Probing Anti-Leukemic Metabolites from Marine-Derived Streptomyces sp. LY1209
Abstract
:1. Introduction
2. Results and Discussion
2.1. Establishment of High-Performance Liquid Chromatography (HPLC)-Based High-Resolution Anti-Proliferative Profile
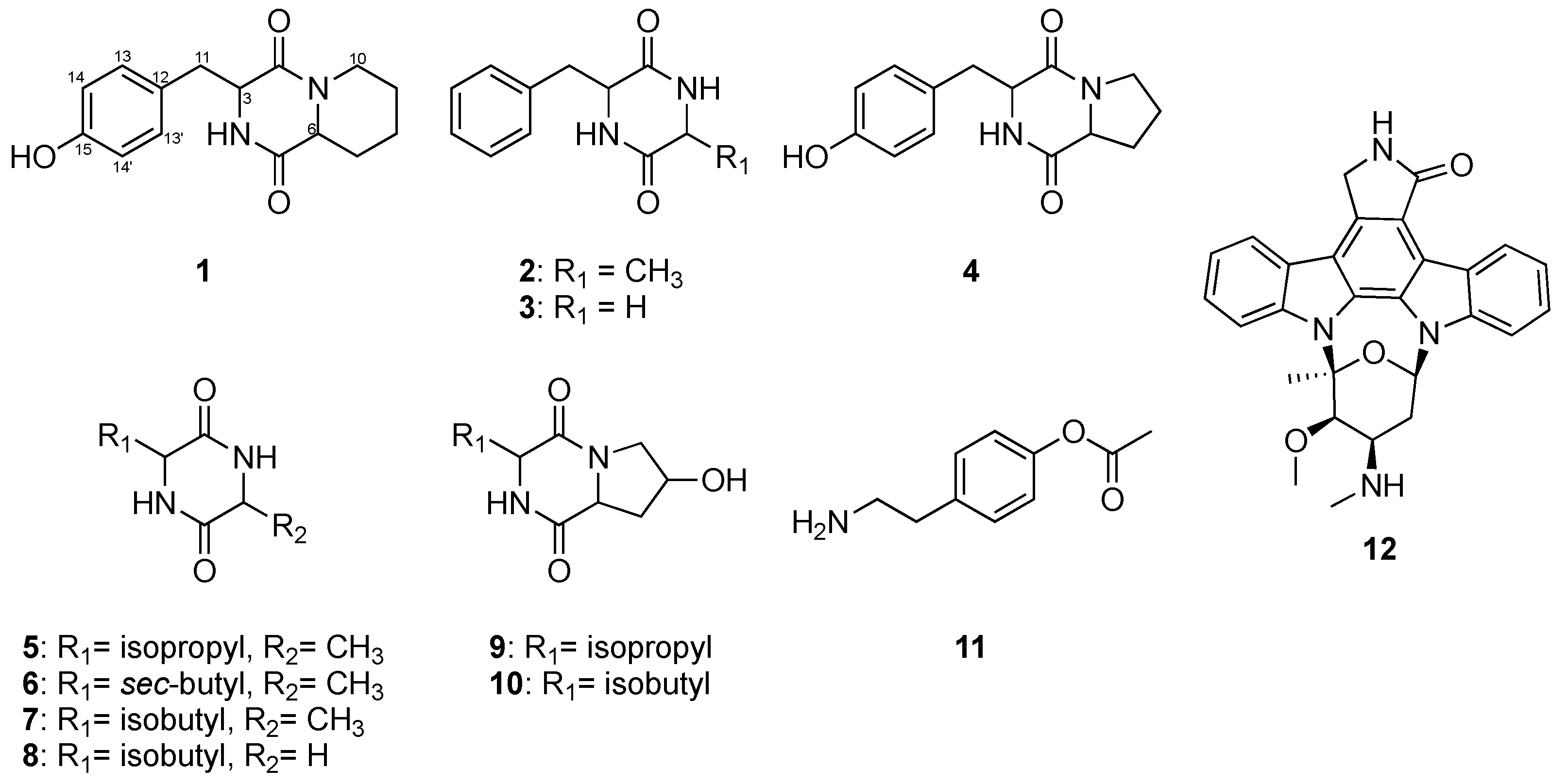
2.2. Structure Elucidation of Diketopiperazines (DKPs) Obtained from Streptomyces sp. LY1209
2.3. Anti-Proliferative Assessments of the Isolated Compounds
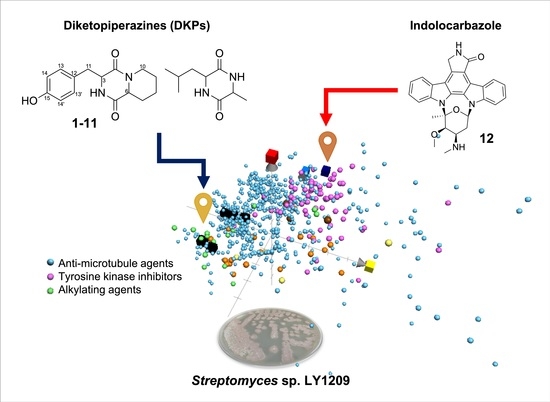
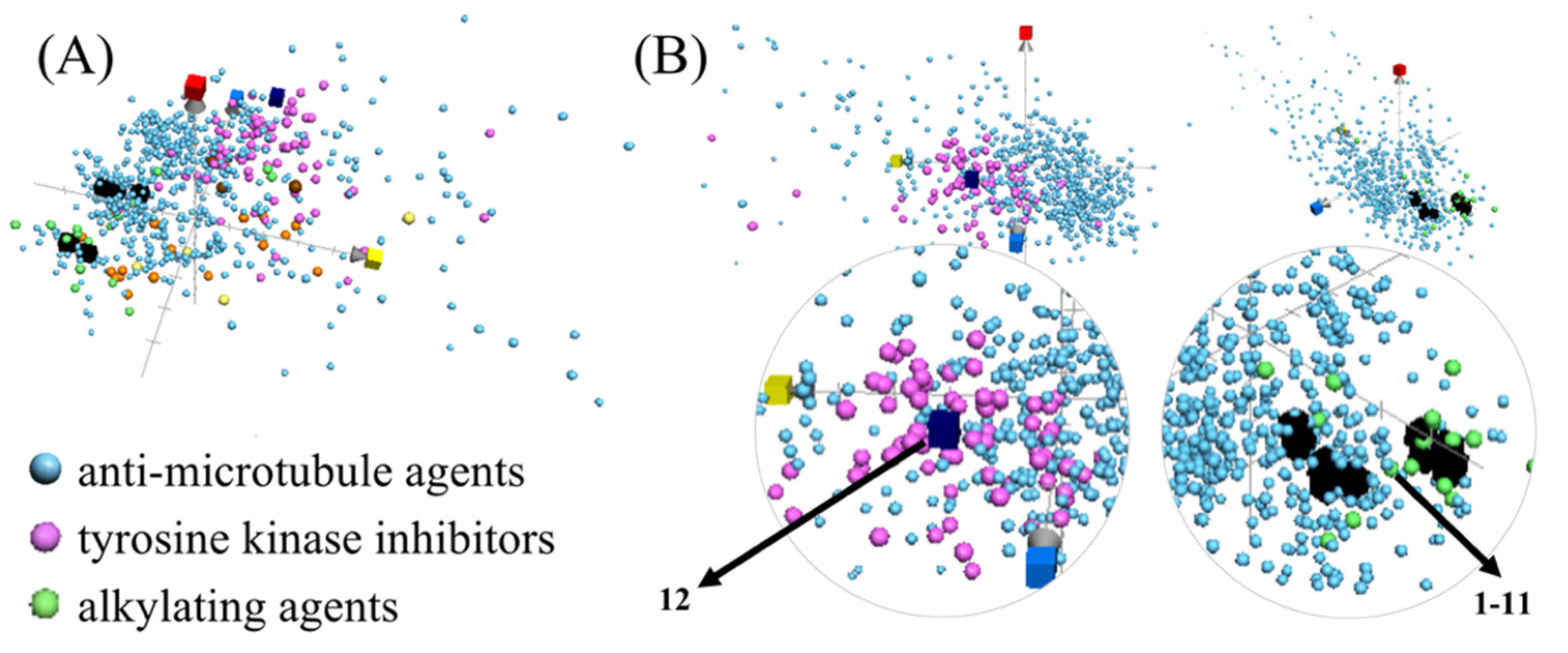
2.4. Navigating Potential Therapeutical Targets of Anti-Proliferative Compounds Using ChemGPS-NP
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Isolation, Culture, and Extraction of Marine Bacteria
3.3. Fraction Preparation for HPLC-Based High-Resolution Anti-Proliferative Assay
3.4. MTT Proliferation Assay
3.5. ChemGPS-NP Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- PDQ Adult Treatment Editorial Board. Adult Acute Lymphoblastic Leukemia Treatment (PDQ(R)): Health Professional Version; PDQ Cancer Information Summaries; National Cancer Institute: Bethesda, MD, USA, 2002.
- Jabbour, E.; Thomas, D.; Cortes, J.; Kantarjian, H.M.; O’Brien, S. Central nervous system prophylaxis in adults with acute lymphoblastic leukemia: Current and emerging therapies. Cancer 2010, 116, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Kraszewska, M.D.; Dawidowska, M.; Szczepanski, T.; Witt, M. T-cell acute lymphoblastic leukaemia: Recent molecular biology findings. Br. J. Haematol. 2012, 156, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Durinck, K.; Goossens, S.; Peirs, S.; Wallaert, A.; Van Loocke, W.; Matthijssens, F.; Pieters, T.; Milani, G.; Lammens, T.; Rondou, P.; et al. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp. Hematol. 2015, 43, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Kuehl, F.A., Jr.; Peck, R.L.; Walti, A.; Folkers, K. Streptomyces antibiotics I crystalline salts of streptomycin and streptothricin. Science 1945, 102, 34–35. [Google Scholar] [CrossRef] [PubMed]
- Alshaibani, M.M.; Mohamad Zin, N.; Jalil, J.; Sidik, N.M.; Ahmad, S.J.; Kamal, N.; Edrada-Ebel, R. Isolation, purification, and characterization of five active diketopiperazine derivatives from endophytic Streptomyces SUK 25 with antimicrobial and cytotoxic activities. J. Microbiol. Biotechnol. 2017, 27, 2074. [Google Scholar] [CrossRef]
- Nishanth Kumar, S.; Mohandas, C.; Siji, J.V.; Rajasekharan, K.N.; Nambisan, B. Identification of antimicrobial compound, diketopiperazines, from a Bacillus sp. N strain associated with a rhabditid entomopathogenic nematode against major plant pathogenic fungi. J. Appl. Microbiol. 2012, 113, 914–924. [Google Scholar] [CrossRef]
- Kohn, H.; Widger, W. The molecular basis for the mode of action of bicyclomycin. Curr. Drug Targets Infect. Disord. 2005, 5, 273–295. [Google Scholar] [CrossRef]
- Kouchaksaraee, R.M.; Li, F.; Nazemi, M.; Farimani, M.M.; Tasdemir, D. Molecular networking-guided isolation of new etzionin-type diketopiperazine hydroxamates from the persian gulf sponge Cliona celata. Mar. Drugs 2021, 19, 439. [Google Scholar] [CrossRef]
- Lucietto, F.R.; Milne, P.J.; Kilian, G.; Frost, C.L.; Van De Venter, M. The biological activity of the histidine-containing diketopiperazines cyclo(His-Ala) and cyclo(His-Gly). Peptides 2006, 27, 2706–2714. [Google Scholar] [CrossRef]
- Stelmasiewicz, M.; Swiatek, L.; Ludwiczuk, A. Phytochemical profile and anticancer potential of endophytic microorganisms from liverwort species, Marchantia polymorpha L. Molecules 2021, 27, 153. [Google Scholar] [CrossRef]
- Tangerina, M.M.P.; Furtado, L.C.; Leite, V.M.B.; Bauermeister, A.; Velasco-Alzate, K.; Jimenez, P.C.; Garrido, L.M.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V.; et al. Metabolomic study of marine Streptomyces sp.: Secondary metabolites and the production of potential anticancer compounds. PLoS ONE 2020, 15, e0244385. [Google Scholar] [CrossRef] [PubMed]
- Jinendiran, S.; Teng, W.; Dahms, H.U.; Liu, W.; Ponnusamy, V.K.; Chiu, C.C.; Kumar, B.S.D.; Sivakumar, N. Induction of mitochondria-mediated apoptosis and suppression of tumor growth in zebrafish xenograft model by cyclic dipeptides identified from Exiguobacterium acetylicum. Sci. Rep. 2020, 10, 13721. [Google Scholar] [CrossRef] [PubMed]
- Garrido Gonzalez, F.P.; Mancilla Percino, T. Synthesis, docking study and inhibitory activity of 2,6-diketopiperazines derived from alpha-amino acids on HDAC8. Bioorg. Chem. 2020, 102, 104080. [Google Scholar] [CrossRef] [PubMed]
- Rhee, K.H. Isolation and characterization of Streptomyces sp. KH-614 producing anti-VRE (vancomycin-resistant enterococci) antibiotics. J. Gen. Appl. Microbiol. 2002, 48, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Schupp, P.; Eder, C.; Proksch, P.; Wray, V.V.; Schneider, B.; Herderich, M.; Paul, V.V. Staurosporine derivatives from the ascidian eudistoma toealensis and its predatory flatworm Pseudoceros sp. J. Nat. Prod. 1999, 62, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wei, N.; Huo, Y.; Kang, X.; Chen, G.; Wen, L. Secondary metabolites of the endophytic fungus Cladosporium sp. CYC38. Chem. Nat. Compd. 2020, 56, 1166–1169. [Google Scholar] [CrossRef]
- Lu, X.; Shen, Y.; Zhu, Y.; Xu, Q.; Liu, X.; Ni, K.; Cao, X.; Zhang, W.; Jiao, B. Diketopiperazine constituents of marine Bacillus subtilis. Chem. Nat. Compd. 2009, 45, 290–292. [Google Scholar] [CrossRef]
- Nalli, Y.; Gupta, S.; Khajuria, V.; Singh, V.P.; Sajgotra, M.; Ahmed, Z.; Thakur, N.L.; Ali, A. TNF-α and IL-6 inhibitory effects of cyclic dipeptides isolated from marine bacteria Streptomyces sp. Med. Chem. Res. 2016, 26, 93–100. [Google Scholar] [CrossRef]
- Zhao, J.Y.; Ding, J.H.; Li, Z.H.; Feng, T.; Zhang, H.B.; Liu, J.K. A new cyclic dipeptide from cultures of Coprinus plicatilis. Nat. Prod. Res. 2018, 32, 972–976. [Google Scholar] [CrossRef]
- Chen, L.; Guo, Q.-f.; Ma, J.-w.; Kang, W.-y. Chemical constituents of Bacillus coagulans LL1103. Chem. Nat. Compd. 2018, 54, 419–420. [Google Scholar] [CrossRef]
- Tian, S.; Yang, Y.; Liu, K.; Xiong, Z.; Xu, L.; Zhao, L. Antimicrobial metabolites from a novel halophilic actinomycete Nocardiopsis terrae YIM 90022. Nat. Prod. Res. 2014, 28, 344–346. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, X.Y.; Deng, S.; Cao, L.; Xue, Q.H.; Gao, J.M. Alpha-glucosidase inhibitors and phytotoxins from Streptomyces xanthophaeus. Nat. Prod. Res. 2017, 31, 2062–2066. [Google Scholar] [CrossRef] [PubMed]
- Harish, V.; Periasamy, M. Enantiomerically pure piperazines via NaBH4/I2 reduction of cyclic amides. Tetrahedron 2017, 28, 175–180. [Google Scholar] [CrossRef]
- Wild, G.P.; Wiles, C.; Watts, P.; Haswell, S.J. The use of immobilized crown ethers as in-situ N-protecting groups in organic synthesis and their application under continuous flow. Tetrahedron 2009, 65, 1618–1629. [Google Scholar] [CrossRef]
- Noel, A.; Ferron, S.; Rouaud, I.; Gouault, N.; Hurvois, J.P.; Tomasi, S. Isolation and structure identification of novel brominated diketopiperazines from Nocardia ignorata—A lichen-associated actinobacterium. Molecules 2017, 22, 371. [Google Scholar] [CrossRef]
- Schrooten, R.; Joshi, S.; Anteunis, M.J.O. A comparative study of the aggregation, rotational side-chain isomerism and side-chain/side-chain interactions in the cyclic dipeptides c/Phe, Pip/and c/Phe, (s)Pip. Bull. Soc. Chim. Belg. 2010, 89, 615–628. [Google Scholar] [CrossRef]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, J.; Lovgren, A.; Kogej, T.; Muresan, S.; Gottfries, J.; Backlund, A. ChemGPS-NP(Web): Chemical space navigation online. J. Comput. Aided Mol. Des. 2009, 23, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.H.; Lu, M.C.; Du, Y.C.; El-Shazly, M.; Wu, T.Y.; Hsu, Y.M.; Henz, A.; Yang, J.C.; Backlund, A.; Chang, F.R.; et al. Cytotoxic lanostanoids from Poria cocos. J. Nat. Prod. 2016, 79, 2805–2813. [Google Scholar] [CrossRef]
- Korinek, M.; Tsai, Y.H.; El-Shazly, M.; Lai, K.H.; Backlund, A.; Wu, S.F.; Lai, W.C.; Wu, T.Y.; Chen, S.L.; Wu, Y.C.; et al. Anti-allergic hydroxy fatty acids from Typhonium blumei explored through ChemGPS-NP. Front. Pharmacol. 2017, 8, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Chai, C.Z.; Yan, Y.; Duan, Y.D.; Henz, A.; Zhang, B.L.; Backlund, A.; Yu, B.Y. Spasmolytic mechanism of aqueous licorice extract on oxytocin-induced uterine contraction through inhibiting the phosphorylation of heat shock protein 27. Molecules 2017, 22, 1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.H.; Lai, K.H.; Su, Y.D.; Chang, Y.C.; Peng, B.R.; Backlund, A.; Wen, Z.H.; Sung, P.J. Briaviolides K-N, new briarane-type diterpenoids from cultured octocoral Briareum violaceum. Mar. Drugs 2018, 16, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henz Ryen, A.; Backlund, A. Charting angiosperm chemistry: Evolutionary perspective on specialized metabolites reflected in chemical property space. J. Nat. Prod. 2019, 82, 798–812. [Google Scholar] [CrossRef] [PubMed]
- Alajlani, M.M.; Backlund, A. Evaluating antimycobacterial screening schemes using chemical global positioning system-natural product analysis. Molecules 2020, 25, 945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, K.H.; Peng, B.R.; Su, C.H.; El-Shazly, M.; Sun, Y.L.; Shih, M.C.; Huang, Y.T.; Yen, P.T.; Wang, L.S.; Su, J.H. Anti-proliferative potential of secondary metabolites from the marine sponge Theonella sp.: Moving from correlation toward causation. Metabolites 2021, 11, 532. [Google Scholar] [CrossRef]
- Alajlani, M.M. The chemical property position of bedaquiline construed by a chemical global positioning system-natural product. Molecules 2022, 27, 753. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Ma, M.; Zhong, C.; Wang, S.; Fu, Z.; Hou, Y.; Liu, Y.; Zhong, L.; Chu, Y.; Li, F.; et al. Development of novel phenoxy-diketopiperazine-type plinabulin derivatives as potent antimicrotubule agents based on the co-crystal structure. Bioorg. Med. Chem. 2020, 28, 115186. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Okuyama, K.; Kaida, A.; Hayashi, Y.; Hayashi, Y.; Harada, K.; Miura, M. KPU-300, a novel benzophenone–diketopiperazine–type anti-microtubule agent with a 2-pyridyl structure, is a potent radiosensitizer that synchronizes the cell cycle in early M phase. PLoS ONE 2015, 10, e0145995. [Google Scholar]
- Tullberg, M.; Grøtli, M.; Luthman, K. Synthesis of functionalized, unsymmetrical 1,3,4,6-tetrasubstituted 2,5-diketopiperazines. J. Org. Chem. 2006, 72, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Vale, N.; Ferreira, A.; Matos, J.; Fresco, P.; Gouveia, M. Amino acids in the development of prodrugs. Molecules 2018, 23, 2318. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Li, C.; Zhu, X. FLT3 inhibitors in acute myeloid leukemia. J. Hematol. Oncol. 2018, 11, 133. [Google Scholar] [CrossRef] [PubMed]


| Position | δH (J in Hz) a | δC (Mult.) b | 1H–1H COSY | HMBC |
|---|---|---|---|---|
| 2 | 167.0 (C) | |||
| 3 | 4.23, td (3.9, 1.3) c | 57.2 (CH) d | H-11 | C-2, C-5 |
| 5 | 169.9 (C) | |||
| 6 | 2.54, dd (11.3, 1.6) | 58.4 (CH) | H-7 | |
| 7 | 1.32, m; 2.09, d (11.0) | 31.6 (CH2) | ||
| 8 | 1.33, m; 1.83, m | 24.8 (CH2) | H-7, H-9 | |
| 9 | 1.33, m; 1.65, m | 25.3 (CH2) | H-7, H-8, H-10 | |
| 10 | 2.20, td (12.9, 3.2); | 43.3 (CH2) | H-8, H-9 | |
| 4.50, dt (13.5, 2.0) | ||||
| 11 | 3.17, dd (14, 1.8) | 40.9 (CH2) | H-3 | C-2, C-3, C-12, C-13 |
| 4.50, dt (13.5, 2.0) | ||||
| 12 | 126.5 (C) | |||
| 13, 13‘ | 6.91, d (8.5)u | 132.5 (CH) | H-14 | C-11, C-15 |
| 14, 14‘ | 6.70, d (8.5) | 116.2 (CH) | H-13 | C-12, C-15 |
| 15 | 158.2 (C) |
| Compounds | Cell Lines (IC50 μM) |
|---|---|
| Molt 4 | |
| 1 | 95.93 ± 2.32 |
| 2 | – a |
| 3 | – a |
| 4 | – a |
| 5 | – a |
| 6 | 56.23 ± 7.52 |
| 7 | – a |
| 8 | – a |
| 9 | – a |
| 10 | – a |
| 11 | – a |
| 12 | 0.01 ± 0.001 |
| Doxorubicin b | 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-Y.; Chen, L.-Y.; Chen, P.-J.; El-Shazly, M.; Peng, B.-R.; Chen, Y.-C.; Su, C.-H.; Su, J.-H.; Sung, P.-J.; Yen, P.-T.; et al. Probing Anti-Leukemic Metabolites from Marine-Derived Streptomyces sp. LY1209. Metabolites 2022, 12, 320. https://doi.org/10.3390/metabo12040320
Chen Y-Y, Chen L-Y, Chen P-J, El-Shazly M, Peng B-R, Chen Y-C, Su C-H, Su J-H, Sung P-J, Yen P-T, et al. Probing Anti-Leukemic Metabolites from Marine-Derived Streptomyces sp. LY1209. Metabolites. 2022; 12(4):320. https://doi.org/10.3390/metabo12040320
Chicago/Turabian StyleChen, You-Ying, Lo-Yun Chen, Po-Jen Chen, Mohamed El-Shazly, Bo-Rong Peng, Yu-Cheng Chen, Chun-Han Su, Jui-Hsin Su, Ping-Jyun Sung, Pei-Tzu Yen, and et al. 2022. "Probing Anti-Leukemic Metabolites from Marine-Derived Streptomyces sp. LY1209" Metabolites 12, no. 4: 320. https://doi.org/10.3390/metabo12040320
APA StyleChen, Y.-Y., Chen, L.-Y., Chen, P.-J., El-Shazly, M., Peng, B.-R., Chen, Y.-C., Su, C.-H., Su, J.-H., Sung, P.-J., Yen, P.-T., Wang, L.-S., & Lai, K.-H. (2022). Probing Anti-Leukemic Metabolites from Marine-Derived Streptomyces sp. LY1209. Metabolites, 12(4), 320. https://doi.org/10.3390/metabo12040320