Abstract
Due to their unique multi-gastric digestion system highly adapted for rumination, dairy livestock has complicated physiology different from monogastric animals. However, the microbiome-based mechanism of the digestion system is congenial for biology approaches. Different omics and their integration have been widely applied in the dairy sciences since the previous decade for investigating their physiology, pathology, and the development of feed and management protocols. The rumen microbiome can digest dietary components into utilizable sugars, proteins, and volatile fatty acids, contributing to the energy intake and feed efficiency of dairy animals, which has become one target of the basis for omics applications in dairy science. Rumen, liver, and mammary gland are also frequently targeted in omics because of their crucial impact on dairy animals’ energy metabolism, production performance, and health status. The application of omics has made outstanding contributions to a more profound understanding of the physiology, etiology, and optimizing the management strategy of dairy animals, while the multi-omics method could draw information of different levels and organs together, providing an unprecedented broad scope on traits of dairy animals. This article reviewed recent omics and multi-omics researches on physiology, feeding, and pathology on dairy animals and also performed the potential of multi-omics on systematic dairy research.
1. Introduction
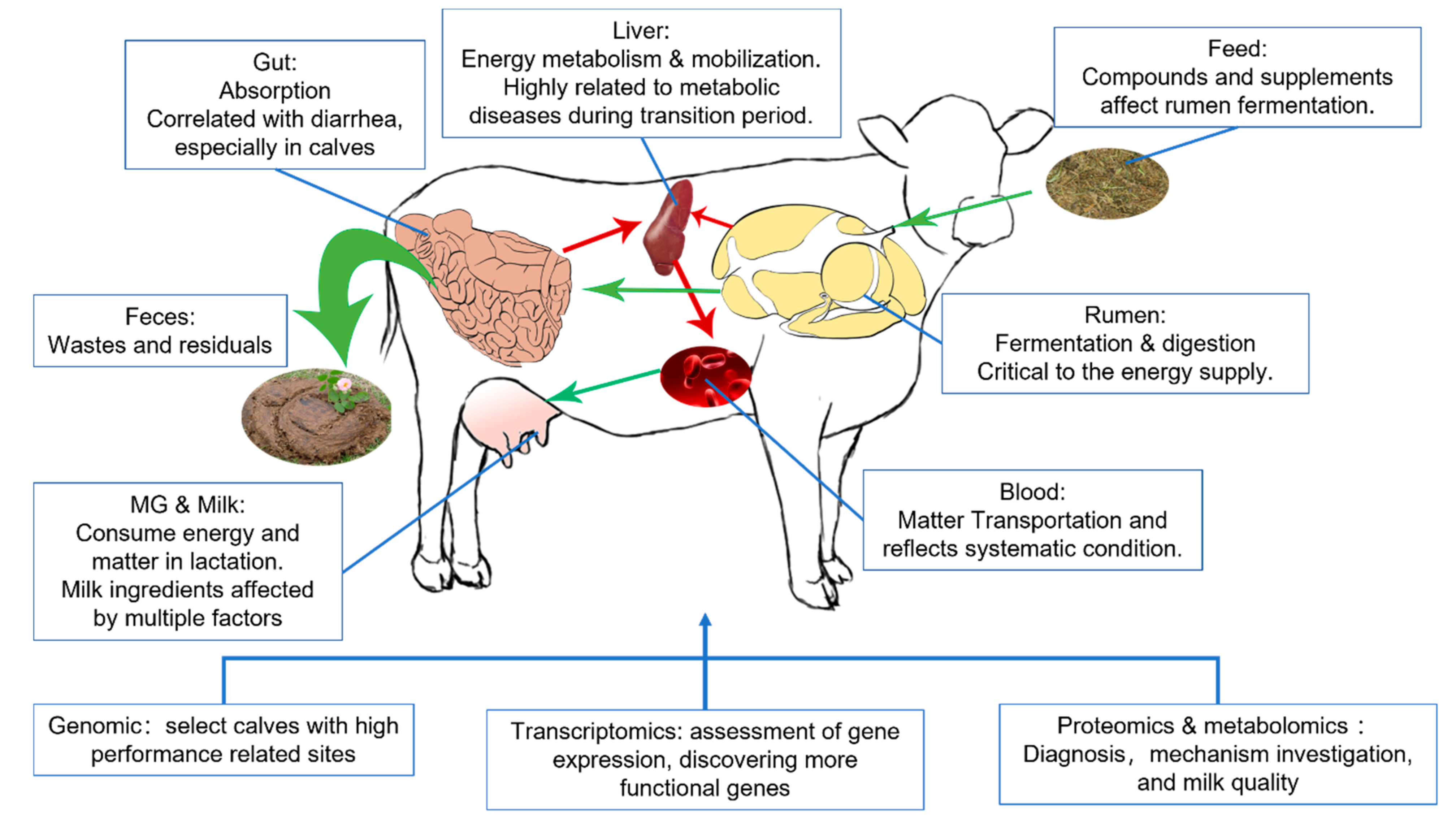
Omics, referring to a field of study in biological sciences that ends with -omics, aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms. The development of an automatic DNA sequencer in the early 1990s made whole-genome sequencing possible [1], announcing the dawn of omics. In the following 30 years, novel omics assays have been set up once a corresponding high-throughput qualifying or quantifying method has been established, such as transcriptomics, proteomics, or metabolomics [2,3]. Transcriptomic could clarify and quantify RNA sequences in the sample, representing a snapshot of cellular metabolism, while proteomic and metabolomic divided by chromatography, then qualifying them by comparing mass spectrometry (MS) or nuclear magnetic resonance (NMR) data with databases to capture the function status of target tissues. Omics methods provide researchers with an expanded vision of all detectable molecules on a certain level. They have become an effective multifunctional tool that has been applied from screening differential molecules to sorting phenotypes [4,5,6]. Indeed, single omics provides systematic information on a certain level, but researchers are always eager to have a broader scope. In ruminant research, especially dairy sciences, the factors usually have to pass through more barriers and biology levels than monogastric animals. Those organs are relatively isolated systems while interacting, weaving a tangled web of connection, and making physiology and pathology studies on a single organ or level hard to acquire certain conclusions [7]. In those cases, the multi-omics investigation will promote the exploration of phenotype-related biomarkers and corresponding mechanisms, like how dietary nutrients affect milk components [8] and the impact of rumen microbiota on lactation performance [9]. Current dairy research mainly applies omics methods to breeding, investigating physiology and pathology, developing new traits, and evaluating feed sources and supplements in widely spread organisms from the rumen to spermatozoa. Among those organisms, rumen, liver, and mammary glands are the critical point research spots, which were suggested as critical organs related to the performance of dairy cows [10], in which enriched rumen could ferment dietary carbohydrates into volatile fatty acid (VFA), and also could convert indigestible forage into nutrients by colonized symbiotic microbiota; the liver plays a critical role in processing absorbed nutrient and other bioactive components, acts as the core of fat mobilization, also modifying the component of mammary gland secretion [11]. This rumen–liver–mammary glands network included the path of nutrient molecules transportation from the very first feed intake and ruminant fermentation to finally milk secretion. Different types of omics research on nutrient and metabolic disease have targeted this network, and recent application of multi-omics reported in dairy science focused on relating those organs altogether or discovering the mechanisms of how those organs were affected (Figure 1) [12,13].
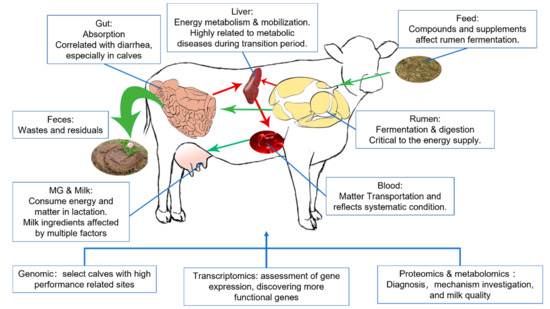
Figure 1.
Organs mainly involved in the lactation physiology and utilities of omics. Green and red arrows stand for the nutrition transport and energy supply separately.
However, as the highly specialized symbiotic fermentation system, many omics studies in dairy sciences are integrated with microbiome analysis of the gastrointestinal tract. Genomic and transcriptomics have discovered more traits related to milk production and feed efficiency; non-target proteome and metabolome are with an increase of their significance in the expanding of biomarkers and have contributed to understanding the mechanisms of mastitis and infertility, which could cause massive economic losses to dairy farmers (Table 1).

Table 1.
Omics applied in the dairy research.
Even multi-omics applications in the dairy sciences have just started in the recent few years, and it has already become a novel hotspot for research. This article reviewed the application of omics techniques from metagenomics to metabolomics and their integration in the dairy research about lactation physiology, fertility, feeding, management, and diseases, emphasizing the significance of systematic view in the dairy research prospected futural multi-omics utilizations for dairy sciences studies.
2. Multi-Omics Studies in Lactation Physiology
Due to their unique multi-gastric digestion system highly adapted for rumination, dairy livestock’s physiology of energy metabolism varied from monogastric animals. Ruminal symbiotic microbes are highly specialized in degrading lignocellulosic biomass into fermentable sugar, finally, fermenting plant-derived carbohydrates into VFAs [63]. Ultimately, VFAs are absorbed through the gastrointestinal tract into the portal vein, utilized by the liver. For dairy animals, rumen, liver, and mammary gland are nodes of the lactation physiology network, corresponding to energy intake, distribution, and output. Because of the vital role of ruminal microbiota in energy metabolism, those organs, with the symbiotic microbiome, provide researchers with a great example of “superorganism” for host–microbial interactions [63].
Rumen and symbiotic microbes directly contribute to rumen metabolites and dietary components and alter the ruminal microbiome by changing fermentation substrates [64]; they are similar to intestinal microbes in monogastric animals in some aspects but have much more impact on the animal body. A high concentrate diet increases the abundance of potentially harmful rumen metabolites like LPS and methylamine in rumen fluid, also higher the risk of rumen acidosis [31]. On the contrary, fresh grass improves microbe colonization, digestion, and microbial protein synthesis and decreases the methane emissions of rumen compared with grass hay [12,35]. Feed composition plays a significant role in shaping the rumen microbiome of calves by modulating initial colonization [65,66]. Proteobacteria is the dominant microbial in the newborn calves, then replaced by Bacteroidetes during the ruminal development [24,67], and ruminants with well-developed ruminal flora are more robust in challenging diarrhea.
Meanwhile, the symbiotic microbiome affects rumen digestion, feed efficiency, and milk production. Cows with different milk yields have significantly altered rumen fluid metabolomic patterns related to protein digestion and absorption, ABC transporters, and unsaturated fatty acid biosynthesis pathways associated with firmicutes, actinobacteria, and synergistetes in the rumen [44]. Metabolomics studies proved that cows with higher feed efficiency showed downregulated amino acid, ruminal linoleic, and alpha-linolenic metabolism [39,45]. Moreover, multiple studies showed that milk performances are related to Prevotella [9,46,68]; cows with higher protein yields have a higher abundance of Prevotella sp. and lower methane-producing microorganisms in the rumen and related to branched-chain amino acid biosynthesis and less methane emission. Cows with higher feed efficiency might have a microbiome with fewer but more efficient metabolic pathways and dropped low-value metabolites production [22,69,70,71]. For instance, protozoa, a member of the rumen microbiome, have a controversial effect on ruminal digestion [70]. Several studies reported that protozoa are not essential, lead to increased ammonia nitrogen and methane emissions, and negatively correlate to nitro utilization and rumen microbial protein synthesis [72,73]. However, recent research found that protozoa have a positive effect by directly contributing to fiber degradation and indirectly consuming ruminal oxygen to maintain anaerobic conditions, especially in high-forage diets [73,74,75]. Due to the lack of protozoa sequence information, there are still obstacles to discovering its role in ruminal digestion [23]. Even in the same nutritional and management condition, cows also perform different milk yields and milk components related to the rumen microbiome. As demonstrated above, Prevotella is correlated with high milk protein yields. Cows with higher saturated fatty acids usually have a higher abundance of lactic acid bacteria (Lactobacillus, Leuconosto, and Weissella) and acetogenic Proteobacteria (Acetobacter and Kozakia) and showed higher concentrations of butyrate, propionate, and tyrosine and lower concentrations of xanthine and hypoxanthine in the rumen, suggesting those cows might be adapted to reduced rumen pH [51].
The liver is the nexus of lipid metabolism and plays a critical role in ruminant physiology. Nearly 70% of circular glucose in the dairy cow is derived from hepatic gluconeogenesis. After calving, the energy consumption elevates rapidly with the initiation of lactation, meanwhile, decreased dry matter intake (DMI) limited the energy supply, the residual feed intake would soon become negative, leading to the negative energy balance (NEB) [76], in this circumstance, sugar storage will soon be exhausted. Because of maintaining physiological functions, the liver becomes the processing center of mobilized body fat. Ketones derived by triacylglycerol hydrolyzation from adipocytes are usually excessive for liver oxidation, inducing hepatic lipid accumulation, and finally, causing fatty liver and ketosis [77]. These effects also alter the lipid composition of the milk; cows with serum BHB (β-hydroxybutyrate) higher than 0.1 mmol/L may have lower C6 (caproic acid), C22:1ω9 (Erucic acid), C22:5ω3 (Decosapentaenoic acid, DPA), and C23 (Tricosanoic acid) [78]. Varied milk LCFA and VLCFA concentrations also reflect the risk of ketosis and metabolic changes in ewes and donkeys [79,80]. Nearly 50% of dairy cows suffer from metabolic diseases in their first month of lactation [81]. However, it seems that the liver has adapted to the metabolism condition before calving under the regulation of transcript factor PPARA and NFE2L2 [82]. Cows with higher lipid mobilization have an altered plasma lipidome [36,83], indicating other metabolic pathways may also be influenced. L-carnitine showed a potential metabolism to promote the effect of Non-esterified fatty acids (NEFA) and reduce lipid accumulation in the liver [84]. However, the L-carnitine does not seem to affect the hepatic transcriptome profile [19]. Until now, a large number of essential hepatic genes, proteins, and metabolites related to lactation physiology have been reported, but it is still hard to build a systematic view of how liver function affects lactation, the differences between physiological and pathological conditions are also waiting to be investigated by a systematic scope.
Milk, the main product of dairy animals, as mentioned above, is closely related to the condition of rumen and liver. It is widely known that milk yield and components are heritable [43]. Genomic research showed that single nucleotide polymorphisms (SNPs) of RAP1A and DGAT1 are correlated with milk protein yield [85,86]. Transcriptome also revealed that more than 33,000 SNPs are associated with lactation, in which expression levels of 31 genes are directly related to milk yield [18]. Apart from heritable reasons, milk production is directly influenced by dietary structure. Alfalfa hay, rice straw, and corn straw could change mammary glands and liver’s transcript and protein profile [37,43]. However, protein expression changes are sometimes not positively correlated with their corresponding mRNAs [38].
Meanwhile, the rumen microbiome and feed components would change lactation performance. Varied rumen microbiome would induce a different milk fatty acid profile with altered rumen fermentation and protein metabolism under the same diet [51]. Different forage sources could also change the function and composition of the rumen microbiome. Cows fed with corn stover have a significantly lower abundance on gene encoding lactaldehyde reductase, glutamine synthetase type I, methylmalonyl-CoA decarboxylase, succinate dehydrogenase, and alpha-xyloside ABC transporter in the rumen microbiome [43]. Mammary glands are the output positions in lactation physiology, also are the most direct factors related to the milk components and yield. Advancing their knowledge would bring researchers more precise methods for mastitis diagnosis and milk quality assessments.
The rumen–liver–mammary gland network is the core of lactation physiology. Each element, including genotypes, feed components, rumen fermentation, liver conditions, mammary gland function, the interaction between symbiotic microbes and host, also different tissues and organs within the host, would influence the final milk production and milk contents. Even though many metagenomic, transcriptomic, and proteomic studies have been administrated in dairy sciences, there is still not enough information about how different organs are connected within physiological conditions. Studies are focused on the changes induced by specific changes but not on the normal condition. Indeed, due to the individual differences, it is hard to define a “normal condition” of dairy animals, but with the expansion of bioinformatics, a widely accepted baseline multi-omics fingerprint map may be established—leading to a novel recognition of the lactation physiology of dairy animals.
3. Multi-Omics Methods for Reproduction Research
The fertility of dairy cows has declined gradually since the 1980s, causing an increased eliminating rate and reduced probable life [87]. Subfertility has become a significant problem with the increasing milk yield of dairy cows and causing tremendous economic losses [88,89]. Genomic research showed that daughter pregnancy rate could become a fertility prediction index [90], and different breeds of dairy cows share few SNPs related to the reproduction traits [16], while bulls have more conservative x chromosomal fertility-related SNPs [91]. However, the clinical evaluation of fertility mainly focuses on morphological features and hormone levels, which are unilateral measures that lack objectivity [82]. Thus, even omics applications in ruminant reproduction are still limited compared with reproduction research in humans, the systematic information acquired by omics approaches becomes significant to expanding our fertility and reproduction knowledge.
Fertility differences in the dairy cattle also performed in the proteome pattern in the gamete. More than 125 proteins significantly differ between bulls with high and low fertility [53]. Those proteins are related to TCA-cycle, ATP concentration, and mitochondria functions [92,93]. Those differences also appear in the transcription level of semen [17]. The expression level of miRNAs also differs in bulls with different fertility and infertility rate [20,94,95]. Pear-shaped sperm is one of the widely known sperm deformity patterns, which has a different protein profile related to reduced antioxidative activities, sperm capacitation, and cytoskeleton [55].
The fertility property of cows is much more complicated compared to bulls. The omics pattern of oocytes is also related to fertility even before ovulation [58]. Even the mechanism is complex. Nevertheless, oxidative stress and inflammation could be the primary reasons for decreased fertility [96]. For instance, annexins, a family of proteins related to anti-inflammation, have higher levels in the uterus fluid in early pregnancy and glutathione-S-transferases concentration during the late estrus [54,97]. Meanwhile, some proteins could perform varied relationships to the fertility in different sections of the reproductive tract, tissue inhibitor of matrix metalloproteinase 2 (TIMP2), an enzyme related to trophoblast invasion and possibly in endometrial remodeling, have an increasing level in uterus fluid with the progress of pregnancy [98]. However, cows with lower fertility have two times higher TIMP2 concentrations in the follicular fluid than regular cows [58], while TIMPs also perform as biomarkers in human reproduction [99]. These may suggest that proteins perform varied functions in the different stages of reproduction. Metabolites in the reproductive tract are also correlated to fertility. Methionine, an amino acid that has been approved that could change rumen microbiota and improve milk components [8], has a different abundance in the follicular fluid between maiden and first parous heifers [33]. Guerreiro et al. [100] discovered the differential metabolites between the follicular fluid in cows with different fertility divided by oocyte production, and found that antioxidative metabolites resveratrol 4′-glucoside, lupinisoflavone N, peonidin acetyl 3,5-diglucoside, 3,3′,4,5′-tetrahydroxy-trans-stilbene, 5,7-dihydroxy-6-methyl-8-prenylflavanone, xanthohumol, and prostaglandin M could become the marker of high fertility.
Omics methods assist researchers in discovering and locating the reproduction-related genes in both cows and bulls, and they also emphasized that oxidation and inflammation are the main factors related to fertility and provide researchers new biomarkers in the screening of high fertility calves. Furthermore, studies that applied the omics approach have expanded our knowledge about the microenvironment reproduction tract and indicated the effect of metabolites on the implantation and pregnancy process. Through the multi-omics methods, researchers may include the reproduction system into the rumen–liver–mammary gland network and develop more intervention protocols to improve the reproduction traits.
4. Multi-Omics Assists Feeding and Management
Improving feed efficiency is a long run for the dairy industry, as a complicated phenotype, the efficacy of feed utilization depends on genotypes, ruminal fermentation, and feeding components [14]. Genomic studies showed that feed efficiency is heritable and optimized via genomic selection in large herds [101]. Lactation and growth performance are also regulated by ruminal and hepatic micro RNAs (miRNA, miR). Dietary components and particle size could affect the expression of feed efficiency and rumen function-associated miRNAs [21,25].
Periparturient (or transition) period, from 3 weeks pre-calving until three weeks post-calving, has a critical impact on dairy production [102]. In this period, significant physiological, metabolic, and nutritional changes occur when most metabolic disorders occur [103]. Understanding the physiological, metabolic, and nutritional changes will help to improve current management for the better welfare of periparturient dairy cows and minimize economic losses. The dry period, a regularly 60-day no-milking phase before expected calving to support fetus development and prepare for the next lactation, is widely accepted in dairy farms [104]. On the one hand, a dry-off phase would increase the infection risk of the mammary gland. However, a continuous milking protocol has been described to reduce health problems but reduces milk protein and change proteomic profile, significantly lowering the concentration of colostrum immunoglobulins by nearly 50% and may weaken adequate passive immune transfer [105]. During the dry period, compositions of mammary gland secretions also alter in response to the drying administration, miRNAs related to gestation, lactation, inflammation, and disease formation significantly changed in the different phases of the dry period [27]. The temperature–humidity control is also an essential part of transition period management, cows exposed to high temperature and humidity environment would have heat stress. According to Skibiel et al. [61], heat stress that occurs in the dry period would affect liver proteome profile, interferes oxidative phosphorylation, mitochondrial function, farnesoid X receptor/retinoid X receptor (FXR/RXR) activation, and the methylmalonyl pathways; reduce ATP production, aggravates oxidative stress; and accelerate hepatic triglycerides and cholesterol accumulation. Heat stress leads to higher susceptibility to transition-related diseases; a whole-genome analysis showed that high-producing cows are more susceptible to heat stress. Hsp90 protein binding, zinc ion binding, and gated channel activity pathways are also related to heat stress, and at least three different genomic regions on BTA5, BTA14, and BTA15 chromosomes are strongly associated with milk production under heat stress conditions [15]. By the integration of multi-omics, we can find the mechanism that related to stress in the management process and develop a more effective protocol that provide more economic benefits for dairy farmers and better welfare for dairy animals.
Effect of dietary component is also an essential aspect of feed formulation, and feed material could provide energy and substrates for physiological function. Ametaj et al. [31], the first group using metabolomics to evaluate the effect of dietary components on the rumen microbiome, found that 30% and more barley grain could increase the concentration of potentially harmful rumen metabolites. Changes in rumen metaproteomic also observed by Snelling and Wallace [60], dairy cows fed by a high concentrate diet, protozoa structural proteins will dominate proteome profile of ruminal digesta, and bacterial proteins are mainly glycolysis related proteins. Different forage sources could also alter the metabolites profile of rumen fluid and milk, serum, and urine in dairy cows, and alfalfa hay-fed cows have a higher N efficacy, amino acids metabolism, and milk performance than corn stover fed cows [106]. Different processing of the same material could also change milk protein content: heat-treated soybean meal could induce higher milk α-casein abundance and lower β-casein, α-lactalbumin, and zinc-alpha-2-glycoprotein than solvent-extracted soybean meals [56]. A recent study by Veshkini et al. [62,107] performed that the supplement of FAs would improve metabolism of xenobiotics by cytochrome P450, drug metabolism—cytochrome P450, retinol metabolism, and steroid hormone biosynthesis in the transition period; the addition of polyunsaturated fatty acid (PUFA)-enriched marine microalgae also affects ruminal microbiome and FA profile of milk, improves rumen fermentation, and increases concentrations of PUFA in milk [68].
Furthermore, the functions of bioactive compounds in materials should be considered in feed development. For instance, malt, a high-starch ingredient with a lactation inhibitory effect, might not be a suitable TMR ingredient in the lactation period but could be a functional supplement in the dry period [108]. Thus, omics methods become a powerful tool in evaluating the effect of feed additives on the physiology of dairy animals. Wang et al. [50] found that the 300 g/d of Perilla frutescens leaf supplementation could upregulate oleanolic acid and nucleotides in milk while downregulating 2-hydroxycaprylic acid and enriching metabolic pathways such as pyrimidine metabolism and biosynthesis of unsaturated fatty acids in both rumen and milk. Rumen-protected nutrients are processed for avoiding the ruminal degradation and final release in the intestine, which have been widely researched in recent years. Elolimy et al. [42] found that rumen-protected (RP) methionine supplement in late pregnancy cows altered their calves’ fecal microbiome and metabolomic profiles to have better growth performance. Further research by Gu et al. [8] showed that RP methionine could increase ruminal Acetobacter and Saccharofermentan abundance and elevate milk α-ketoglutaric acid concentration and milk fat may explain the beneficial effect to their offspring. Phenotype research also proved that RP methionine supplements improved the oxidative status of dairy cows [109]. Other RP amino acids also had an effect on the rumen metabolomic profile and lactation performance: RP lysine could improve milk production in corn-fed dairy cows but decreased their oxidative stability [110]; supplement RP arginine to pregnant sheep not only alleviated the nutrient restriction during pregnancy, but also altered the amino acid, carbohydrate, and metabolic pattern in umbilical venous blood of fetus [40]. Apart from amino acids, glucose and alkaloids after the RP process also showed altered digestive parameters: 200 g/d of RP glucose supplement increased rumen bacterial richness and diversity, elevated cellulolytic bacteria abundance, and changed rumen fermentation, increased the concentrations of acetate, propionate, butyrate, and total volatile fatty acid [52]; additional RP betaine increased milk yield and milk protein and influenced pathways related to the synthesis of arginine and cyanoamino acid, also the degradation of proline; however, RP betaine had no significant difference on growth performance comparing with unprotected betaine [47,111].
As mentioned above, the rumen microbiota are related to metabolism, lactation, fertility, and feed utilization. While rumen microbiota also relate to the variation of performance among individual cows under the same feeding and management conditions, Xue et al. [9] found that cows with higher milk protein yield showed different microbial compositions of bacteria and archaea, especially Prevotella sp. the altered microbiome pattern also performed in their metabolites, rumen fluid of cows with higher milk and milk protein yield have a higher concentration of amino acids, carboxylic acids, and fatty acid. The changes in rumen microbiota also reflect amino acid (glycine, serine, threonine, alanine, aspartate, glutamate, cysteine, and methionine) metabolism. Further analysis found that rumen microbial components, functions, metabolites, and serum metabolites of the host are all related to the phenotype of milk protein yields. The metabolites of the rumen microbiome and host serum have a similar contribution ratio to the milk protein yields. Although rumen flora plays a crucial part in nearly their performances, it is also the main reason for methane emission [112]. Meta-transcriptomics research demonstrates that bacterial, archaeal, and eukaryotic biomass, methane emission, and VFA concentration increased rapidly in the first hour after feed intake, with corresponding changes of carbohydrate-active enzyme transcripts [26]. Furthermore, this process could be recognized as a phenotype with individual varieties. Those rumen microbiome-related phenotypes may be adjusted or improved by probiotics and direct feed microbial (DFMs). Ogunade et al. [113] found that additional 15 g/d live yeast (S. cerevisiae) increases eight cellulolytic bacterial genera while optimizing the utilization of oxygen and lactic acid and inhibits the growth of pathogenic Salmonella. Supplementation of 0.1% live Enterococcus faecium in dietary could significantly increase the propionate concentration in the rumen fluid while inhibiting the emission of methane, and the dose of E. faecium supplement has a different impact on the rumen microbiomes [114]. The effect of complex DFMs with multiple microorganism species and their fermentation products on rumen function and host serum metabolomics is also evaluated by Ogunade et al. [46], although two different DFMs showed varied impacts on the rumen microbiome, both DFMs elevated serum glucose, total VFA, propionate, isovalerate, and valerate concentrations in the rumen also showed similar effects on VFA profile and energy status. The multi-omics combines the responses of different levels and organs, provides a whole vision for the impact of feed components to dairy animals, would become a better way for novel feed sources developments
Omics methods help researchers develop novel feed resources and management protocols, evaluate the effects and mechanisms of feed supplements, and use nutrient interventions to improve milk contents. Meanwhile, those methods assist dairy farmers in adjusting the dietary formula and optimizing management to elevate production efficiency and prevent transition diseases. Furthermore, they provide potential solutions to decrease the emission of greenhouse gas. However, limited by sampling time points and current technologies, omics analysis still could not provide a timely response as a monitoring technique but has already shown the advantages in detecting molecular indicators and predicting functional compounds. With the improvement, multi-omics would become a capable tool for feeding and management assessment and herds’ health observation.
5. Multi-Omics Promotes Revealing Dairy Diseases
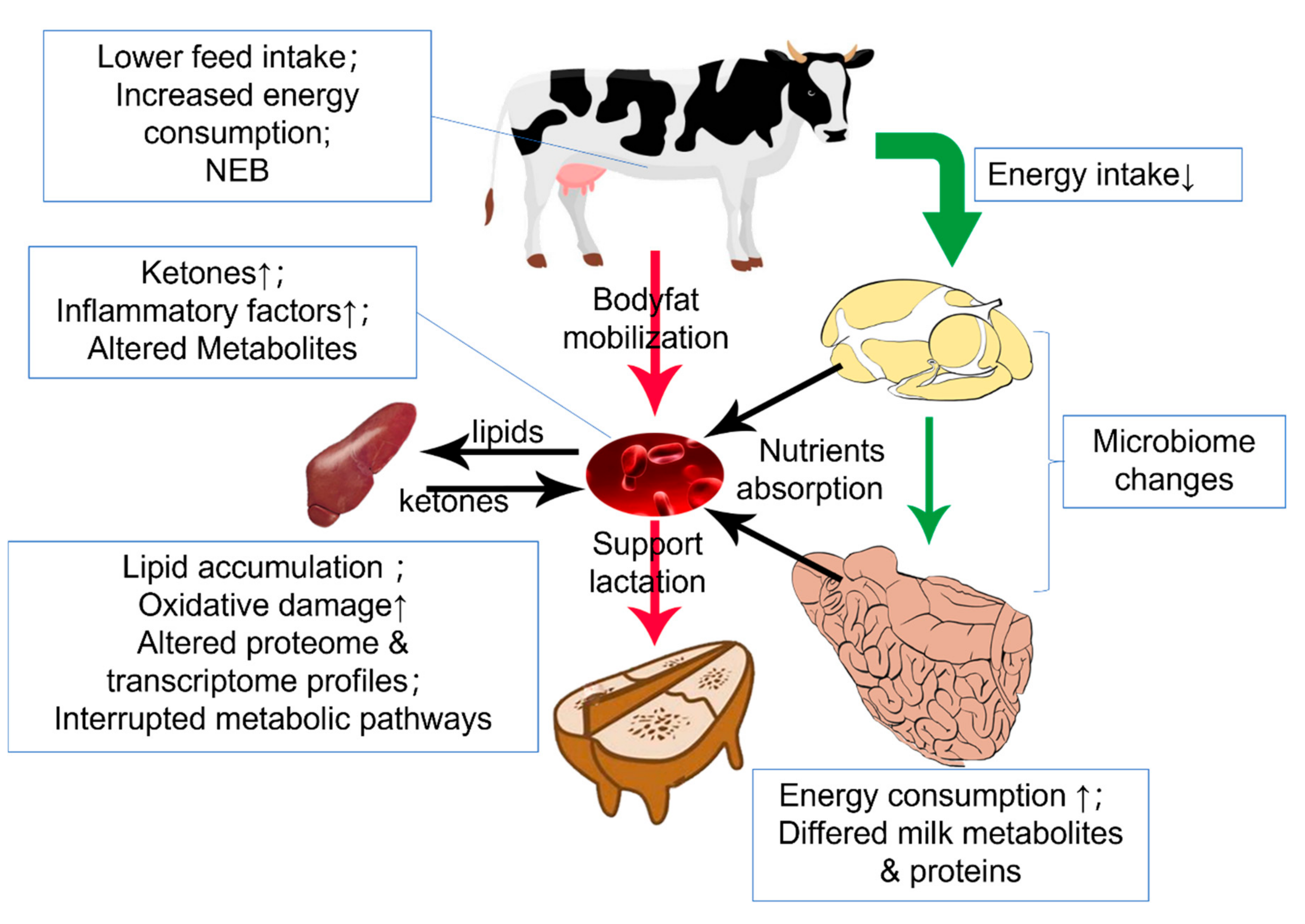
The unique rumen fermentation mechanism allows ruminants to consume plant fiber as a regular diet and degrades indigestible fiber by symbiotic microorganisms [115]. This digest mechanism forms the base of complete digestion and harvesting energy from the ingested feed, making the energy metabolism of ruminants rely on the rumen flora to maintain function. Rumen commensal microbiota reflects the dietary components, and the microbiota metabolites will determine the body’s energy metabolism. Once the fermentation feature is biased from the average level, cows will risk suffering diseases [116,117]. Apart from the rumen, the liver is also a critical organ in metabolic disease, especially in the periparturient period, where NEB usually occurs. Cows in NEB condition will mobilize their body fat into NEFA for β-oxidation in the liver. This process will elevate serum ketone concentrations and hepatic lipid accumulation, directionally leading to ketosis and fatty liver (Figure 2) [118]. At the same time, the different urea cycle and plasma AAs during the late gestation and early lactation may also involve in this process [119]. Researchers have clarified the general etiology of those diseases but still lack systematic knowledge on the molecular level [120].
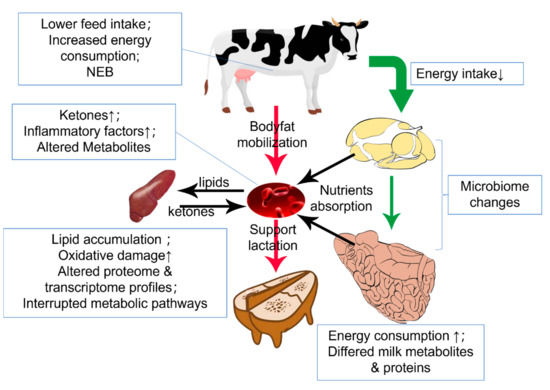
Figure 2.
Metabolic condition during the transition period. Down arrow(↓) means decrease and up arrow(↑) means increase. In the transition period, matter intake decreases while the initialed lactation demands more energy. Hence, body fat is mobilized and oxidated into ketone in the liver. The metabolic burden of liver induces oxidative stress and inflammation.
Without proper feeding and management, energy consumption could finally exhaust glycogen storage during the transition period. To compensate for the negative energy balance, cows mobilize their body fat, and serum adipokine levels are changed to adapt to this process [121]. Therefore, ketones, including NEFA and BHBA (beta-hydroxybutyrate acid), are produced as the intermediate metabolites of lipid metabolization and act as an alternative energy source of glucose. However, this process would interfere with the multiple metabolic pathways and burden the liver. Current research performed that the high concentration of BHB is related to altered serum metabolic profile, anaerobic rumen fermentation, lipid metabolism, and oxidative stress further proved this point [49,79]. Even ketosis and fatty liver are metabolic diseases closely related to lactation physiology, the incidence and severity of those diseases are still heritable [122,123]. Cows with ketosis and fatty liver show lower expression levels of genes related to glycolysis, gluconeogenesis, and tricarboxylic acid (TCA) cycle, especially oxidative phosphorylation, protein ubiquitination, and ubiquinone synthesis, but have predominant activation of selenoamino acid metabolism, ribosome and replication, and repair [124,125]. The expression level of FGF21 and APOBR in the liver are tightly related to NEB condition, and ketosis becomes a potential biomarker [123,126,127]. Furthermore, Soares et al. [128] found that PPARA and ACACA also have varying expression levels in different metabolic conditions, and there are 24 ketosis-related SNPs located in seven chromosomes. The pathogenesis of those diseases is similar to the non-alcoholic fatty liver disease (NAFLD) in humans. A study found that knock-downed Fasn, Thrsp, Pklr, and Chchd6 could alleviate steatosis and insulin resistance in mice with NAFLD by downregulating mitochondrial respiration, indicating mitochondria dysfunction might be the key to NAFLD [129]. While Xu et al. [130] found that the plasma concentrations of neurosecretory protein FGA, c1 inhibitor (C1INH), serum amyloid A(SAA), transthyretin (TTR), hepcidin, apoprotein C III (APoCIII), amyloid precursor protein (APP), cystatin C (CysC), osteopontin (OpN) are significantly decreased in cows with fatty liver, not only proved the mitochondria dysfunction of NAFLD. Not only BHBA, the golden standard for ketosis diagnosis, is one of the most altered components in the metabolome profile of ketosis cows [48], but also serum 4-hydroxy-6-methyl-2-pyrone and cinnamoyl glycine show their potential as ketosis biomarkers [131]. Ketosis and fatty liver are high susceptive diseases for transition dairy animals and similar to NAFLD, which have been widely researched by omics methods. With the references of human studies, multi-omics studies for these diseases may produce great progress.
Mastitis, inflammation of the mammary gland, is the most common and costly disease of dairy cattle, which could induce by breast injury, environmental microorganisms (Enterobacteriaceae, Streptococcus spp., Lactococcus spp., Prototheca spp., etc.), and contagious pathogens (Streptococcus lactis, Streptococcus agalactiae, and Staphylococcus aureus). There are two types of mastitis, clinical or subclinical, depending on milk SCC and properties [132,133,134]. Clinical mastitis has symptoms including redness and swelling udder, decreased milk yield and quality with an SCC higher than 500,000 cells/mL (or 400,000 cells/mL in Europe), while subclinical mastitis lacks diagnosis signs in the milk or udder [135]. Research using omics has made a great progress in screening diagnose indices for subclinical mastitis. Thomas et al. [34,57,59] integrated metabolomics, peptidomics, and proteomics to investigate Streptococcus uberis mastitis and found that top abundance proteins change from caseins, β-lactoglobulin, and α-lactalbumin to albumin, lactoferrin, and IgG after challenge, and acute-phase protein (APP), mammary-associated serum amyloid A 3 (M-SAA3), haptoglobin, and C-reaction protein could be potential infectious mastitis indicators. At the metabolites level, the carbohydrates and nucleic acid concentration in milk dropped significantly in the acute phase. In contrast, lipid metabolites and peptides levels, especially the bile acid-nuclear receptor FXR signaling pathway, have been significantly elevated in the Streptococcus uberis challenged cows. In addition, S. aureus and E. coli mastitis have different cytokines reactions and lead to varying severity [134], indicating bacterial mastitis might have different biomarkers corresponding to Gram-positive and -negative pathogens or different pathogenesis pathways, which still need revealing.
Subacute ruminal acidosis (SARA), a metabolic disease mainly caused by feeding rumen microbiota fermentable carbohydrates (like corn and wheat) highly and induced consequent accumulation of organic acids, affects cows behavior, rumen fermentation, and metabolism, leading to systemic symptoms [136]. Most research about SARA aimed to detect specific indices or the change of microorganisms but lacks screening and analysis of the pathogenesis with a broader scope [137]. In recent years, more cognition about the SARA mechanism has been brought by omics techniques. Single-omics studies revealed changes in composition, transcription, and metabolites of rumen microbiota but still could not explain the mechanism of SARA-induced production decrease [30,138]. An in vitro study of Murovec et al. [41] showed that the inhibited fermentation reactor has an altered abundance of acetate, caprylate, trimethylamine, thymine, pyruvate, alanine, xanthine, and succinate. Zhang [32] combined rumen microbiome, metabolomics, epithelial genomic, and milk microbiota analysis, and found that SARA could disturb normal ruminal symbiotic flora and biosynthesis, especially valine, leucine, and isoleucine synthesis pathways (p < 0.05), elevating the level of toxic and proinflammatory bacterial metabolites (p < 0.05), meanwhile, the expression level of proinflammatory cytokines (IL-1β, IL-2, IL-22, etc.) increased (p < 0.05) and anti-inflammatory(IL-6) ones decreased(p < 0.05), in addition, SARA will increase milk somatic cell count (SCC) with a dropped milk protein and lipid component (p < 0.05). Li et al. [28,29] found that young calves with high starch induced rumen acidosis performs a different rumen epithelial transcriptome and meta-trascriptome profile with correlated rumen microbiome, liver transcriptome pattern also involved in this process, the abundance of Olsenella, Desulfovibri, and Fusobacterium necrophorum increased significantly in the rumen fluid, and 95 genes in the liver changed with differed microbiome rRNA expression, among them, 77 genes are enriched in the pathways of membrane-bounded organelle and transferase activity. Six-hundred-and-seventy-two epithelial genes related to cell signaling and morphogenesis showed significantly altered expression, in which, 12 genes (COX5B, KRT78, KRT15, ATP5I, ATP5L, ATP5G2, COX8B, COX8A, UBC, DSP, ITM2B, and C10H15orf48) related to hydrogen ion transmembrane transport only exists in the acidosis group; other differentially expressed genes are mostly involved in cell division and growth, like membrane-bounded organelle, cytoplasm, cellular component organization or biogenesis.
Based on clinical studies, omics analysis could give researchers a systematic cognition of certain diseases and evaluate the mechanism of clinical manifestations in different organs and levels, enhancing the knowledge about how diseases affect dairy production for more specific diagnosis and therapy. Although diary research is relatively hard to have a large quantity of biology replication to ensure the statistically robust and overcome obstacles in distinguishing pathology and physiology process, omics-based research on dairy veterinary still has increased contributions to the knowledge about the pathogenesis and corresponding genetic information of diseases in recent years. Multi-omics could combine the data from multiple organs and levels to demonstrate a complete diagram of diseases, clarifying the response of the rumen–liver–mammary gland network to pathogenetic factors, providing more effective solutions for prevention, diagnosis, and treatments.
6. Conclusions and Prospects
This review summarized recent genomics, transcriptomics, proteomics, and metabolomics research about physiology, feeding and management, and veterinary in dairy animals. In researching dairy animals, we must remind ourselves that the physiology and etiology are complicated phenomena based on heritable and acquired traits in multiple organs and levels. Those factors may be relatively isolated and work as independent systems while they are connected and weaved a tangled web of regulation. Each factor that affects any organ level may have a systematic influence, some of them are physiological responses, and others may become a part of the pathological process. From feed intake to milk yield and components, only metabolites themselves need to pass through multiple organs, while more regulation processes are involved in transcription and expression levels, needless to say, the complicated rumen fermentation. In dairy research, investigations on production performance show the effectiveness of factors; studies on certain levels and indices indicate the mechanism of factors; omics studies would reveal the response of the organs to factors; multi-omics research would connect them, demonstrate how the factor, directly and indirectly, affects the body, and how the body reacts to the factor. Because the ruminal fermentation mechanism is still not completely discovered, non-targeted proteomics and metabolomics have provided researchers with a systematic scope on crucial genes, proteins, and metabolites that regulates the metabolic pathways and the mechanisms of breeding selection, nutritional management, and diseases prevention. However, there are still limits on the single omics to combine multiple levels and organs and build a panoramic view of certain factors’ impact on the dairy animals and tracking the mechanism of its impact on their performance. With the assistance of multi-omics methods, researchers could screen more genes related to heritable traits, clarifying mechanisms of lactation physiology as well as the pathology of metabolic diseases. These promising methods would draw all organs and levels together to construct a whole vision of dairy production and establish novel directions for dairy research.
Author Contributions
Y.Z. Drafted the manuscript; L.M. and D.B. revised this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by the National Key Research and Development Program of China (2018YFE0101400), the Key Research and Development Program of the Ningxia Hui Autonomous Region (2021BEF02018), the Scientific Research Project for Major Achievements of The Agricultural Science and Technology Innovation Program (ASTIP) (No.ASTIPIAS07, XTCX2016011-01), and Beijing Dairy Industry Innovation Team (BAIC06-2021).
Acknowledgments
The authors wish to acknowledge Stafford Vigors, University College Dublin, for his advice in drafting.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vailati-Riboni, M.; Palombo, V.; Loor, J.J. What Are Omics Sciences? In Periparturient Diseases of Dairy Cows, 1st ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–7. [Google Scholar]
- Anderson, N.L.; Anderson, N.G. Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis 1998, 19, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Shao, W.; Huang, Z.; Tang, H.; Zhang, J.; Ding, Z.; Huang, K. MOGONET integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nat. Commun. 2021, 12, 3445. [Google Scholar] [CrossRef]
- Dimitrakopoulos, L.; Prassas, I.; Diamandis, E.P.; Charames, G.S. Onco-proteogenomics: Multi-omics level data integration for accurate phenotype prediction. Crit. Rev. Clin. Lab. Sci. 2017, 54, 414–432. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Guan, L.L. Gut microbiome and omics: A new definition to ruminant production and health. Anim. Front. 2016, 6, 8–12. [Google Scholar] [CrossRef]
- Ametaj, B. A Systems Veterinary Approach in Understanding Transition Cow Diseases: Metabolomics. In Proceedings of the 4th International Symposium on Dairy Cow Nutrition and Milk Quality, Session 1, Advances in Fundamental Research, Beijing, China, 8–10 May 2015. [Google Scholar]
- Gu, F.F.; Liang, S.L.; Zhu, S.L.; Liu, J.X.; Sun, H.Z. Multi-omics revealed the effects of rumen-protected methionine on the nutrient profile of milk in dairy cows. Food Res. Int. 2021, 149, 110682. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Liu, J.X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef]
- Burd, N.A.; Hamer, H.M.; Pennings, B.; Pellikaan, W.F.; Senden, J.M.G.; Gijsen, A.P.; van Loon, L.J.C. Substantial Differences between Organ and Muscle Specific Tracer Incorporation Rates in a Lactating Dairy Cow. PLoS ONE 2013, 8, e68109. [Google Scholar] [CrossRef] [PubMed]
- Belanche, A.; Kingston-Smith, A.H.; Newbold, C.J. An Integrated Multi-Omics Approach Reveals the Effects of Supplementing Grass or Grass Hay with Vitamin E on the Rumen Microbiome and Its Function. Front. Microbiol. 2016, 7, 905. [Google Scholar] [CrossRef]
- Mamuad, L.L.; Lee, S.S.; Lee, S.S. Recent insight and future techniques to enhance rumen fermentation in dairy goats. Asian Austral. J. Anim. 2019, 32, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.E.; Wales, W.J.; de Haas, Y.; Veerkamp, R.F.; Hayes, B.J. Genomic selection for feed efficiency in dairy cattle. Animal 2014, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, A.; Abdollahi-Arpanahi, R.; Aguilar, I.; Peñagaricano, F. Whole Genome Mapping Reveals Novel Genes and Pathways Involved in Milk Production Under Heat Stress in US Holstein Cows. Front. Genet. 2019, 10, 928. [Google Scholar] [CrossRef]
- Tarekegn, G.M.; Strandberg, E.; Andonov, S.; Båge, R.; Ask-Gullstrand, P.; Rius-Vilarrasa, E.; Christensen, J.M.; Berglund, B. Single-step genome-wide association study uncovers known and novel candidate genomic regions for endocrine and classical fertility traits in Swedish Red and Holstein dairy cows. Livest. Sci. 2021, 253, 104731. [Google Scholar] [CrossRef]
- Feugang, J.M.; Rodriguez-Osorio, N.; Kaya, A.; Wang, H.; Page, G.; Ostermeier, G.C.; Topper, E.K.; Memili, E. Transcriptome analysis of bull spermatozoa: Implications for male fertility. Reprod. Biomed. Online 2010, 21, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, A.; Rincon, G.; Islas-Trejo, A.; Wickramasinghe, S.; Medrano, J.F. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mamm. Genome 2010, 21, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Akbar, H.; Bionaz, M.; Carlson, D.B.; Rodriguez-Zas, S.L.; Everts, R.E.; Lewin, H.A.; Drackley, J.K.; Loor, J.J. Feed restriction, but not l-carnitine infusion, alters the liver transcriptome by inhibiting sterol synthesis and mitochondrial oxidative phosphorylation and increasing gluconeogenesis in mid-lactation dairy cows. J. Dairy Sci. 2013, 96, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Fagerlind, M.; Stalhammar, H.; Olsson, B.; Klinga-Levan, K. Expression of miRNAs in Bull Spermatozoa Correlates with Fertility Rates. Reprod. Domest. Anim. 2015, 50, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liang, G.; Wang, B.; Sun, H.; Liu, J.; Guan, L.L. Systematic microRNAome profiling reveals the roles of microRNAs in milk protein metabolism and quality: Insights on low-quality forage utilization. Sci. Rep. 2016, 6, 21194. [Google Scholar] [CrossRef]
- Li, F.; Guan, L.L. Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle. Appl. Environ. Microb. 2017, 83, e00061-17. [Google Scholar] [CrossRef] [PubMed]
- Comtet-Marre, S.; Parisot, N.; Lepercq, P.; Chaucheyras-Durand, F.; Mosoni, P.; Peyretaillade, E.; Bayat, A.R.; Shingfield, K.J.; Peyret, P.; Forano, E. Metatranscriptomics Reveals the Active Bacterial and Eukaryotic Fibrolytic Communities in the Rumen of Dairy Cow Fed a Mixed Diet. Front. Microbiol. 2017, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Malmuthuge, N.; Steele, M.A.; Guan, L.L. Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol. Ecol. 2017, 94, fix179. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, D.; Wu, X.; Cai, J.; Liu, M.; Huang, X.; Wu, J.; Liu, J.; Guan, L. Effects of dietary physical or nutritional factors on morphology of rumen papillae and transcriptome changes in lactating dairy cows based on three different forage-based diets. BMC Genom. 2017, 18, 353. [Google Scholar] [CrossRef] [PubMed]
- Sollinger, A.; Tveit, A.T.; Poulsen, M.; Noel, S.J.; Bengtsson, M.; Bernhardt, J.; Hellwing, A.L.F.; Lund, P.; Riedel, K.; Schleper, C.; et al. Holistic Assessment of Rumen Microbiome Dynamics through Quantitative Metatranscriptomics Reveals Multifunctional Redundancy during Key Steps of Anaerobic Feed Degradation. Msystems 2018, 3, e00038-18. [Google Scholar] [CrossRef] [PubMed]
- Putz, E.J.; Putz, A.M.; Jeon, H.; Lippolis, J.D.; Ma, H.; Reinhardt, T.A.; Casas, E. MicroRNA profiles of dry secretions through the first three weeks of the dry period from Holstein cows. Sci. Rep. 2019, 9, 19658. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gelsinger, S.; Edwards, A.; Riehle, C.; Koch, D. Changes in meta-transcriptome of rumen epimural microbial community and liver transcriptome in young calves with feed induced acidosis. Sci. Rep. 2019, 9, 18967. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gelsinger, S.; Edwards, A.; Riehle, C.; Koch, D. Transcriptome analysis of rumen epithelium and meta-transcriptome analysis of rumen epimural microbial community in young calves with feed induced acidosis. Sci. Rep. 2019, 9, 4744. [Google Scholar] [CrossRef] [PubMed]
- Ogunade, I.; Pech-Cervantes, A.; Schweickart, H. Metatranscriptomic Analysis of Sub-Acute Ruminal Acidosis in Beef Cattle. Animals 2019, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Ametaj, B.N.; Zebeli, Q.; Saleem, F.; Psychogios, N.; Lewis, M.J.; Dunn, S.M.; Xia, J.G.; Wishart, D.S. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 2010, 6, 583–594. [Google Scholar] [CrossRef]
- Zhang, R. Omics Based Approaches to Assess the Effects of Subacute Ruminal Acidosis on Rumen Microbiota Metabolism and Epithelial Function in Dairy Cows; Nanjing Agricultural University: Nanjing, China, 2015. [Google Scholar]
- Forde, N.; O’Gorman, A.; Whelan, H.; Duffy, P.; O’Hara, L.; Kelly, A.K.; Havlicek, V.; Besenfelder, U.; Brennan, L.; Lonergan, P. Lactation-induced changes in metabolic status and follicular-fluid metabolomic profile in postpartum dairy cows. Reprod. Fertil. Dev. 2016, 28, 1882. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.C.; Mudaliar, M.; Tassi, R.; McNeilly, T.N.; Burchmore, R.; Burgess, K.; Herzyk, P.; Zadoks, R.N.; Eckersall, P.D. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 3. Untargeted metabolomics. Mol. Biosyst. 2016, 12, 2762–2769. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, B.; Newbold, C.J.; Wanchang, L.; Pauline, R.S.; Kingston-Smith, A.H. A Systems Biology Approach Reveals Differences in the Dynamics of Colonization and Degradation of Grass vs. Hay by Rumen Microbes with Minor Effects of Vitamin E Supplementation. Front. Microbiol. 2017, 8, 1456. [Google Scholar]
- Humer, E.; Khol-Parisini, A.; Metzler-Zebeli, B.U.; Gruber, L.; Zebeli, Q. Alterations of the Lipid Metabolome in Dairy Cows Experiencing Excessive Lipolysis Early Postpartum. PLoS ONE 2016, 11, e0158633. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, B.; Wang, J.; Liu, H.; Liu, J. Biomarker and pathway analyses of urine metabolomics in dairy cows when corn stover replaces alfalfa hay. J. Anim. Sci. Biotechnol. 2016, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Chen, Q.; Wang, Q.; White, R.R.; Liu, J.; Liu, H. Complementary transcriptomic and proteomic analyses reveal regulatory mechanisms of milk protein production in dairy cows consuming different forages. Sci. Rep. 2017, 7, 44234. [Google Scholar] [CrossRef] [PubMed]
- Artegoitia, V.M.; Foote, A.P.; Lewis, R.M.; Freetly, H.C. Rumen Fluid Metabolomics Analysis Associated with Feed Efficiency on Crossbred Steers. Sci. Rep. 2017, 7, 2864. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, H.; Fan, Y.; Guo, Y.; Zhang, G.; Nie, H.; Wang, F. Metabolomic profiling in umbilical venous plasma reveals effects of dietary rumen-protected arginine or N-carbamylglutamate supplementation in nutrient-restricted Hu sheep during pregnancy. Reprod. Domest. Anim. 2017, 52, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Murovec, B.; Makuc, D.; Repinc, S.K.; Prevorsek, Z.; Zavec, D.; Sket, R.; Pecnik, K.; Plavec, J.; Stres, B. 1H NMR metabolomics of microbial metabolites in the four MW agricultural biogas plant reactors: A case study of inhibition mirroring the acute rumen acidosis symptoms. J. Environ. Manag. 2018, 222, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Elolimy, A.; Alharthi, A.; Zeineldin, M.; Parys, C.; Helmbrecht, A.; Loor, J.J. Supply of Methionine During Late-Pregnancy Alters Fecal Microbiota and Metabolome in Neonatal Dairy Calves Without Changes in Daily Feed Intake. Front. Microbiol. 2019, 10, 2159. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Z.; Zhou, M.; Wang, O.; Chen, Y.H.; Liu, J.X.; Guan, L.L. Multi-omics reveals functional genomic and metabolic mechanisms of milk production and quality in dairy cows. Bioinformatics 2020, 36, 2530–2537. [Google Scholar] [CrossRef]
- Zhang, H.; Tong, J.J.; Zhang, Y.H.; Xiong, B.H.; Jiang, L.S. Metabolomics reveals potential biomarkers in the rumen fluid of dairy cows with different levels of milk production. Asian-Australas. J. Anim. Sci. 2020, 33, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Clemmons, B.A.; Powers, J.B.; Campagna, S.R.; Seay, T.B.; Embree, M.M.; Myer, P.R. Rumen fluid metabolomics of beef steers differing in feed efficiency. Metabolomics 2020, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Ogunade, I.M.; McCoun, M.; Idowu, M.D.; Peters, S.O. Comparative effects of two multispecies direct-fed microbial products on energy status, nutrient digestibility, and ruminal fermentation, bacterial community, and metabolome of beef steers. J. Anim. Sci. 2020, 98, skaa201. [Google Scholar] [CrossRef]
- Wang, C.; Liu, H.; Wang, C.; Liu, J.; Liu, H. Effects of Dietary Rumen-Protected Betaine on Lactation Performance and Serum Metabolites of Mid-lactation Holstein Dairy Cows. J. Agric. Food Chem. 2020, 68, 13154–13159. [Google Scholar] [CrossRef] [PubMed]
- Luke, T.D.W.; Pryce, J.E.; Elkins, A.C.; Wales, W.J.; Rochfort, S.J. Use of Large and Diverse Datasets for H-1 NMR Serum Metabolic Profiling of Early Lactation Dairy Cows. Metabolites 2020, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Lisuzzo, A.; Laghi, L.; Faillace, V.; Zhu, C.; Contiero, B.; Morgante, M.; Mazzotta, E.; Gianesella, M.; Fiore, E. Differences in the serum metabolome profile of dairy cows according to the BHB concentration revealed by proton nuclear magnetic resonance spectroscopy (1H-NMR). Sci. Rep. 2022, 12, 2525. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Z.Q.; Tu, Y.; Si, B.W.; Liu, Y.L.; Yang, L.; Luo, H.L.; Yu, Z. Untargeted metabolomic investigate milk and ruminal fluid of Holstein cows supplemented with Perilla frutescens leaf. Food Res. Int. 2021, 140, 110017. [Google Scholar] [CrossRef]
- Stergiadis, S.; Cabeza-Luna, I.; Mora-Ortiz, M.; Stewart, R.D.; Dewhurst, R.J.; Humphries, D.J.; Watson, M.; Roehe, R.; Auffret, M.D. Unravelling the Role of Rumen Microbial Communities, Genes, and Activities on Milk Fatty Acid Profile Using a Combination of Omics Approaches. Front. Microbiol. 2021, 11, 590441. [Google Scholar] [CrossRef]
- Wang, Y.; Nan, X.; Zhao, Y.; Wang, Y.; Jiang, L.; Xiong, B. Ruminal Degradation of Rumen-Protected Glucose Influences the Ruminal Microbiota and Metabolites in Early-Lactation Dairy Cows. Appl. Environ. Microbiol. 2021, 87, e01908-20. [Google Scholar] [CrossRef] [PubMed]
- Peddinti, D.; Nanduri, B.; Kaya, A.; Feugang, J.M.; Burgess, S.C.; Memili, E. Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst. Biol. 2008, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Ledgard, A.M.; Berg, M.C.; Mcmillan, W.H.; Smolenski, G.; Peterson, A.J. Effect of asynchronous transfer on bovine embryonic development and relationship with early cycle uterine proteome profiles. Reprod. Fertil. Dev. 2012, 24, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Saadi, H.A.S.; van Riemsdijk, E.; Dance, A.L.; Rajamanickam, G.D.; Kastelic, J.P.; Thundathil, J.C. Proteins associated with critical sperm functions and sperm head shape are differentially expressed in morphologically abnormal bovine sperm induced by scrotal insulation. J. Proteom. 2013, 82, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Shen, J.S.; Ren, D.X.; Liu, J.X. Effects of the processing methods of corn grain and soybean meal on milk protein expression profiles in dairy cows. Animal 2015, 9, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.C.; Mullen, W.; Tassi, R.; Ramirez-Torres, A.; Mudaliar, M.; McNeilly, T.N.; Zadoks, R.N.; Burchmore, R.; Eckersall, P.D. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 1. High abundance proteins, acute phase proteins and peptidomics. Mol. Biosyst. 2016, 12, 2735–2747. [Google Scholar] [CrossRef]
- Zachut, M.; Sood, P.; Livshitz, L.; Kra, G.; Levin, Y.; Moallem, U. Proteome dataset of pre-ovulatory follicular fluids from less fertile dairy cows. Data Brief 2016, 7, 1515–1518. [Google Scholar] [CrossRef][Green Version]
- Mudaliar, M.; Tassi, R.; Thomas, F.C.; McNeilly, T.N.; Weidt, S.K.; McLaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.D.; et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef] [PubMed]
- Snelling, T.J.; Wallace, R.J. The rumen microbial metaproteome as revealed by SDS-PAGE. BMC Microbiol. 2017, 17, 9. [Google Scholar] [CrossRef]
- Skibiel, A.L.; Zachut, M.; do Amaral, B.C.; Levin, Y.; Dahl, G.E. Liver proteomic analysis of postpartum Holstein cows exposed to heat stress or cooling conditions during the dry period. J. Dairy Sci. 2018, 101, 705–716. [Google Scholar] [CrossRef]
- Veshkini, A.; Bonnet, M.; Vogel, L.; Trscher, A.; Sauerwein, H. liver proteomics of dairy cows supplied with essential fatty acids and conjugated linoleic acids. In Proceedings of the 71st Annual Meeting of the European Federation of Animal Science, Porto, Portugal, 31 August–4 September 2020. [Google Scholar]
- Morgavi, D.P.; Kelly, W.J.; Janssen, P.H.; Attwood, G.T. Rumen microbial (meta)genomics and its application to ruminant production. Animal 2012, 7, 184–201. [Google Scholar] [CrossRef] [PubMed]
- Tapio, I.; Fischer, D.; Blasco, L.; Tapio, M.; Wallace, R.J.; Bayat, A.R.; Ventto, L.; Kahala, M.; Negussie, E.; Shingfield, K.J.; et al. Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. PLoS ONE 2017, 12, e0180260. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef] [PubMed]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Munoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges Through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Zhang, Y.; Choi, S.H.; Nogoy, K.M.; Liang, S. Review: The development of the gastrointestinal tract microbiota and intervention in neonatal ruminants. Animal 2021, 15, 100316. [Google Scholar] [CrossRef] [PubMed]
- Mavrommatis, A.; Skliros, D.; Flemetakis, E.; Tsiplakou, E. Changes in the Rumen Bacteriome Structure and Enzymatic Activities of Goats in Response to Dietary Supplementation with Schizochytrium spp. Microorganisms 2021, 9, 1528. [Google Scholar] [CrossRef]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Gruninger, R.J.; Ribeiro, G.O.; Cameron, A.; McAllister, T.A. Invited review: Application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal 2019, 13, 1843–1854. [Google Scholar] [CrossRef] [PubMed]
- Alves, K.L.G.C.; Granja-Salcedo, Y.T.; Messana, J.D.; de Souza, V.C.; Ganga, M.J.G.; Colovate, P.H.D.; Kishi, L.T.; Berchielli, T.T. Rumen bacterial diversity in relation to nitrogen retention in beef cattle. Anaerobe 2021, 67, 102316. [Google Scholar] [CrossRef] [PubMed]
- Guyader, J.; Eugene, M.; Noziere, P.; Morgavi, D.P.; Doreau, M.; Martin, C. Influence of rumen protozoa on methane emission in ruminants: A meta-analysis approach. Animal 2014, 8, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; de la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The Role of Ciliate Protozoa in the Rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.; Tamminga, S. Simulation of the Effects of Diet on the Contribution of Rumen Protozoa to Degradation of Fiber in the Rumen. Br. J. Nutr. 1995, 74, 617–634. [Google Scholar] [CrossRef] [PubMed]
- Belanche, A.; de la Fuente, G.; Newbold, C.J. Effect of progressive inoculation of fauna-free sheep with holotrich protozoa and total-fauna on rumen fermentation, microbial diversity and methane emissions. FEMS Microbiol. Ecol. 2015, 91, fiu026. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.W. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J. Anim. Sci. 1995, 2804–2819. [Google Scholar] [CrossRef]
- Drackley, J.K. Biology of dairy cows during the transition period: The final frontier? J. Dairy Sci. 1999, 82, 2259–2273. [Google Scholar] [CrossRef]
- Fiore, E.; Blasi, F.; Morgante, M.; Cossignani, L.; Badon, T.; Gianesella, M.; Contiero, B.; Berlanda, M. Changes of milk fatty acid composition in four lipid classes as biomarkers for the diagnosis of bovine ketosis using bioanalytical Thin Layer Chromatography and Gas Chromatographic techniques (TLC-GC). J. Pharmaceut. Biomed. 2020, 188, 113372. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; Lisuzzo, A.; Tessari, R.; Spissu, N.; Moscati, L.; Morgante, M.; Gianesella, M.; Badon, T.; Mazzotta, E.; Berlanda, M.; et al. Milk Fatty Acids Composition Changes According to β-Hydroxybutyrate Concentrations in Ewes during Early Lactation. Animals 2021, 11, 1371. [Google Scholar] [CrossRef] [PubMed]
- Lisuzzo, A.; Bonelli, F.; Sgorbini, M.; Nocera, I.; Cento, G.; Mazzotta, E.; Turini, L.; Martini, M.; Salari, F.; Morgante, M.; et al. Differences of the Plasma Total Lipid Fraction from Pre-Foaling to Post-Foaling Period in Donkeys. Animals 2022, 12, 304. [Google Scholar] [CrossRef]
- Bobe, G.; Young, J.W.; Beitz, D.C. Invited review: Pathology, etiology, prevention, and treatment of fatty liver in dairy cows. J. Dairy Sci. 2004, 87, 3105–3124. [Google Scholar] [CrossRef]
- Ametaj, B.; Ametaj, B.N.; Herold. Periparturient Diseases of Dairy Cows; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Gerspach, C.; Imhasly, S.; Gubler, M.; Naegeli, H.; Ruetten, M.; Laczko, E. Altered plasma lipidome profile of dairy cows with fatty liver disease. Res. Vet. Sci. 2017, 110, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Carlson, D.B.; McFadden, J.W.; D’Angelo, A.; Woodworth, J.C.; Drackley, J.K. Dietary L-carnitine affects periparturient nutrient metabolism and lactation in multiparous cows. J. Dairy Sci. 2007, 90, 3422–3441. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Zinder, M.; Donthu, R.; Larkin, D.M.; Kumar, C.G.; Rodriguez-Zas, S.L.; Andropolis, K.E.; Oliveira, R.; Lewin, H.A. Multisite haplotype on cattle chromosome 3 is associated with quantitative trait locus effects on lactation traits. Physiol. Genom. 2011, 43, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Colombani, C.; Legarra, A.; Fritz, S.; Guillaume, F.; Croiseau, P.; Ducrocq, V.; Robert-Granie, C. Application of Bayesian least absolute shrinkage and selection operator (LASSO) and BayesCpi methods for genomic selection in French Holstein and Montbeliarde breeds. J. Dairy Sci. 2013, 96, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.R.; Burke, C.R.; Crookenden, M.A.; Heiser, A.; Loor, J.L.; Meier, S.; Mitchell, M.D.; Phyn, C.V.C.; Turner, S.A. Fertility and the transition dairy cow. Reprod. Fertil. Dev. 2018, 30, 85. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, S. Assessing the Association of the Level of Milk Production with Reproductive Performance in Dairy Cattle. J. Reprod. Dev. 2010, 56, S1–S7. [Google Scholar] [CrossRef] [PubMed]
- Aranciaga, N.; Morton, J.D.; Berg, D.K.; Gathercole, J.L. Proteomics and metabolomics in cow fertility: A systematic review. Reproduction 2020, 160, 639–658. [Google Scholar] [CrossRef] [PubMed]
- Lima, F.S.; Silvestre, F.T.; Penagaricano, F.; Thatcher, W.W. Early genomic prediction of daughter pregnancy rate is associated with improved fertility outcomes in Holstein dairy cows. J. Dairy Sci. 2019, 102, 114. [Google Scholar]
- Pacheco, H.A.; Rezende, F.M.; Peñagaricano, F. Gene mapping and genomic prediction of bull fertility using sex chromosome markers. J. Dairy Sci. 2020, 103, 3304–3311. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Kwon, W.S.; Oh, S.A.; Pang, M.G. Fertility-Related Proteomic Profiling Bull Spermatozoa Separated by Percoll. J. Proteome Res. 2012, 11, 4162–4168. [Google Scholar] [CrossRef]
- Soggiu, A.; Piras, C.; Hussein, H.A.; De Canio, M.; Gaviraghi, A.; Galli, A.; Urbani, A.; Bonizzia, L.; Roncada, P. Unravelling the bull fertility proteome. Mol. Biosyst. 2013, 9, 1188–1195. [Google Scholar] [CrossRef]
- Govindaraju, A.; Dogan, S.; Rodriguez-Osorio, N.; Grant, K.; Kaya, A.; Memili, E. Delivering value from sperm proteomics for fertility. Cell Tissue Res. 2012, 349, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, A.; Uzun, A.; Robertson, L.; Atli, M.O.; Kaya, A.; Topper, E.; Crate, E.A.; Padbury, J.; Perkins, A.; Memili, E. Dynamics of microRNAs in bull spermatozoa. Reprod. Biol. Endocrin. 2012, 10, 82. [Google Scholar] [CrossRef]
- Gilbert, R.O. Symposium review: Mechanisms of disruption of fertility by infectious diseases of the reproductive tract. J. Dairy Sci. 2019, 102, 3754–3765. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, S.; Elia, G.; O’Boyle, P.; Dunn, M.; Morris, D. Composition of the bovine uterine proteome is associated with stage of cycle and concentration of systemic progesterone. Proteomics 2013, 13, 3333–3353. [Google Scholar] [CrossRef] [PubMed]
- Forde, N.; Simintiras, C.A.; Sturmey, R.; Mamo, S.; Kelly, A.K.; Spencer, T.E.; Bazer, F.W.; Lonergan, P. Amino Acids in the Uterine Luminal Fluid Reflects the Temporal Changes in Transporter Expression in the Endometrium and Conceptus during Early Pregnancy in Cattle. PLoS ONE 2014, 9, e100010. [Google Scholar] [CrossRef] [PubMed]
- Macklon, N.S.; van der Gaast, M.H.; Hamilton, A.; Fauser, B.C.J.M.; Gludice, L.C. The impact of ovarian stimulation with recombinant FSH in combination with GnRH antagonist on the endometrial transcriptome in the window of implantation. Reprod. Sci. 2008, 15, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, T.M.; Goncalves, R.F.; Melo, C.F.O.R.; de Oliveira, D.N.; Lima, E.D.; Visintin, J.A.; de Achilles, M.A.; Catharino, R.R. A Metabolomic Overview of Follicular Fluid in Cows. Front. Vet. Sci. 2018, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Oliveira, H.R.; Houlahan, K.; Fonseca, P.A.S.; Lam, S.; Butty, A.M.; Seymour, D.J.; Vargas, G.; Chud, T.C.S.; Silva, F.F.; et al. Genetic mechanisms underlying feed utilization and implementation of genomic selection for improved feed efficiency in dairy cattle. Can. J. Anim. Sci. 2020, 100, 587–604. [Google Scholar] [CrossRef]
- Grummer, R.R. Impact of Changes in Organic Nutrient Metabolism on Feeding the Transition Dairy-Cow. J. Anim. Sci. 1995, 73, 2820–2833. [Google Scholar] [CrossRef] [PubMed]
- Redfern, E.A.; Sinclair, L.A.; Robinson, P.A. Dairy cow health and management in the transition period: The need to understand the human dimension. Res. Vet. Sci. 2021, 137, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.R.; Rastani, R.R. Why Reevaluate Dry Period Length? J. Dairy Sci. 2004, 87, E77–E85. [Google Scholar] [CrossRef]
- Verweij, J.J.; Koets, A.P.; Eisenberg, S.W.F. Effect of continuous milking on immunoglobulin concentrations in bovine colostrum. Vet. Immunol. Immunopathol. 2014, 160, 225–229. [Google Scholar] [CrossRef]
- Sun, H.-Z.; Wang, D.-M.; Wang, B.; Wang, J.-K.; Liu, H.-Y.; Guan, L.L.; Liu, J.-X. Metabolomics of Four Biofluids from Dairy Cows: Potential Biomarkers for Milk Production and Quality. J. Proteome Res. 2015, 14, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Veshkini, A.; Hammon, H.M.; Sauerwein, H.; Tröscher, A.; Viala, D.; Delosière, M.; Ceciliani, F.; Déjean, S.; Bonnet, M. Longitudinal liver proteome profiling in dairy cows during the transition from gestation to lactation: Investigating metabolic adaptations and their interactions with fatty acids supplementation via repeated measurements ANOVA-simultaneous component analy. J. Proteom. 2022, 252, 104435. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Chang, G.T. A Literature Review on the Delactation Effect of Malt. Adv. Integr. Med. 2019, 6, S110–S111. [Google Scholar] [CrossRef]
- Lobos, N.E.; Wattiaux, M.A.; Broderick, G.A. Effect of rumen-protected lysine supplementation of diets based on corn protein fed to lactating dairy cows. J. Dairy Sci. 2021, 104, 6620–6632. [Google Scholar] [CrossRef] [PubMed]
- Mavrommatis, A.; Mitsiopoulou, C.; Christodoulou, C.; Kariampa, P.; Simoni, M.; Righi, F.; Tsiplakou, E. Effects of Supplementing Rumen-Protected Methionine and Lysine on Milk Performance and Oxidative Status of Dairy Ewes. Antioxidants 2021, 10, 654. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.Q.; Jiang, B.B.; Wang, H.R. Growth performance, meat quality and lipid metabolism in finishing lambs fed diets containing rumen-unprotected and rumen-protected betaine. Ital. J. Anim. Sci. 2021, 20, 2041–2050. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Ogunade, I.; Schweickart, H.; McCoun, M.; Cannon, K.; McManus, C. Integrating 16S rRNA Sequencing and LC-MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals 2019, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Mamuad, L.L.; Kim, S.H.; Biswas, A.A.; Yu, Z.; Cho, K.-K.; Kim, S.-B.; Lee, K.; Lee, S.S. Rumen fermentation and microbial community composition influenced by live Enterococcus faecium supplementation. AMB Express 2019, 9, 123. [Google Scholar] [CrossRef]
- Choudhury, P.K.; Sirohi, S.K.; Puniya, A.K.; Chaudhary, P.P. Harnessing the Diversity of Rumen Microbes using Molecular Approaches. In Livestock Green House Gases: Emission and Options for Mitigation; Satish Serial Publishing House: Delhi, India, 2012. [Google Scholar]
- Xu, Q.B.; Qiao, Q.Q.; Gao, Y.; Hou, J.X.; Hu, M.Y.; Du, Y.F.; Zhao, K.; Li, X. Gut Microbiota and Their Role in Health and Metabolic Disease of Dairy Cow. Front. Nutr. 2021, 8, 701511. [Google Scholar] [CrossRef]
- Krause, K.M.; Oetzel, G.R. Understanding and preventing subacute ruminal acidosis in dairy herds: A review. Anim. Feed. Sci. Technol. 2006, 126, 215–236. [Google Scholar] [CrossRef]
- Oetzel, G.R. Understanding the Impact of Subclinical Ketosis. In Proceedings of the 2012 Cornell Nutrition Conference for Feed Manufacturers; University of Wisconsin: Madison, WI, USA, 2012; pp. 15–26. [Google Scholar]
- Luo, Z.Z.; Shen, L.H.; Jiang, J.; Huang, Y.X.; Bai, L.P.; Yu, S.M.; Yao, X.P.; Ren, Z.H.; Yang, Y.X.; Cao, S.Z. Plasma metabolite changes in dairy cows during parturition identified using untargeted metabolomics. J. Dairy Sci. 2019, 102, 4639–4650. [Google Scholar] [CrossRef]
- Piazzolla, V.A.; Mangia, A. Noninvasive Diagnosis of NAFLD and NASH. Cells 2020, 9, 1005. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhu, Y.; Xiao, J.; Qian, B.; You, L.; Zhang, Y.; Yu, S.; Zong, X.; Cao, S. Serum adipokines play different roles in type I and II ketosis. Asian-Australas. J. Anim. Sci. 2019, 33, 1930. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Chang, Y.M.; Gianola, D.; Klemetsdal, G. Genetic analysis of clinical mastitis, milk fever, ketosis, and retained placenta in three lactations of Norwegian Red cows. J. Dairy Sci. 2005, 88, 3273–3281. [Google Scholar] [CrossRef]
- Tetens, J.; Heuer, C.; Heyer, I.; Klein, M.S.; Gronwald, W.; Junge, W.; Oefner, P.J.; Thaller, G.; Krattenmacher, N. Polymorphisms within the APOBR gene are highly associated with milk levels of prognostic ketosis biomarkers in dairy cows. Physiol. Genom. 2015, 47, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Loor, J.J.; Everts, R.E.; Bionaz, M.; Dann, H.M.; Morin, D.E.; Oliveira, R.; Rodriguez-Zas, S.L.; Drackley, J.K.; Lewin, H.A. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol. Genom. 2007, 32, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, K. Systems Physiology and Nutrition in Dairy Cattle: Applications of Omics and Bioinformatics to Better Understand the Hepatic Metabolomics and Transcriptomics Adaptations in Transition Dairy Cows; University of Illinois at Urbana-Champaign: Champaign, IL, USA, 2017. [Google Scholar]
- Akbar, H.; Batistel, F.; Drackley, J.K.; Loor, J.J. Alterations in Hepatic FGF21, Co-Regulated Genes, and Upstream Metabolic Genes in Response to Nutrition, Ketosis and Inflammation in Peripartal Holstein Cows. PLoS ONE 2015, 10, e0139963. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Sun, L.-W.; Xia, C.; Zhang, H.-Y.; Zheng, J.-S.; Wang, J.-S. 1H-Nuclear Magnetic Resonance-Based Plasma Metabolic Profiling of Dairy Cows with Fatty Liver. Asian-Australas. J. Anim. Sci. 2015, 29, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.A.N.; Vargas, G.; Muniz, M.M.M.; Soares, M.A.M.; Cánovas, A.; Schenkel, F.; Squires, E.J. Differential gene expression in dairy cows under negative energy balance and ketosis: A systematic review and meta-analysis. J. Dairy Sci. 2021, 104, 602–615. [Google Scholar] [CrossRef]
- Chella Krishnan, K.; Kurt, Z.; Barrere-Cain, R.; Sabir, S.; Das, A.; Floyd, R.; Vergnes, L.; Zhao, Y.; Che, N.; Charugundla, S.; et al. Integration of Multi-omics Data from Mouse Diversity Panel Highlights Mitochondrial Dysfunction in Non-alcoholic Fatty Liver Disease. Cell Syst. 2018, 6, 103–115.e107. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhu, K.-L.; Chen, Y.-Y.; Yang, W.; Xia, C.; Zhang, H.-Y.; Wu, L.; Shen, T.-Y.; Yu, H.; Xu, Q.-S.; et al. Isolation Identification and Bioinformatics of Differences Protein in Plasma of Cows Suffer from Fatty Liver with SELDI-TOF-MS Techniques. Sci. Agric. Sin. 2016, 49, 1585–1598. [Google Scholar]
- Wu, Z.L.; Chen, S.Y.; Hu, S.Q.; Jia, X.B.; Wang, J.; Lai, S.J. Metabolomic and Proteomic Profiles Associated With Ketosis in Dairy Cows. Front. Genet. 2020, 11, 551587. [Google Scholar] [CrossRef]
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.C. Common Mammary Pathogens and Factors in Infection and Mastitis. J. Dairy Sci. 1979, 62, 128–134. [Google Scholar] [CrossRef]
- Wellnitz, O.; Bruckmaier, R.M. The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 2012, 192, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed]
- Giger-Reverdin, S. Recent advances in the understanding of subacute ruminal acidosis (SARA) in goats, with focus on the link to feeding behaviour. Small Rumin. Res. 2018, 163, 24–28. [Google Scholar] [CrossRef]
- Dill-McFarland, K.A.; Breaker, J.D.; Suen, G. Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. Sci. Rep. 2017, 7, 40864. [Google Scholar] [CrossRef]
- Qi, W.P.; Mu, Y.Y.; Zhang, T.; Zhang, J.Y.; Mao, S.Y. Plasma biochemical indexes and metabolomics profile changes of dairy cows with subacute ruminal acidosis. Acta Pratacult. Sin. 2021, 30, 141–150. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).