3,3′-Diindolylmethane Enhances Fluorouracil Sensitivity via Inhibition of Pyrimidine Metabolism in Colorectal Cancer
Abstract
:1. Introduction
2. Results
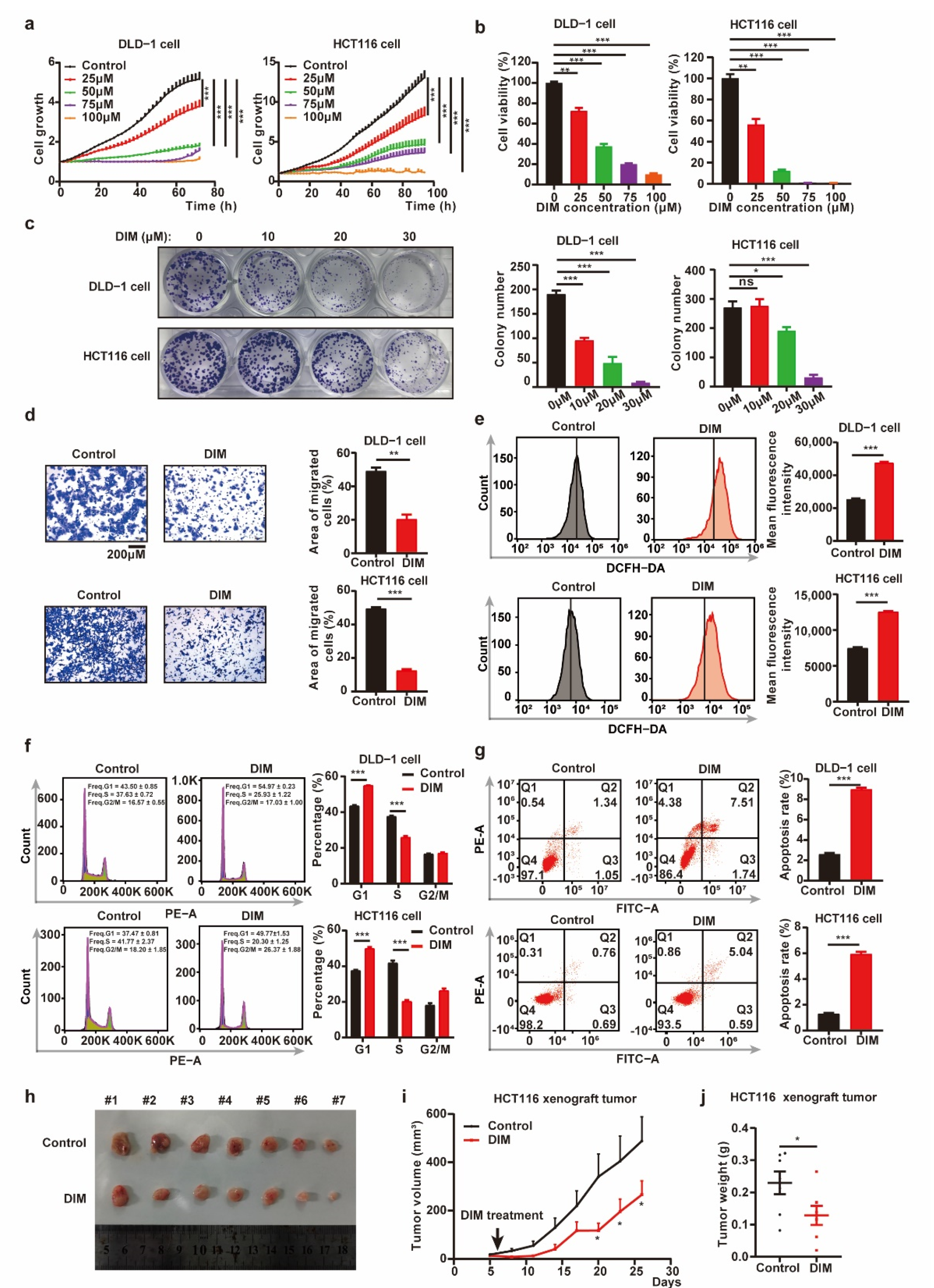
2.1. DIM inhibits CRC Progression In Vitro and In Vivo
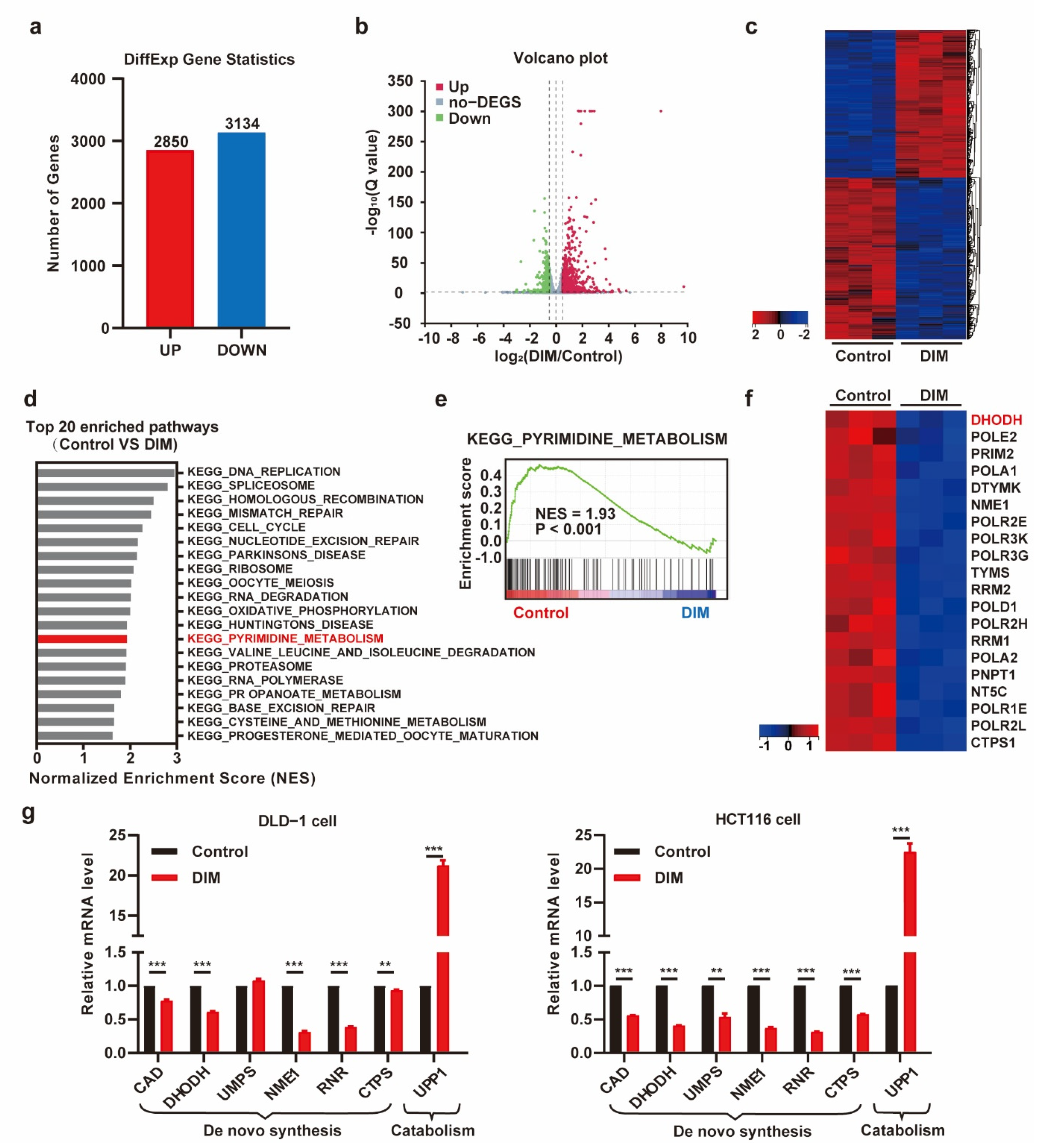
2.2. DIM Treatment Attenuates Pyrimidine Metabolism in CRC Cells
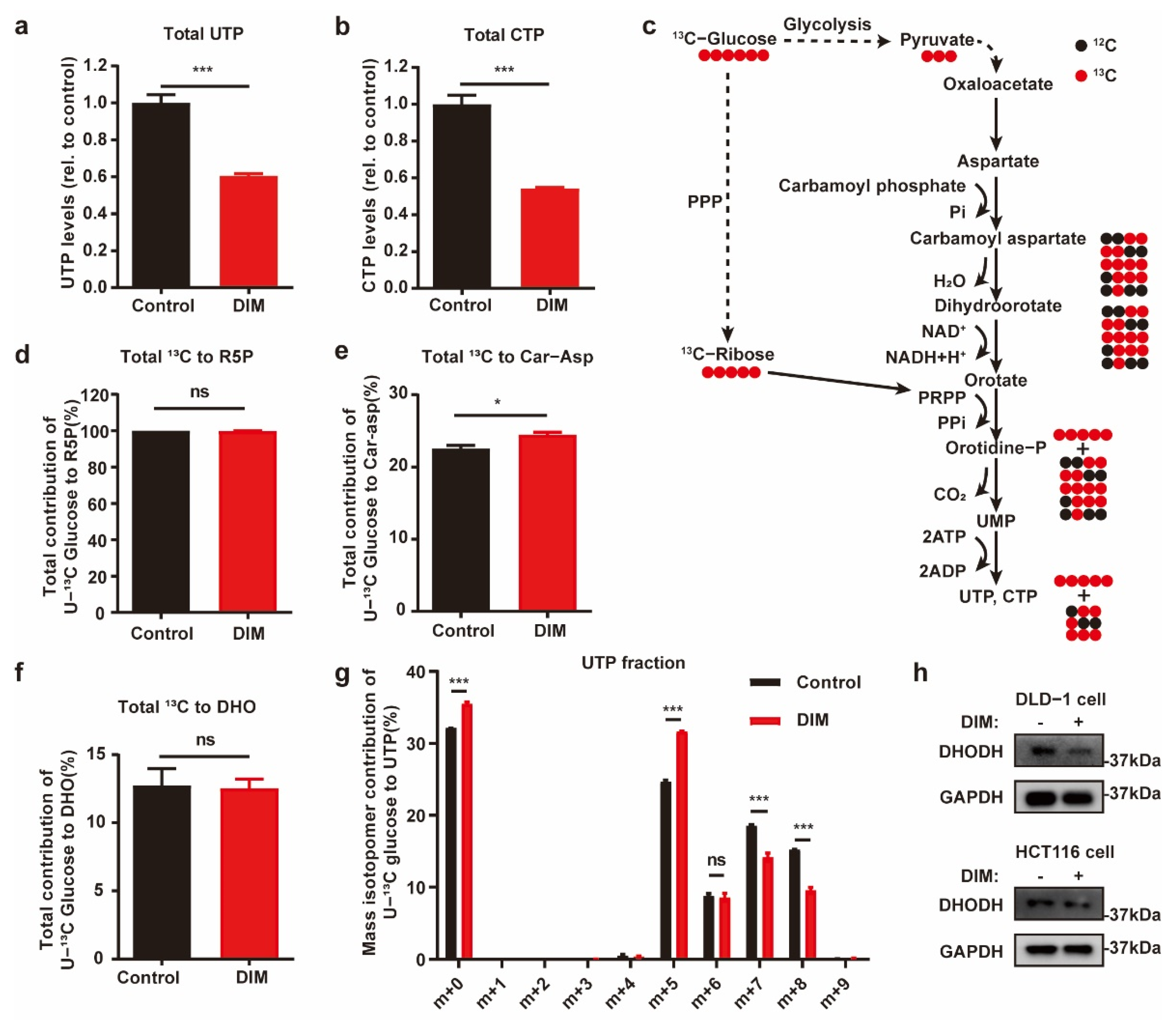
2.3. Metabolic Profiling Reveals de Novo Pyrimidine Biosynthesis Alteration in CRC under DIM Treatment
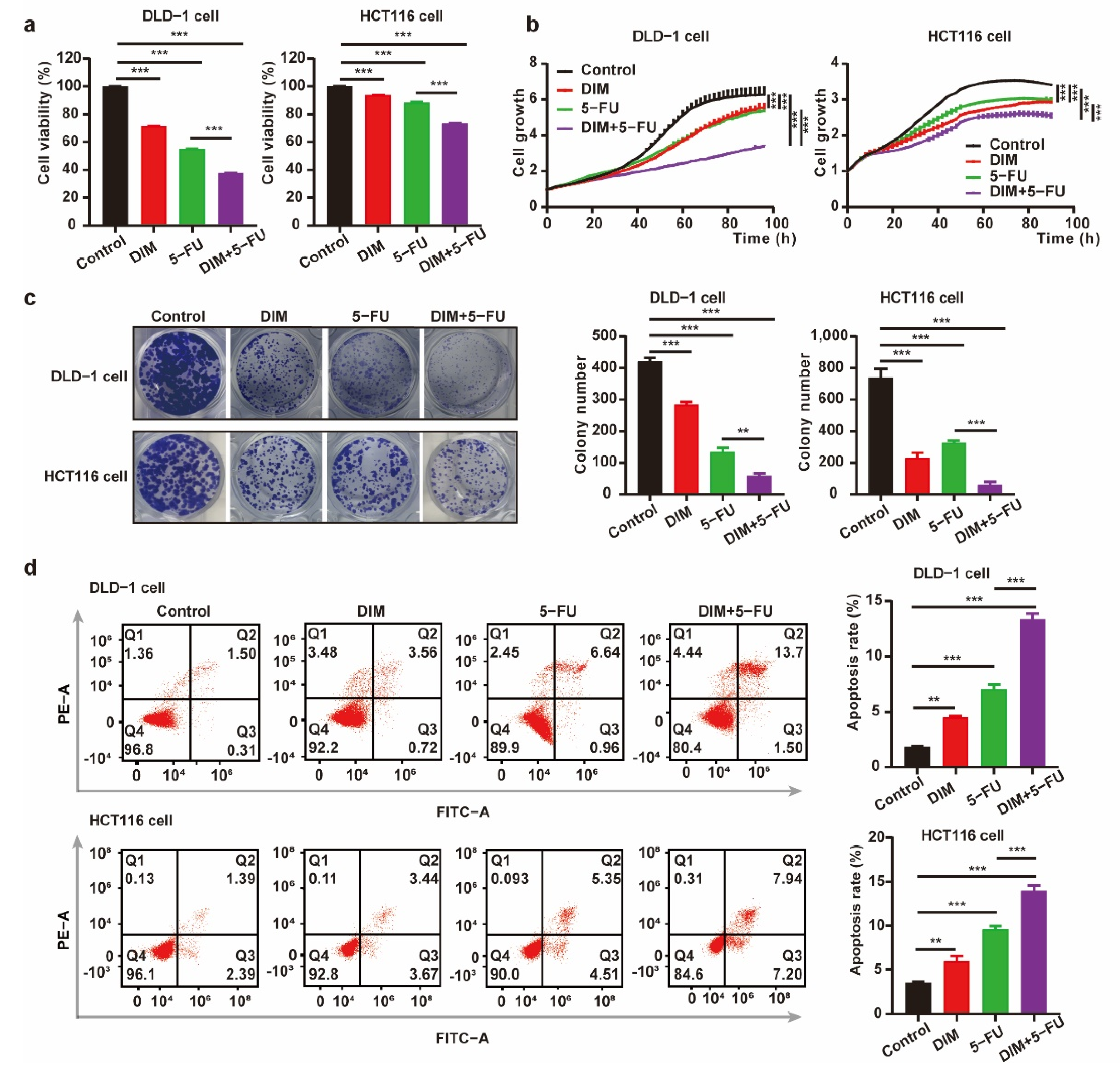
2.4. DIM Promotes Chemosensitivity of 5-FU and Potentiates 5-FU-Induced Suppressive Effects on CRC Cells
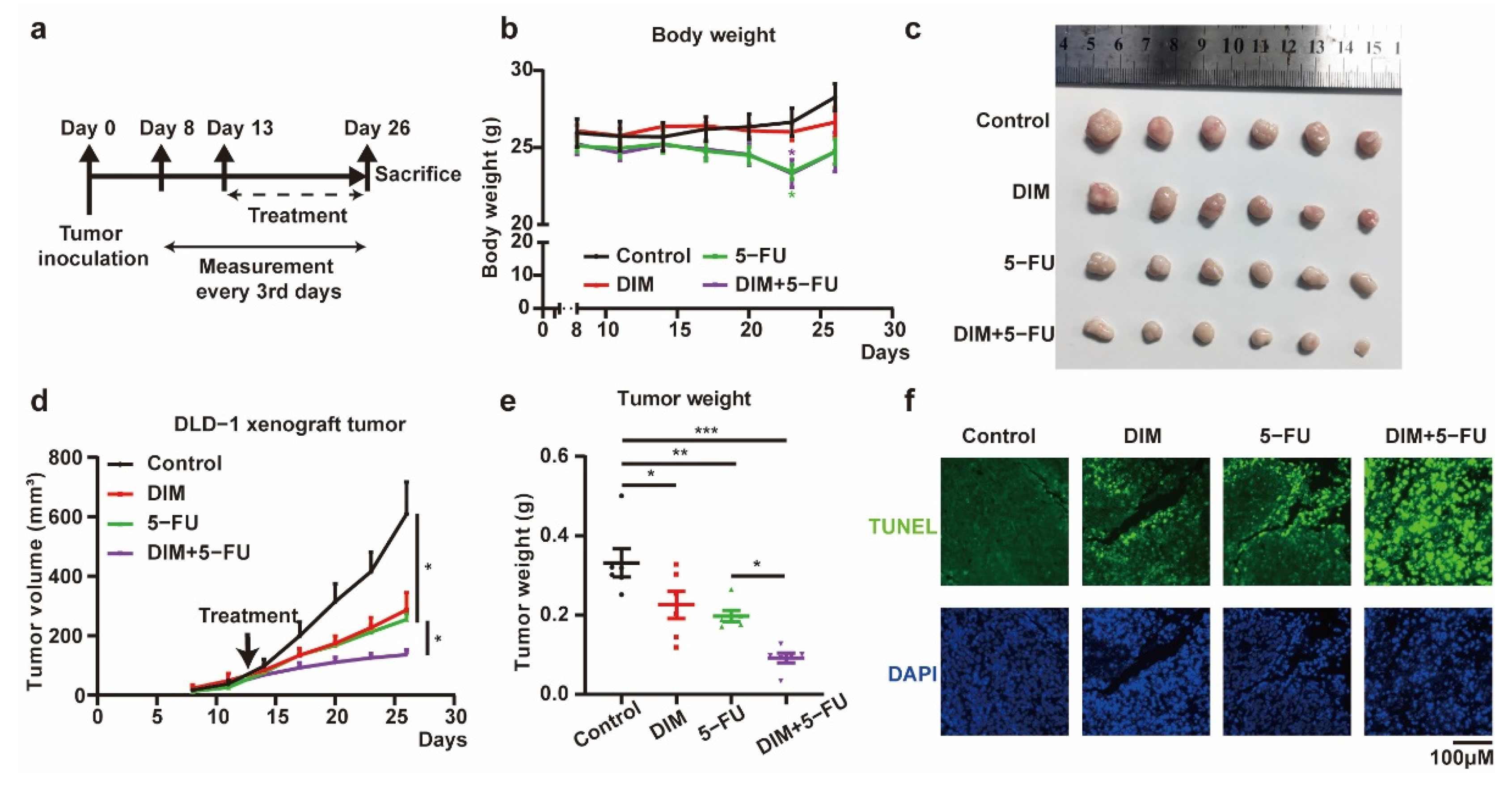
2.5. DIM Enhances the Effects of 5-FU-Based Chemotherapy In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reagents and Antibodies
4.3. Western Blotting
4.4. CCK-8 Assay
4.5. Growth Curve
4.6. Colony Formation
4.7. Cell Migration Assay
4.8. RNA Extraction and Quantitation
4.9. Flow Cytometry
4.10. TdT-Mediated dUTP Nick-End Labeling (TUNEL) Assay
4.11. RNA Sequencing
4.12. Metabolite Profiling Isotope Analysis
4.13. In Vivo Studies Using Xenograft CRC Model and Treatment
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Colorectal cancer | CRC |
| 3,3′-diindolylmethane | DIM |
| 5-fluorouracil | 5-FU |
| Gene set enrichment analysis | GSEA |
| Dihydroorotate | DHO |
| Dihydroorotate dehydrogenase | DHODH |
| Cytidine triphosphate | CTP |
| Uridine triphosphate | UTP |
| Carbamoyl phosphate | Car-Asp |
| Fetal bovine serum | FBS |
References
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Shi, L.; He, X.; Luo, Y. Gastrointestinal cancers in China, the USA, and Europe. Gastroenterol. Rep. 2021, 9, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Huang, J.; Luo, H.; Luo, C.; Zhu, X.; Bu, F.; Xiao, H.; Xiao, L.; Zhu, Z. Development of a prognostic index and screening of potential biomarkers based on immunogenomic landscape analysis of colorectal cancer. Aging 2020, 12, 5832–5857. [Google Scholar] [CrossRef]
- Fang, L.; Lu, W.; Choi, H.H.; Yeung, S.C.; Tung, J.Y.; Hsiao, C.D.; Fuentes-Mattei, E.; Menter, D.; Chen, C.; Wang, L.; et al. ERK2-Dependent Phosphorylation of CSN6 Is Critical in Colorectal Cancer Development. Cancer Cell 2015, 28, 183–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, L.; Yang, Z.; Zhang, M.; Meng, M.; Feng, J.; Chen, C. Clinical characteristics and survival analysis of colorectal cancer in China: A retrospective cohort study with 13,328 patients from southern China. Gastroenterol. Rep. 2021, 9, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Fang, L.; Yang, Z.; Zhou, J.; Tung, J.-Y.; Hsiao, C.-D.; Wang, L.; Deng, Y.; Wang, P.; Wang, J.; Lee, M.-H. Circadian Clock Gene CRY2 Degradation Is Involved in Chemoresistance of Colorectal Cancer. Mol. Cancer Ther. 2015, 14, 1476–1487. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Wu, J.L.; Qin, B.; Fan, Z.; Tang, Q.; Lu, W.; Zhang, H.; Xing, F.; Meng, M.; Zou, S.; et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 2020, 30, 163–178. [Google Scholar] [CrossRef]
- Geijsen, A.; Kok, D.; van Zutphen, M.; Keski-Rahkonen, P.; Achaintre, D.; Gicquiau, A.; Gsur, A.; Kruyt, F.; Ulrich, C.; Weijenberg, M.; et al. Diet quality indices and dietary patterns are associated with plasma metabolites in colorectal cancer patients. Eur. J. Nutr. 2021, 60, 3171–3184. [Google Scholar] [CrossRef]
- Puzzono, M.; Mannucci, A.; Grannò, S.; Zuppardo, R.; Galli, A.; Danese, S.; Cavestro, G. The Role of Diet and Lifestyle in Early-Onset Colorectal Cancer: A Systematic Review. Cancers 2021, 13, 5933. [Google Scholar] [CrossRef]
- Yammine, A.; Namsi, A.; Vervandier-Fasseur, D.; Mackrill, J.; Lizard, G.; Latruffe, N. Polyphenols of the Mediterranean Diet and Their Metabolites in the Prevention of Colorectal Cancer. Molecules 2021, 26, 3483. [Google Scholar] [CrossRef] [PubMed]
- Rejhová, A.; Opattová, A.; Čumová, A.; Slíva, D.; Vodička, P. Natural compounds and combination therapy in colorectal cancer treatment. Eur. J. Med. Chem. 2018, 144, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, J.; Kanwar, S.; Yu, Y.; Majumdar, A. Combination of dasatinib and curcumin eliminates chemo-resistant colon cancer cells. J. Mol. Signal. 2011, 6, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Song, J.; Hwang, S.; Choi, J.; Song, G.; Lim, W. Apigenin enhances apoptosis induction by 5-fluorouracil through regulation of thymidylate synthase in colorectal cancer cells. Redox. Biol. 2021, 47, 102144. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Ha, J.; Park, I.; Lee, S.; Baik, H.; Kim, Y.; Park, O. Apoptotic effect of EGCG in HT-29 colon cancer cells via AMPK signal pathway. Cancer Lett. 2007, 247, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, K.; Zu, X.; Ma, F.; Li, Z.; Lee, M.; Chen, H.; Li, Y.; Zhao, Y.; Liu, F.; et al. 3,3’-Diindolylmethane inhibits patient-derived xenograft colon tumor growth by targeting COX1/2 and ERK1/2. Cancer Lett 2019, 448, 20–30. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Guo, B. Chemopreventive agent 3,3’-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res 2010, 70, 646–654. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Jiang, M.; Zhu, C.; He, J.; Fan, S. Amelioration of whole abdominal irradiation-induced intestinal injury in mice with 3,3’-Diindolylmethane (DIM). Free Radic. Biol. Med. 2019, 130, 244–255. [Google Scholar] [CrossRef]
- Lane, A.N.; Fan, W.M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef] [Green Version]
- Villa, E.; Ali, E.S.; Sahu, U.; Ben-Sahra, I. Cancer Cells Tune the Signaling Pathways to Empower de Novo Synthesis of Nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef] [Green Version]
- Ben-Sahra, I.; Howell Asara, J.M.; Manning, B.D. Stimulation of de Novo Pyrimidine Synthesis by Growth Signaling Through mTOR and S6K1. Science 2013, 339, 1323–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornell, R.B.; Ridgway, N.D. CTP:phosphocholine cytidylyltransferase: Function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog. Lipid Res. 2015, 59, 147–171. [Google Scholar] [CrossRef]
- Egger, S.; Chaikuad, A.; Kavanagh, K.L.; Oppermann, U.; Nidetzky, B. Structure and Mechanism of Human UDP-glucose 6-Dehydrogenase. J. Biol. Chem. 2011, 286, 23877–23887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peneff, C.; Ferrari, P.; Charrier, V.; Taburet, Y.; Monnier, C.; Zamboni, V.; Winter, J.; Harnois, M.; Fassy, F.; Bourne, Y. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: Role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. Embo J. 2014, 20, 6191–6202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Sun, F.; Wang, H.; Hu, Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol. Ther. 2021, 224, 107827. [Google Scholar] [CrossRef]
- Gao, X.; Liu, J.; Cho, K.B.; Kedika, S.; Guo, B. Chemopreventive Agent 3,3′-Diindolylmethane Inhibits MDM2 in Colorectal Cancer Cells. Int. J. Mol. Sci. 2020, 21, 4642. [Google Scholar] [CrossRef]
- Zha, S.; Li, T.; Zheng, Q.; Li, L. Whether Patients With Stage/Colorectal Cancer Benefit From Adjuvant Chemotherapy: A Modeling Analysis of Literature Aggregate Data. Front Pharm. 2022, 13, 826785. [Google Scholar] [CrossRef]
- Blondy, S.; David, V.; Verdier, M.; Mathonnet, M.; Perraud, A.; Christou, N. 5-Fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. [Google Scholar] [CrossRef]
- Ahmad, A.; Sakr, W.; Rahman, K. Anticancer properties of indole compounds: Mechanism of apoptosis induction and role in chemotherapy. Curr. Drug Targets 2010, 11, 652–666. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, L.; Liu, Y.; Wang, J.; Jiang, W. Indole-3-Carbinol (I3C) and its Major Derivatives: Their Pharmacokinetics and Important Roles in Hepatic Protection. Curr. Drug Metab. 2016, 17, 401–409. [Google Scholar] [CrossRef]
- Sethy, C.; Kundu, C.N. 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed. Pharm. 2021, 137, 111285. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Hu, W.; Yin, S.; Huang, Z.; Zhu, Q.; Chen, J.; Zang, Y.; Dong, L.; Zhang, J. 3,3′-Diindolymethane ameliorates adriamycin-induced cardiac fibrosis via activation of a BRCA1-dependent anti-oxidant pathway. Pharmacol. Res. 2013, 70, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Riby, J.E.; Firestone, G.L.; Bjeldanes, L. 3,3′-Diindolylmethane reduces levels of HIF-1α and HIF-1 activity in hypoxic cultured human cancer cells. Biochem. Pharmacol. 2008, 75, 1858–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.J.; Park, S.Y.; Shin, H.K.; Kwon, D.Y.; Surh, Y.J.; Park, J. Activation of caspase-8 contributes to 3,3’-Diindolylmethane-induced apoptosis in colon cancer cells. J. Nutr. 2007, 137, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, C.A.; Emily, H.; Strom, M.B. Chemopreventive properties of 3,3’-diindolylmethane in breast cancer: Evidence from experimental and human studies. Nutr. Rev. 2016, 74, 432–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Ser, Z.; Gao, X.; Johnson, C.; Mehrmohamadi, M.; Liu, X.; Li, S.; Locasale, J. Targeting One Carbon Metabolism with an Antimetabolite Disrupts Pyrimidine Homeostasis and Induces Nucleotide Overflow. Cell Rep. 2016, 15, 2367–2376. [Google Scholar] [CrossRef] [Green Version]
- Mao, C.; Liu, X.; Zhang, Y.; Lei, G.; Yan, Y.; Lee, H.; Koppula, P.; Wu, S.; Zhuang, L.; Fang, B.; et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 2021, 593, 586–590. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Weinberg, E.M.; Nguyen, A.; Liberti, M.V.; Goodarzi, H.; Janjigian, Y.Y.; Paty, P.B.; Saltz, L.B.; Kingham, T.P.; Loo, J.M.; et al. PCK1 and DHODH drive colorectal cancer liver metastatic colonization and hypoxic growth by promoting nucleotide synthesis. Elife 2019, 8, e52135. [Google Scholar] [CrossRef]
- Peters, G.; Kraal, I.; Pinedo, H. In vitro and in vivo studies on the combination of Brequinar sodium (DUP-785; NSC 368390) with 5-fluorouracil; effects of uridine. Br. J. Cancer 1992, 65, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, M.; Dousset, L.; Michon, P.; Mahfouf, W.; Rezvani, H.R. UVB-induced DHODH upregulation, which is driven by STAT3, is a promising target for chemoprevention and combination therapy of photocarcinogenesis. Oncogenesis 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.T.; Zou, S.; Chen, M.; Xiong, F.; Lee, M.H.; Fang, L.; Tang, B. Tuning Push-Pull Electronic Effects of AIEgens to Boost the Theranostic Efficacy for Colon Cancer. J. Am. Chem. Soc. 2020, 142, 11442–11450. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Kremer, D.M.; Huang, H.; Breitkopf, S.B.; Ben-Sahra, I.; Manning, B.D.; Lyssiotis, C.A.; Asara, J. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat Protoc. 2019, 14, 313–330. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| CAD | CCATGCACTAGACAGCCAAGA | CGGCTCAGTGTGGATACGAC |
| DHODH | CCACGGGAGATGAGCGTTTC | CAGGGAGGTGAAGCGAACA |
| UMPS | TCTCGACCGCGTCTTCTGA | ACACACGGTGTCAAAACTGAT |
| NME1 | AAGGAGATCGGCTTGTGGTTT | CTGAGCACAGCTCGTGTAATC |
| CTPS | CCTGGGTAACTATGAGCGGTT | ACAACTTGGACAGTTTTCCCC |
| RNR | ACTTCGGCTTTAAGACGCTAGA | GCATGAGTAAACCACCTCTCAGA |
| UPP1 | GGTGCTCCAACGTCACTATCA | TTGAAGCAGGTATCCACTGCC |
| Actin Beta | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zou, S.; Zhang, Y.; Yang, Z.; Wang, W.; Meng, M.; Feng, J.; Zhang, P.; Xiao, L.; Lee, M.-H.; et al. 3,3′-Diindolylmethane Enhances Fluorouracil Sensitivity via Inhibition of Pyrimidine Metabolism in Colorectal Cancer. Metabolites 2022, 12, 410. https://doi.org/10.3390/metabo12050410
Zhang J, Zou S, Zhang Y, Yang Z, Wang W, Meng M, Feng J, Zhang P, Xiao L, Lee M-H, et al. 3,3′-Diindolylmethane Enhances Fluorouracil Sensitivity via Inhibition of Pyrimidine Metabolism in Colorectal Cancer. Metabolites. 2022; 12(5):410. https://doi.org/10.3390/metabo12050410
Chicago/Turabian StyleZhang, Jieping, Shaomin Zou, Yijing Zhang, Ziqing Yang, Wencong Wang, Manqi Meng, Junyan Feng, Peng Zhang, Lishi Xiao, Mong-Hong Lee, and et al. 2022. "3,3′-Diindolylmethane Enhances Fluorouracil Sensitivity via Inhibition of Pyrimidine Metabolism in Colorectal Cancer" Metabolites 12, no. 5: 410. https://doi.org/10.3390/metabo12050410
APA StyleZhang, J., Zou, S., Zhang, Y., Yang, Z., Wang, W., Meng, M., Feng, J., Zhang, P., Xiao, L., Lee, M.-H., & Fang, L. (2022). 3,3′-Diindolylmethane Enhances Fluorouracil Sensitivity via Inhibition of Pyrimidine Metabolism in Colorectal Cancer. Metabolites, 12(5), 410. https://doi.org/10.3390/metabo12050410






