.65 ± 0.45 | 13.31 ± 0.60 | 13.30 ± 0.51 | |
| 88.83 ± 3.79 | 93.45 ± 3.63 | 95.45 ± 4.80 | 44.90 ± 8.09 *** | ||
| Time: Day 270 | |||||
| (b) | Uranium concentration in kidney (ng U/g) | 10.15 ± 0.56 | 13.09 ± 1.05 * | 12.84 ± 1.03 * | 75.96 ± 14.64 *** |
| Kidney weight (g) | 1.93 ± 0.05 | 2.03 ± 0.06 | 1.86 ± 0.05 | 2.00 ± 0.05 | |
| Body weight (g) | 636.83 ± 10.89 | 653.13 ± 12.91 | 612.13 ± 12.84 | 658.00 ± 12.60 | |
| Urine analysis | |||||
| Urine volume (g/24 h) | 11.60 ± 0.75 | 10.86 ± 0.62 | 11.00 ± 0.50 | 13.59 ± 1.30 | |
| Albumin (mg) | 4.16 ± 0.90 | 4.82 ± 0.84 | 4.12 ± 0.81 | 6.63 ± 1.14 | |
| Chlorine (mmol) | 2.96 ± 0.19 | 2.96 ± 0.16 | 2.70 ± 0.19 | 2.93 ± 0.17 | |
| Creatinine (µmol) | 115.36 ± 4.96 | 120.35 ± 3.31 | 114.88 ± 3.11 | 127.41 ± 3.50 | |
| Glucose (mmol) | 15.75 ± 0.86 | 15.57 ± 0.49 | 14.96 ± 0.51 | 18.50 ± 1.93 | |
| Magnesium (mmol) | 0.18 ± 0.01 | 0.18 ± 0.01 | 0.17 ± 0.01 | 0.21 ± 0.01 | |
| Potassium (mmol) | 1.57 ± 0.11 | 1.60 ± 0.09 | 1.50 ± 0.05 | 1.73 ± 0.10 | |
| Sodium (mmol) | 1.08 ± 0.09 | 1.01 ± 0.07 | 0.93 ± 0.05 | 1.00 ± 0.07 | |
| Total proteins (mg) | 58.06 ± 17.85 | 80.82 ± 24.73 | 51.38 ± 14.45 | 46.79 ± 10.04 | |
| Urea (mmol) | 11.53 ± 0.56 | 11.73 ± 0.40 | 11.27 ± 0.35 | 12.38 ± 0.49 | |
| Uric acid (µmol) | 19.39 ± 1.06 | 19.85 ± 0.80 | 19.70 ± 0.91 | 22.53 ± 0.86 * | |
| Clearance (mL/min) | 1.62 ± 0.11 | 1.76 ± 0.11 | 1.75 ± 0.10 | 1.83 ± 0.134 | |
| Plasma analysis | |||||
| Creatinine (µmol) | 48.74 ± 1.46 | 47.47 ± 1.57 | 46.09 ± 1.93 | 48.82 ± 1.79 | |
| Urea (mmol) | 4.71 ± 0.16 | 4.89 ± 0.24 | 4.71 ± 0.14 | 4.96 ± 0.13 |
| Model | Individuals of Model 1 and Model 2 (24 h, 48 h, 5 d, 15 d) | Metabolite | FDR | Fold Change | Boxplot | |
|---|---|---|---|---|---|---|
| Validation parameters | R2Y(cum) = 84.2% Q2(cum) = 81.9% p value = 5.41155 × 10−34 | Very good permutation test | M137T39 | 1.1906 × 10−10 | 20.262 |  |
| Composite score equation | Score = (1.10234 × 10−10 × M184T138) + (−5.44577 × 10−11 × M137T39) + (−2.31758 × 10−9 × M254T148) + (−1.40745 × 10−8 × M236T148) + (9.49838 × 10−12 × M153T134) + (−2.37778 × 10−8 × M276T148) + (2.55785 × 10−9 × M136T133) + (−4.47195 × 10−9 × M366T259) + (1.52073 × 10−9 × M108T133) + 0.747992 | M236T148 | 4.0973 × 10−21 | 15.462 |  | |
| M254T148 | 4.0973 × 10−21 | 14.841 |  | |||
| M276T148 | 3.4084 × 10−19 | 11.878 |  | |||
| ROC curve | 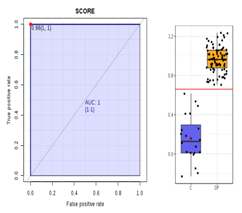 | AUC = 1 | M366T259 | 5.2434 × 10−14 | 8.2975 |  |
| M136T133 | 7.2673 × 10−7 | 0.4324 |  | |||
| M153T134 | 3.8045 × 10−6 | 0.60654 |  | |||
| M108T133 | 1.529 × 10−5 | 0.651 |  | |||
| M184T138 | 2.9739 × 10−3 | 0.69533 |  | |||
| Biological Sample/Masse (g·mol−1/Retention Time (s)) | Primary Name | KEGG ID | CAS | HMDB/YMDB ID |
|---|---|---|---|---|
| 6 common discriminant variables between "M1", "M2" and "M3" (24 h to 5 days and 48 h to 5 days and 5 days to 30 days) | ||||
| Urine_CP_M137T39 | 1-Methylnicotinamide | C02918 | 1005-24-9 | HMDB00699 |
| Urine_CP_M90T38 | Beta-alanine | C00099 | 107-95-9 | HMDB00056 |
| Urine_CN_M221T43 | D-glucurunolactone | C02670 | 32449-92-6 | HMDB06355 |
| Urine_CP_M104T39_1 | N,N-dimethylglycine | C01026 | 1118-68-9 | HMDB0000092 |
| Urine_CN_M209T40 | Saccharate | C00818 | 576-42-1 | HMDB29881 |
| Plasma_CP_M166T208 and CP_M120T208 and CP_M149T208 or Urine_CP_M166T209 and CP_M120T209 and CP_M149T209 | L-Phenylalanine | C00079 | 63-91-2 | HMDB0000159 |
| 5 common discriminant variables between "M1" and "M2" (24 h to 5 days and 48 h to 5 days) | ||||
| Urine_CN_M145T258 and CN_M101T259 | Adipate | C06104 | 124-04-9 | HMDB00448 |
| Urine_CN_M133T46 and CN_M115T46 | Malate | C00149 | 97-67-6 | HMDB00156 |
| Plasma_HP_M424T124 | 5b-cholanic acid-3a,12a-diol-7-one | C04643 | 911-40-0 | HMDB0000391 |
| Plasma_HP_M355T137 | 5b-cholanic acid-3a-ol-12-one | No id. | 5130-29-0 | HMDB0000328 |
| Plasma_CN_M475T491 and CN_M443T491 and CN_M407T491 and CN_M453T491 | Cholate | C00695 | 81-25-4 | HMDB00619 |
| 3 common discriminant variables between "M1" and "M3" (24 h to 5 days and 5 days to 30 days) | ||||
| Urine_CP_M118T53 | 5-aminopentoate | C00431 | 660-88-8 | HMDB03355 |
| Urine_CN_M159T299 and CN_M115T300 | 6-carboxyhexnoate | C02656 | 111-16-0 | HMDB00857 |
| Urine_CP_M144T297 or Plasma_CP_M144T264 | Tryptamine | C00398 | 61-54-1 | HMDB00303 |
| 22 common discriminant variables between "M2" and "M3" (48 h to 5 days and 5 days to 30 days) | ||||
| Urine_HP_M96T134 | 2-Hydroxypyridine | C02502 | 142-08-5 | HMDB13751 |
| Urine_HN_M165T118 and HN_M147T118 | 3-(2-hydroxyphenyl propanoate | C01198 | 495-78-3 | HMDB33752 |
| Urine_CN_M183T294 | 3-Hydroxybenzoate | C00587 | 99-06-9 | HMDB02466 |
| Urine_CP_M134T291 | 5-Hydroxyindole | No id. | 1953-54-4 | HMDB59805 |
| Urine_CP_M126T46_2 | 5-Methylcytosine | C02376 | 58366-64-6 | HMDB02894 |
| Urine_CP_M118T40 | Betaine | C00719 | 107-43-7 | HMDB00043 |
| Urine_CP_M112T40 | Cytosine | C00380 | 71-30-7 | HMDB00630 |
| Urine_CP_M209T375 | dl-benzylsuccinic acid | C09816 | 884-33-3 | HMDB0142179 |
| Urine_CN_M217T39_2 and CN_M227T38 and CN_M181T38 | Sorbitol | C00749 | 50-70-4 | HMDB00247 |
| Urine_HP_M110T838 | Hypotaurine | C00519 | 300-84-5 | HMDB00965 |
| Urine_CP_M176T374 and CP_M130T374 | Indole-3-acetate | C00954 | 6505-45-9 | HMDB00197 |
| Urine_CN_M185T40 | Pentose | No id. | No id. | No id. |
| Urine_CP_M166T209 and CP_M120T209 and CP_M149T209 | L-phenylalanine | C00079 | 63-91-2 | HMDB0000159 |
| Urine_CP_M182T82_1 | L-Threo-3-Phenylserine (DL-3-Phenylserine) | C03290 | 6254-48-4 | HMDB0002184 |
| Urine_CN_M308T40 | N-acetylneuraminic acid | C00270 | 131-48-6 | HMDB0000230 |
| Urine_CN_M206T343 | N-acetylphenylalanine | C03519 | 2018-61-3 | HMDB00512 |
| Urine_CP_M247T361 | N-acetyltryptophan | C03137 | 87-32-1 | HMDB0013713 |
| Urine_CP_M116T42 | Proline | C16435 | 147-85-3 | HMDB00162 |
| Urine_CN_M166T56 and CN_M122T56 | Quinolinate | C03722 | 89-00-9 | HMDB00232 |
| Urine_HP_M205T683 and HP_M188T684 | Tryptophan | C00525 | 153-94-6 | HMDB13609 |
| Plasma_CN_M157T39 | Allantoin | C01551 | 97-59-6 | HMDB00462 |
| Plasma_CP_M130T52 and CP_M84T51 | Pipecolate | C00408 | 3105-95-1 | HMDB00716 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grison, S.; Habchi, B.; Gloaguen, C.; Kereselidze, D.; Elie, C.; Martin, J.-C.; Souidi, M. Early Metabolomic Markers of Acute Low-Dose Exposure to Uranium in Rats. Metabolites 2022, 12, 421. https://doi.org/10.3390/metabo12050421
Grison S, Habchi B, Gloaguen C, Kereselidze D, Elie C, Martin J-C, Souidi M. Early Metabolomic Markers of Acute Low-Dose Exposure to Uranium in Rats. Metabolites. 2022; 12(5):421. https://doi.org/10.3390/metabo12050421
Chicago/Turabian StyleGrison, Stéphane, Baninia Habchi, Céline Gloaguen, Dimitri Kereselidze, Christelle Elie, Jean-Charles Martin, and Maâmar Souidi. 2022. "Early Metabolomic Markers of Acute Low-Dose Exposure to Uranium in Rats" Metabolites 12, no. 5: 421. https://doi.org/10.3390/metabo12050421
APA StyleGrison, S., Habchi, B., Gloaguen, C., Kereselidze, D., Elie, C., Martin, J.-C., & Souidi, M. (2022). Early Metabolomic Markers of Acute Low-Dose Exposure to Uranium in Rats. Metabolites, 12(5), 421. https://doi.org/10.3390/metabo12050421






