The Role of the Metabolome and Non-Coding RNA on Pheochromocytomas and Paragangliomas: An Update
Abstract
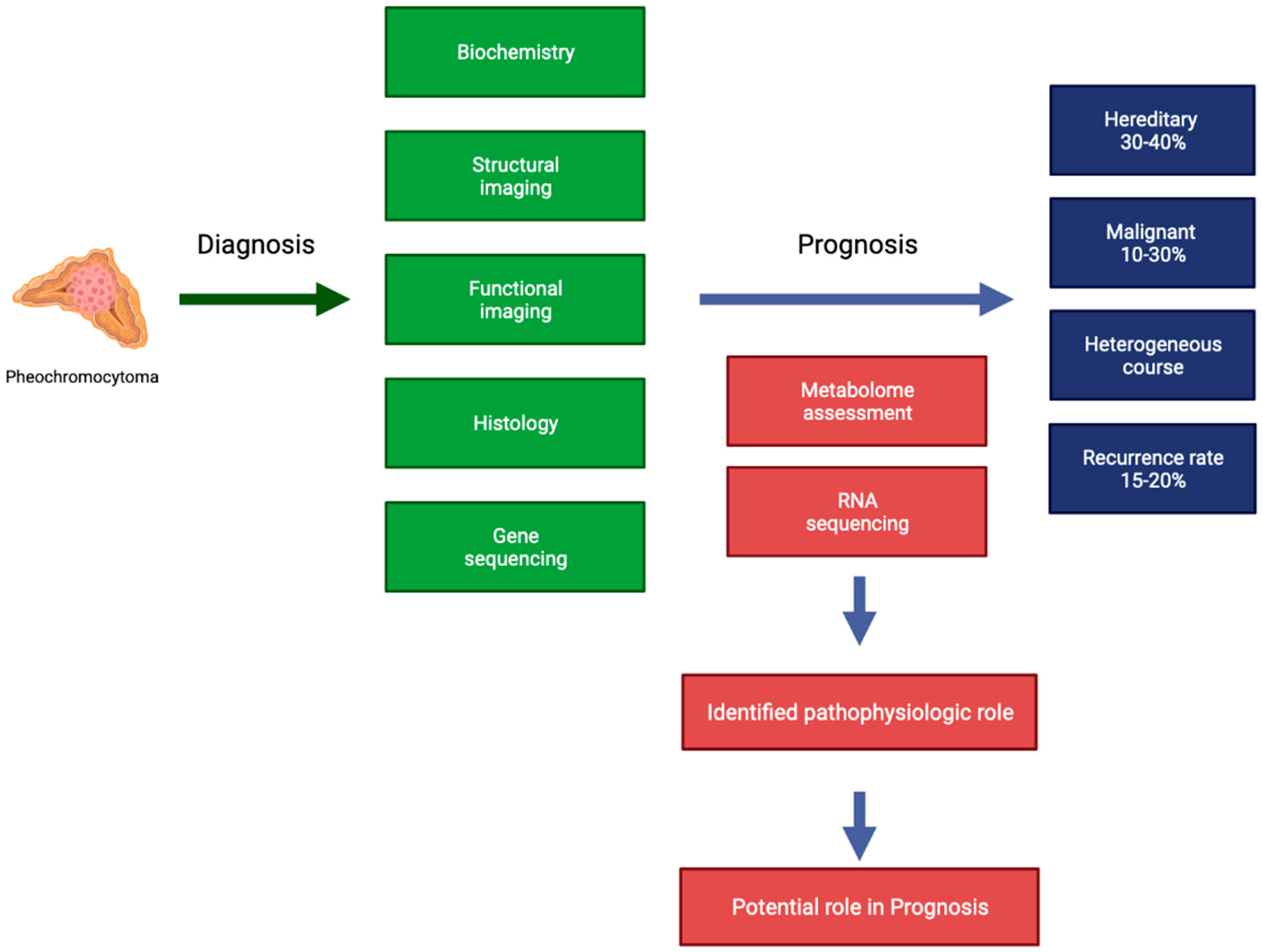
:1. Introduction
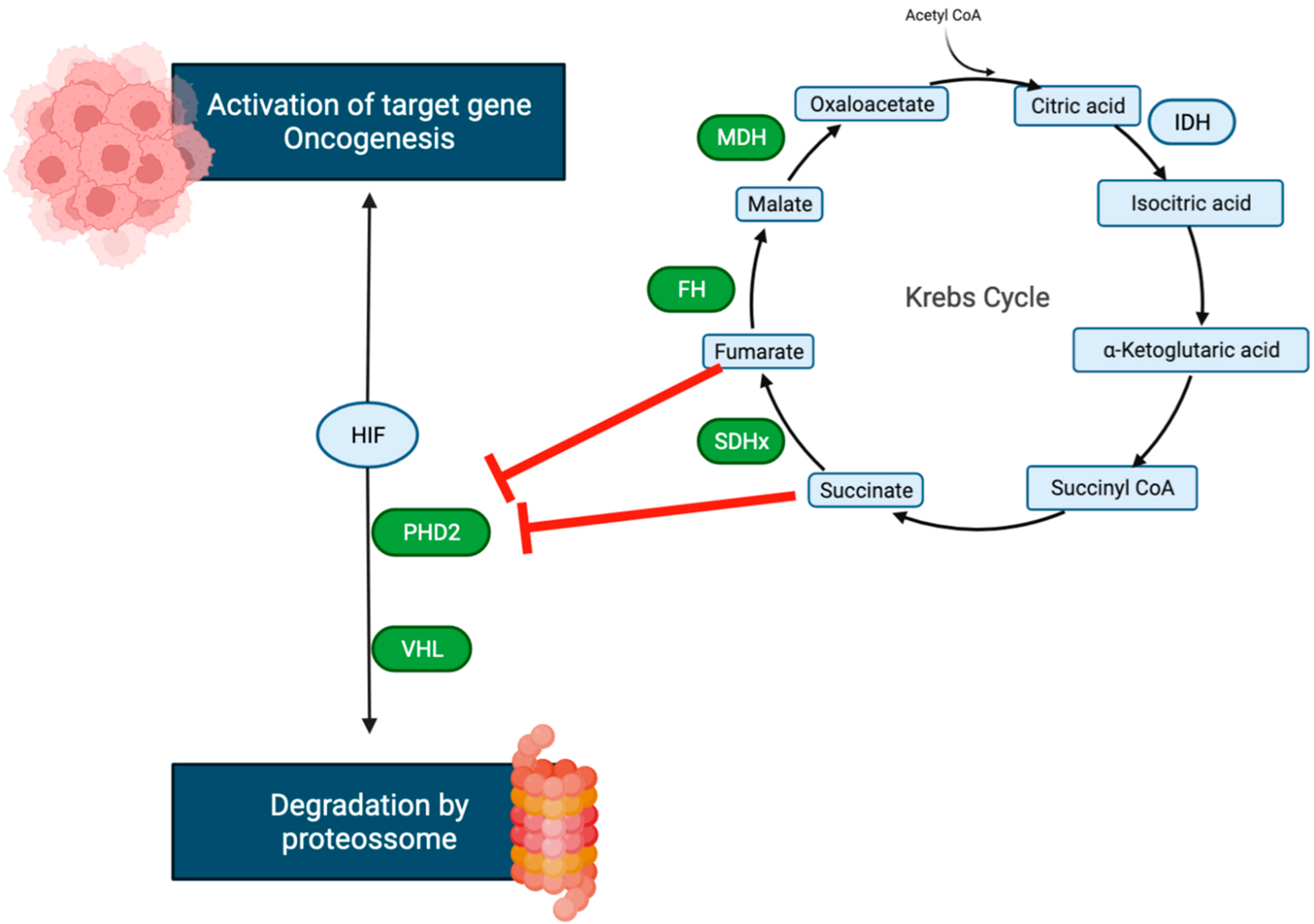
2. Metabolome
3. RNA
3.1. MicroRNA (miRNA)
3.2. Long Non-Coding RNAs (lncRNA)
3.3. Circular RNA (circRNA)
4. Conclusions
Funding
Conflicts of Interest
References
- Pappachan, M.J.; Raskauskiene, D.; Sriraman, R.; Edavalath, M.; Hanna, W.F. Diagnosis and management of phe-ochromocytoma: A practical guide to clinicians. Curr. Hypertens. Rep. 2014, 16, 442. [Google Scholar] [CrossRef]
- Turchini, J.; Cheung, V.K.Y.; Tischler, A.S.; De Krijger, R.R.; Gill, A.J. Pathology and genetics of phaeochromocytoma and paraganglioma. Histopathology 2017, 72, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roman-Gonzalez, A.; Jimenez, C. Malignant pheochromocytoma-paraganglioma: Pathogenesis, TNM staging, and current clinical trials. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Corssmit, E.P.; Snel, M.; Kapiteijn, E. Malignant pheochromocytoma and paraganglioma: Management options. Curr. Opin. Oncol. 2020, 32, 20–26. [Google Scholar] [CrossRef]
- Wachtel, H.; Hutchens, T.; Baraban, E.; Schwartz, L.E.; Montone, K.; Baloch, Z.; LiVolsi, V.; Krumeich, L.; Fraker, D.L.; Nathanson, K.L.; et al. Predicting metastatic potential inpheochromo-cytoma and paraganglioma: A comparison of PASS and GAPPscoring systems. J. Clin. Endocrinol. Metab. 2020, 105, e4661–e4670. [Google Scholar] [CrossRef] [PubMed]
- Stenman, A.; Zedenius, J.; Juhlin, C.C. The value of histological algorithms to predict the malignancy potential of pheo-chromocytomas and abdominal paragangliomas—A meta-analysis and systematic review of the literature. Cancers 2019, 11, 225. [Google Scholar] [CrossRef] [Green Version]
- Antonio, K.; Valdez, M.M.N.; Mercado-Asis, L.; Taïeb, D.; Pacak, K. Pheochromocytoma/paraganglioma: Recent updates in genetics, biochemistry, immunohistochemistry, metabolomics, imaging and therapeutic options. Gland. Surg. 2020, 9, 105–123. [Google Scholar] [CrossRef]
- Dwight, T.; Kim, E.; Novos, T.; Clifton-Bligh, R.J. Metabolomics in the Diagnosis of Pheochromocytoma and Paraganglioma. Horm. Metab. Res. 2019, 51, 443–450. [Google Scholar] [CrossRef]
- Li, Z.; Yu, X.; Shen, J. The role of miRNAs in the pheochromocytomas. Tumor Biol. 2016, 37, 4235–4239. [Google Scholar] [CrossRef]
- Li, Z.; Yu, X.; Shen, J.; Liu, Y.; Chan, M.T.; Wu, W.K. MicroRNA dysregulation in rhabdomyosarcoma: A new player enters the game. Cell Prolif. 2015, 48, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, X.; Shen, J.; Wu, W.K.K.; Chan, M. MicroRNA expression and its clinical implications in Ewing’s sarcoma. Cell Prolif. 2015, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, Z. MicroRNAs regulate vascular smooth muscle cell functionsin atherosclerosis (review). Int. J. Mol. Med. 2014, 34, 923–933. [Google Scholar] [CrossRef] [Green Version]
- Jochmanová, I.; Yang, C.; Zhuang, Z.; Pacak, K. Hypoxia-Inducible Factor Signaling in Pheochromocytoma: Turning the Rudder in the Right Direction. JNCI J. Natl. Cancer Inst. 2013, 105, 1270–1283. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.; Gopalan, V.; Smith, R.A.; Lam, A.K.-Y. Updates on the genetics and the clinical impacts on phaeochromocytoma and paraganglioma in the new era. Crit. Rev. Oncol. 2016, 100, 190–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imperiale, A.; Moussallieh, F.-M.; Sebag, F.; Brunaud, L.; Barlier, A.; Elbayed, K.; Bachellier, P.; Goichot, B.; Pacak, K.; Namer, I.-J.; et al. A New Specific Succinate-Glutamate Metabolomic Hallmark in Sdhx-Related Paragangliomas. PLoS ONE 2013, 8, e80539. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.U.; Engelke, U.F.; Rodenburg, R.J.; Weavers, R.A.; Pacak, K.; Eisenhofer, G.; Qin, N.; Kusters, B.; Goudswaard, A.G.; Lenders, J.W.M.; et al. Genotype-specific abnormalities in mitochondrial function associate with distinct profiles of energy metabolism and catecholamine content in pheochromocytoma and paraganglioma. Clin. Cancer Res. 2013, 19, 3787–3795. [Google Scholar] [CrossRef] [Green Version]
- Imperial, A.; Moussallieh, F.M.; Roche, P.; Battini, S.; Cicek, A.E.; Sebag, F.; Brunaud, L.; Barlier, A.; Elbayed, K.; Loundou, A.; et al. Metabolome profiling by HRMAS NMR spectroscopy of pheochromocytomas and paragangliomas detects SDH deficiency: Clinical and pathophysiological implications. Neoplasia 2015, 17, 55–65. [Google Scholar] [CrossRef]
- Lendvai, N.; Pawlosky, R.; Bullova, P.; Eisenhofer, G.; Patocs, A.; Veech, R.L.; Pacak, K. Succinate-to-Fumarate Ratio as a New Metabolic Marker to Detect the Presence of SDHB/D-related Paraganglioma: Initial Experimental and Ex Vivo Findings. Endocrinology 2014, 155, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Richter, S.; Peitzsch, M.; Rapizzi, E.; Lenders, J.W.; Qin, N.; De Cubas, A.A.; Schiavi, F.; Rao, J.U.; Beuschlein, F.; Quinkler, M.; et al. Krebs Cycle Metabolite Profiling for Identification and Stratification of Pheochromocytomas/Paragangliomas due to Succinate Dehydrogenase Deficiency. J. Clin. Endocrinol. Metab. 2014, 99, 3903–3911. [Google Scholar] [CrossRef] [Green Version]
- Letouzé, E.; Martinelli, C.; Loriot, C.; Burnichon, N.; Abermil, N.; Ottolenghi, C.; Janin, M.; Menara, M.; Nguyen, A.T.; Benit, P.; et al. SDH Mutations Establish a Hypermethylator Phenotype in Paraganglioma. Cancer Cell 2013, 23, 739–752. [Google Scholar] [CrossRef] [Green Version]
- Castro-Vega, L.J.; Buffet, A.; De Cubas, A.A.; Cascón, A.; Menara, M.; Khalifa, E.; Amar, L.; Azriel, S.; Bourdeau, I.; Chabre, O.; et al. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum. Mol. Genet. 2013, 23, 2440–2446. [Google Scholar] [CrossRef] [Green Version]
- Kruspig, B.; Zhivotovsky, B.; Gogvadze, V. Mitochondrial substrates in cancer: Drivers or passengers? Mitochondrion 2014, 19, 8–19. [Google Scholar] [CrossRef]
- Mercado-Asis, L.B.; Wolf, K.I.; Jochmanova, I.; Taïeb, D. Pheochromocytoma: A Genetic And Diagnostic Update. Endocr. Pract. 2018, 24, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Turai, P.; Nyírő, G.; Butz, H.; Patócs, A.; Igaz, P. MicroRNAs, Long Non-Coding RNAs, and Circular RNAs: Potential Biomarkers and Therapeutic Targets in Pheochromocytoma/Paraganglioma. Cancers 2021, 13, 1522. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A Class of Long Noncoding RNAs Synthesized at Enhancers: Figure 1. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [Green Version]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2014, 4, 5150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids 2014, 3, e188. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Tömböl, Z.; Éder, K.; Kovács, A.; Szabó, P.M.; Kulka, J.; Likó, I.; Zalatnai, A.; Rácz, G.; Tóth, M.; Patócs, A.; et al. MicroRNA expression profiling in benign (sporadic and hereditary) and recurring adrenal pheochromocytomas. Mod. Pathol. 2010, 23, 1583–1595. [Google Scholar] [CrossRef] [Green Version]
- Gimm, O.; DeMicco, C.; Perren, A.; Giammarile, F.; Walz, M.K.; Brunaud, L. Malignant pheochromocytomas and paragangliomas: A diagnostic challenge. Langenbecks Arch. Surg. 2012, 397, 155–177. [Google Scholar] [CrossRef] [PubMed]
- Chrisoulidou, A.; Kaltsas, G.; Ilias, I.; Grossman, A.B. The diagnosis and management of malignant phaeochromocytoma and paraganglioma. Endocr. Relat. Cancer 2007, 14, 569–585. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Rochow, G.Y.; Jackson, N.E.; Conaglen, J.V.; Whittle, D.E.; Kunnimalaiyaan, M.; Chen, H.; Westin, G.; Sandgren, J.; Stålberg, P.; Khanafshar, E.; et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocr. Relat. Cancer 2010, 17, 835–846. [Google Scholar] [CrossRef]
- Calsina, B.; Castro-Vega, L.J.; Torres-Pérez, R.; Inglada-Pérez, L.; Currás-Freixes, M.; Roldán-Romero, J.M.; Mancikova, V.; Letón, R.; Remacha, L.; Santos, M.; et al. Integrative multi-omics analysis identifies a prognostic miRNA signature and a targetable miR-21-3p/TSC2/ mTOR axis in metastatic pheochromocytoma/ paraganglioma. Theranostics 2019, 9, 4946–4958. [Google Scholar] [CrossRef] [PubMed]
- de Cubas, A.A.; Leandro-Garcia, L.J.; Schiavi, F.; Mancikova, V.; Comino-Mendez, I.; Inglada-Perez, L.; Perez-Martinez, M.; Ibarz, N.; Ximenez-Embun, P.; Lopez-Jimenez, E.; et al. Integrative analysis of miRNA and mRNA expression profiles in pheochromocytoma and paraganglioma identifies genotype-specific markers and potentially regulated pathways. Endocr. Relat. Cancer 2013, 20, 477–493. [Google Scholar] [CrossRef] [Green Version]
- Patterson, E.; Webb, R.; Weisbrod, A.; Bian, B.; He, M.; Zhang, L.; Holloway, A.K.; Krishna, R.; Nilubol, N.; Pacak, K.; et al. The microRNA expression changes associated with malignancy and SDHB mutation in pheochromocytoma. Endocr. Relat. Cancer 2012, 19, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, Y.; Zhong, Y.; Wang, Y.; Peng, M. Comprehensive Analysis of Aberrantly Expressed Competitive Endogenous RNA Network and Identification of Prognostic Biomarkers in Pheochromocytoma and Paraganglioma. OncoTargets Ther. 2020, 13, 11377–11395. [Google Scholar] [CrossRef]
- Liang, Y.-C.; Wu, Y.-P.; Chen, D.-N.; Chen, S.-H.; Li, X.-D.; Sun, X.-L.; Wei, Y.; Ning, X.; Xue, X.-Y. Building a Competing Endogenous RNA Network to Find Potential Long Non-Coding RNA Biomarkers for Pheochromocytoma. Cell. Physiol. Biochem. 2018, 51, 2916–2924. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosal, S.; Das, S.; Pang, Y.; Gonzales, M.K.; Huynh, T.; Yang, Y.; Taieb, D.; Crona, J.; Shankavaram, U.T.; Pacak, K. Long intergenic noncoding RNA profiles of pheochromocytoma and paraganglioma: A novel prognostic biomarker. Int. J. Cancer 2020, 146, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Job, S.; Georges, A.; Burnichon, N.; Buffet, A.; Amar, L.; Bertherat, J.; Bouatia-Naji, N.; De Reyniès, A.; Drui, D.; Lussey-Lepoutre, C.; et al. Transcriptome Analysis of lncRNAs in Pheochromocytomas and Paragangliomas. J. Clin. Endocrinol. Metab. 2020, 105, 898–907. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell. Mol. Life Sci. 2017, 75, 1071–1098. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.; Li, M.; Xing, C.; Chen, D.; Wang, C.; Xiao, Q.; Zhang, L.; Pang, Y.; Wang, Y.; Zu, X.; et al. A Comprehensive Analysis Identified the Key Differentially Expressed Circular Ribonucleic Acids and Methylation-Related Function in Pheochromocytomas and Paragangliomas. Front. Genet. 2020, 11, 1–12. [Google Scholar] [CrossRef]

| Micro RNA | Expression Alteration and Possible Role |
|---|---|
| miRNA 15 [36] and miRNA 16 [36] | Underexpression in metastatic pheochromocytoma Tumor suppressor–promotes cell death |
| miRNA 21-3p [37] | Associated with higher sensitivity to rapamycin |
| miRNA 96 [38] | Overexpression in SDHB type |
| miRNA 101 [39] | Overexpression in SDHB type and metastatic PPGL |
| miRNA 133 [33,38] | Overexpression in VHL type |
| miRNA 137 [33] | Overexpression in most PPGL |
| miRNA 139-3p [33,38] | Overexpression in VHL type |
| miRNA 148-3p [40] | Associated with good prognosis |
| miRNA 183 [39] | Overexpression in SDHB type |
| miRNA 193 and miRNA 195 [41] | Downregulated in PPGL |
| miRNA 210 [9] | Overexpression in SDH and VHL types Possibly associated with more aggressive disease |
| miRNA 338-3p [40] | Associated with good prognosis |
| miRNA 375 [41] | Overexpression in most PPGL |
| miRNA 382 [38] | Overexpression in VHL, SDHB, SDHD, and RET mutated PPGL |
| miRNA 483-5p [36,39] | Overexpression in metastatic PPGL Associated with worse survival rate |
| miRNA 488 [38] | Overexpression in MEN2 associated PPGL |
| miRNA 497 and miRNA 508 [41] | Downregulated in PPGL |
| miRNA 541 and miRNA 765 [33] | Overexpression in VHL type |
| miRNA 885 [33] | Overexpression in MEN2 associated PPGL |
| miRNA 1225-3p [33] | Overexpression in recurrent PPGL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouça, B.; Bogalho, P.; Rizzo, M.; Silva-Nunes, J. The Role of the Metabolome and Non-Coding RNA on Pheochromocytomas and Paragangliomas: An Update. Metabolites 2022, 12, 131. https://doi.org/10.3390/metabo12020131
Bouça B, Bogalho P, Rizzo M, Silva-Nunes J. The Role of the Metabolome and Non-Coding RNA on Pheochromocytomas and Paragangliomas: An Update. Metabolites. 2022; 12(2):131. https://doi.org/10.3390/metabo12020131
Chicago/Turabian StyleBouça, Bruno, Paula Bogalho, Manfredi Rizzo, and José Silva-Nunes. 2022. "The Role of the Metabolome and Non-Coding RNA on Pheochromocytomas and Paragangliomas: An Update" Metabolites 12, no. 2: 131. https://doi.org/10.3390/metabo12020131
APA StyleBouça, B., Bogalho, P., Rizzo, M., & Silva-Nunes, J. (2022). The Role of the Metabolome and Non-Coding RNA on Pheochromocytomas and Paragangliomas: An Update. Metabolites, 12(2), 131. https://doi.org/10.3390/metabo12020131








