Bioinformatics Modelling and Metabolic Engineering of the Branched Chain Amino Acid Pathway for Specific Production of Mycosubtilin Isoforms in Bacillus subtilis
Abstract
1. Introduction
2. Results
2.1. Purification and Determination of the Antifungal Activity of Mycosubtilin Isoforms
2.2. Effect of Amino Acid Feeding during B. subtilis ATCC 6633 Cultures
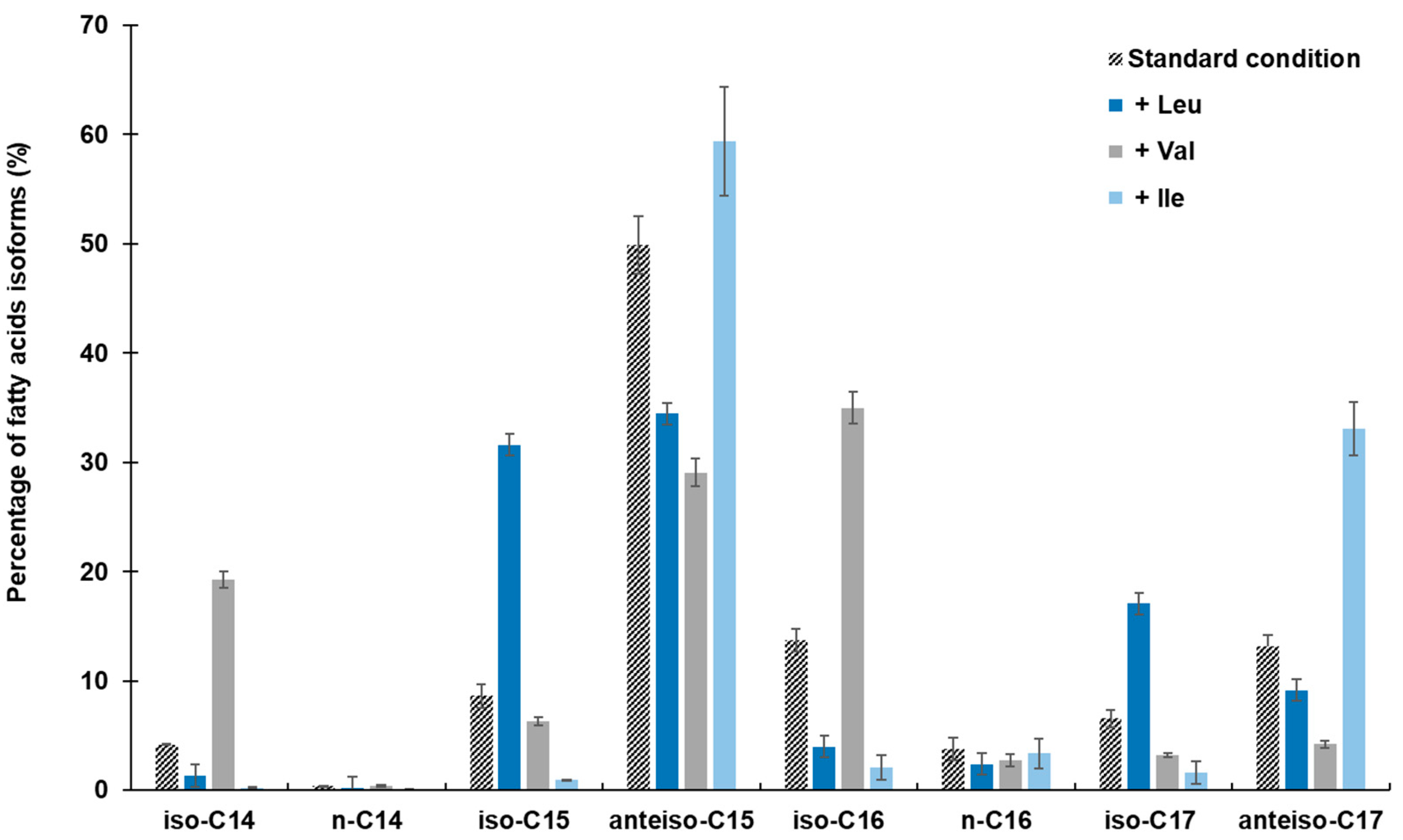
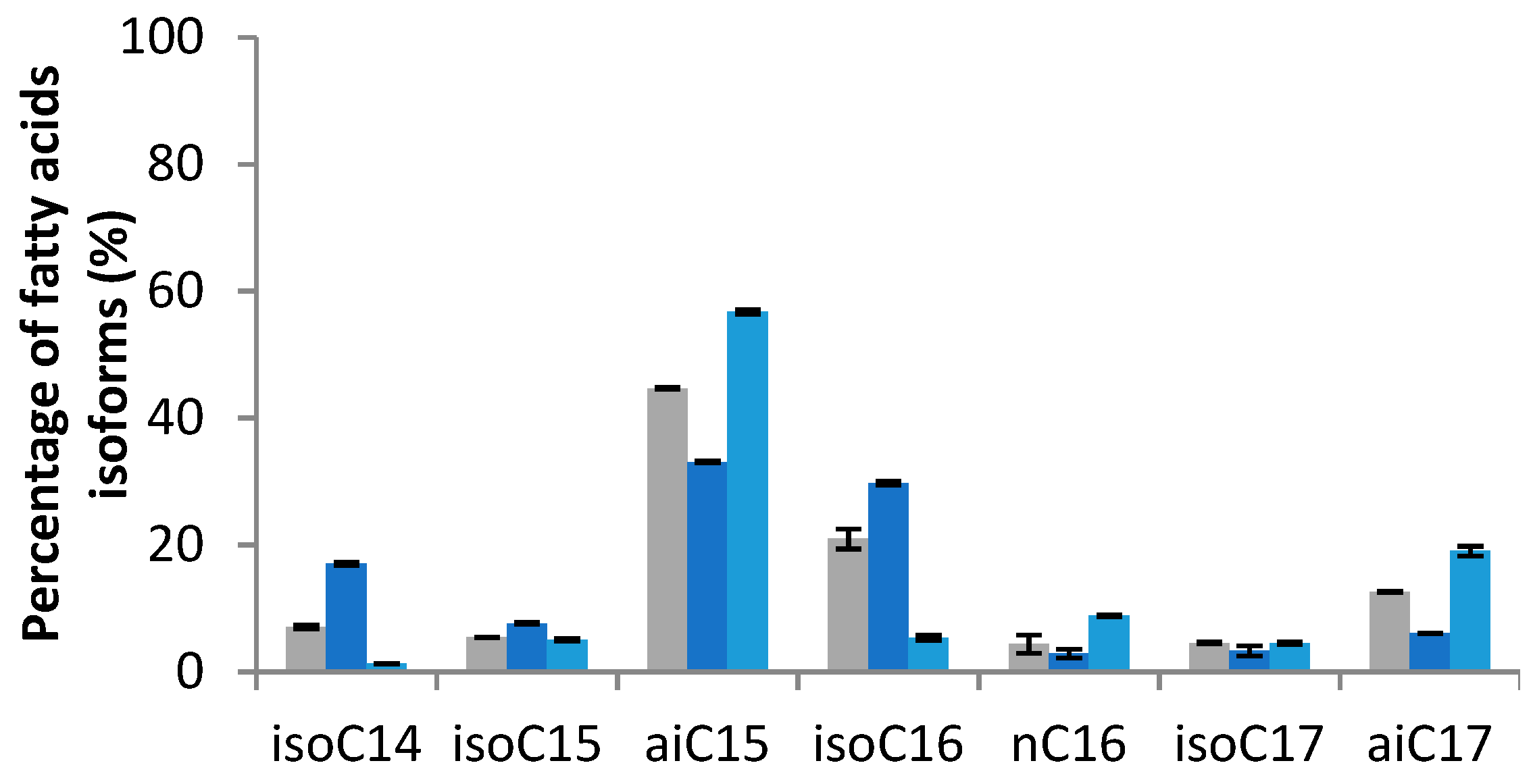
2.2.1. Effect on Cellular Fatty Acids Pattern
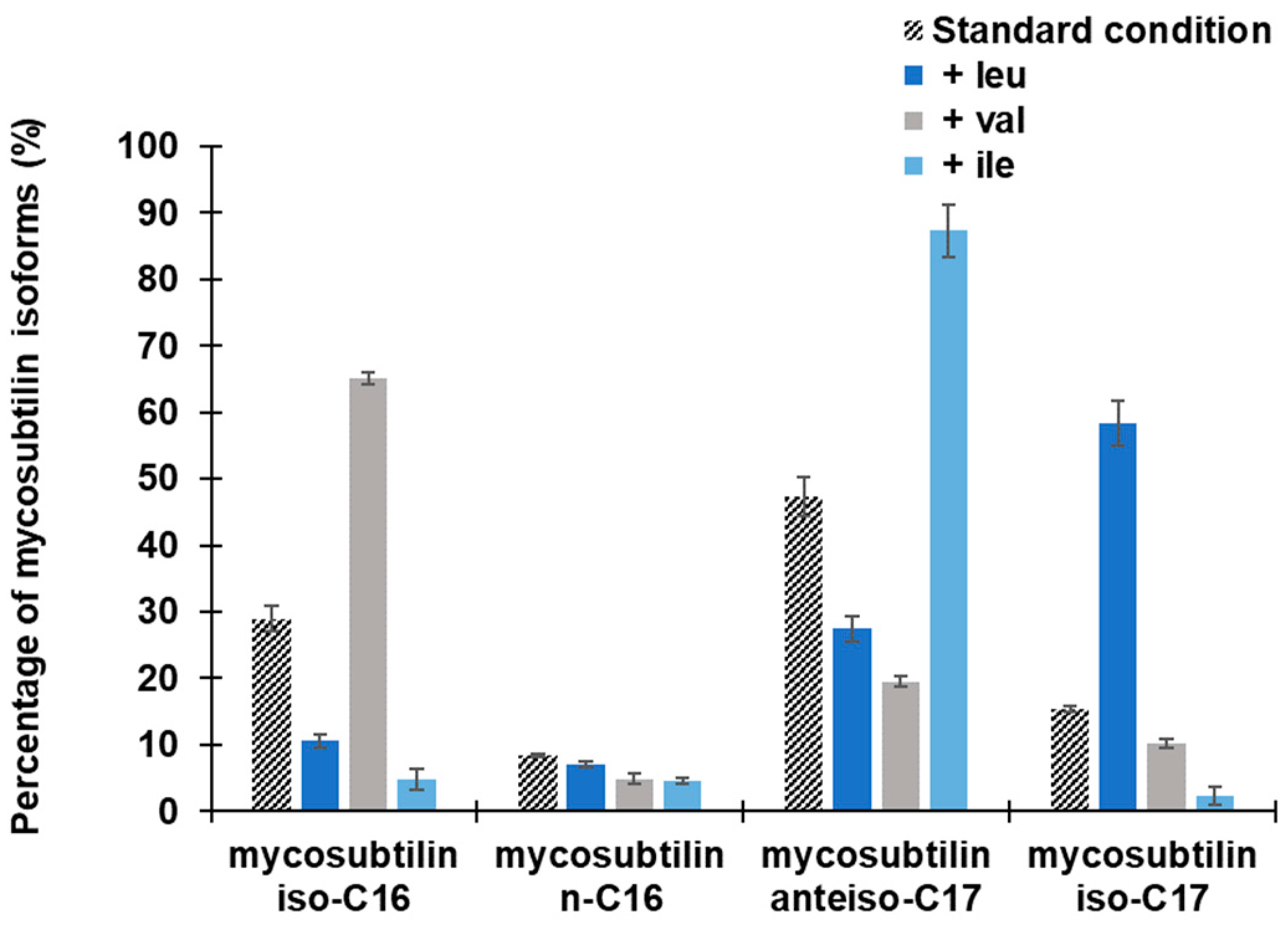
2.2.2. Effect on Mycosubtilin Pattern
2.3. Identify Genetic Targets for Specific Production of Mycosubtilin Isoforms
2.3.1. Transcriptomic Study
2.3.2. Model-Based Prediction for the anteiso Fatty Acid Precursor Overproduction
2.4. Genetic Engineering of Predicted Gene Targets
3. Discussion
3.1. Purification and Determination of the Antifungal Activity of Mycosubtilin Isoforms
3.2. Effect of Amino Acid Feeding during B. subtilis ATCC 6633 Cultures
3.3. Modification of the Selective Synthesis of anteiso-C17 and iso-C16 Mycosubtilin Isoforms
3.4. Identifying Genetic Targets to Increase the Specific Production of Mycosubtilin Isoforms
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Mutants of B. subtilis ATCC 6633 Used in This Study
4.3. RNA Isolation, Reverse Transcription and Microarrays
4.4. Model-Based Change Prediction for the Anteiso Fatty Acid Precurssor Overproduction
4.5. Lipopeptide HPLC Analysis
4.6. Lipopeptide Preparation
4.6.1. Lipopeptide Tandem Mass Spectrometry MS-MS Analysis
4.6.2. Determination of the Mycosubtilin Isoforms by GC
4.7. Cellular Fatty Acid Analysis
4.8. Bacterial Dry Weight Analysis
4.9. Determination of Minimal Inhibitory Concentrations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Jacques, P. Surfactin and Other Lipopeptides from Bacillus spp. In Biosurfactants: From Genes to Applications; Soberón-Chávez, G., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2011; pp. 57–91. ISBN 978-3-642-14490-5. [Google Scholar]
- Nasir, M.N.; Benichou, E.; Guez, J.S.; Jacques, P.; Brevet, P.-F.; Besson, F. Second Harmonic Generation to Monitor the Interactions of the Antimicrobial Mycosubtilin with Membrane-Mimicking Interfacial Monolayers. BioNanoScience 2012, 2, 108–112. [Google Scholar] [CrossRef]
- Latoud, C.; Peypoux, F.; Michel, G. Action of iturin A, an antifungal antibiotic from Bacillus subtilis, on the yeast Saccharomyces cerevisiae: Modifications of membrane permeability and lipid composition. J. Antibiot. 1987, 40, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Maget-Dana, R.; Ptak, M. Iturin lipopeptides: Interactions of mycosubtilin with lipids in planar membranes and mixed monolayers. Biochim. Biophys. Acta (BBA)-Biomembr. 1990, 1023, 34–40. [Google Scholar] [CrossRef]
- Besson, F.; Michel, G. Action of mycosubtilin, an antifungal antibiotic of Bacillus subtilis, on the cell membrane of Saccharomyces cerevisiae. Microbios 1989, 59, 113–121. [Google Scholar]
- Fickers, P.; Guez, J.-S.; Damblon, C.; Leclère, V.; Béchet, M.; Jacques, P.; Joris, B. High-Level Biosynthesis of the Anteiso-C 17 Isoform of the Antibiotic Mycosubtilin in Bacillus subtilis and Characterization of Its Candidacidal Activity. Appl. Environ. Microbiol. 2009, 75, 4636–4640. [Google Scholar] [CrossRef] [PubMed]
- Kourmentza, K.; Gromada, X.; Michael, N.; Degraeve, C.; Vanier, G.; Ravallec, R.; Coutte, F.; Karatzas, K.A.; Jauregi, P. Antimicrobial Activity of Lipopeptide Biosurfactants against Foodborne Pathogen and Food Spoilage Microorganisms and Their Cytotoxicity. Front. Microbiol. 2021, 11, 3398. [Google Scholar] [CrossRef] [PubMed]
- Béchet, M.; Castéra-Guy, J.; Guez, J.-S.; Chihib, N.-E.; Coucheney, F.; Coutte, F.; Fickers, P.; Leclère, V.; Wathelet, B.; Jacques, P. Production of a novel mixture of mycosubtilins by mutants of Bacillus subtilis. Bioresour. Technol. 2013, 145, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Desmyttere, H.; Deweer, C.; Muchembled, J.; Sahmer, K.; Jacquin, J.; Coutte, F.; Jacques, P. Antifungal Activities of Bacillus subtilis Lipopeptides to Two Venturia inaequalis Strains Possessing Different Tebuconazole Sensitivity. Front. Microbiol. 2019, 10, 2327. [Google Scholar] [CrossRef] [PubMed]
- Mejri, S.; Siah, A.; Coutte, F.; Magnin-Robert, M.; Randoux, B.; Tisserant, B.; Krier, F.; Jacques, P.; Reignault, P.; Halama, P. Biocontrol of the wheat pathogen Zymoseptoria tritici using cyclic lipopeptides from Bacillus subtilis. Environ. Sci. Pollut. Res. 2018, 25, 29822–29833. [Google Scholar] [CrossRef]
- Deravel, J.; Lemière, S.; Coutte, F.; Krier, F.; Van Hese, N.; Béchet, M.; Sourdeau, N.; Höfte, M.; Lepretre, A.; Jacques, P. Mycosubtilin and surfactin are efficient, low ecotoxicity molecules for the biocontrol of lettuce downy mildew. Appl. Microbiol. Biotechnol. 2014, 98, 6255–6264. [Google Scholar] [CrossRef] [PubMed]
- Besson, F.; Peypoux, F.; Michel, G.; Delcambe, L. Antifungal activity upon Saccharomyces cerevisiae of iturin A, mycosubtilin, bacillomycin L and of their derivatives; Inhibition of this antifungal activity by lipid antagonists. J. Antibiot. 1979, 32, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Kohli, R.M.; Trauger, J.W.; Schwarzer, D.; Marahiel, M.A.; Walsh, C.T. Generality of Peptide Cyclization Catalyzed by Isolated Thioesterase Domains of Nonribosomal Peptide Synthetases. Biochemistry 2001, 40, 7099–7108. [Google Scholar] [CrossRef]
- Genest, M.; Marion, D.; Caille, A.; Ptak, M. Modelling and refinement of the conformation of mycosubtilin in solution from two-dimensional NMR data. J. Biol. Inorg. Chem. 1987, 169, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Marion, D.; Genest, M.; Caille, A.; Peypoux, F.; Michel, G.; Ptak, M. Conformational study of bacterial lipopeptides: Refinement of the structure of iturin A in solution by two-dimensional1H-nmr and energy calculations. Biopolym. Orig. Res. Biomol. 1986, 25, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Razafindralambo, H.; Popineau, Y.; Deleu, M.; Hbid, C.; Jacques, P.; Thonart, P.; Paquot, M. Foaming Properties of Lipopeptides Produced by Bacillus subtilis: Effect of Lipid and Peptide Structural Attributes. J. Agric. Food Chem. 1998, 46, 911–916. [Google Scholar] [CrossRef]
- Hbid, C. Contribution à l’étude de la Relation Entre la Structure des Lipopeptides de Bacillus subtilis et Leurs activités hémolytique et Antifongique. Ph.D. Thesis, University of Liège, Liège, Belgium, 1996. [Google Scholar]
- Eshita, S.M.; Roberto, N.H.; Beale, J.M.; Mamiya, B.M.; Workman, R.F. Bacillomycin Lc, a New Antibiotic of the Iturin Group: Isolation, Structures, and Antifungal Activities of the Congeners. J. Antibiot. 1995, 48, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Duitman, E.H.; Hamoen, L.W.; Rembold, M.; Venema, G.; Seitz, H.; Saenger, W.; Bernhard, F.; Reinhardt, R.; Schmidt, M.; Ullrich, C.; et al. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: A multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 1999, 96, 13294–13299. [Google Scholar] [CrossRef]
- Hansen, D.B.; Bumpus, S.B.; Aron, Z.; Kelleher, A.N.L.; Walsh, C.T. The Loading Module of Mycosubtilin: An Adenylation Domain with Fatty Acid Selectivity. J. Am. Chem. Soc. 2007, 129, 6366–6367. [Google Scholar] [CrossRef] [PubMed]
- Aron, Z.D.; Dorrestein, P.C.; Blackhall, J.R.; Kelleher, A.N.L.; Walsh, C.T. Characterization of a New Tailoring Domain in Polyketide Biogenesis: The Amine Transferase Domain of MycA in the Mycosubtilin Gene Cluster. J. Am. Chem. Soc. 2005, 127, 14986–14987. [Google Scholar] [CrossRef] [PubMed]
- Weissman, K.J.; Leadlay, P. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Genet. 2005, 3, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Suutari, M.; Laakso, S. Unsaturated and branched chain-fatty acids in temperature adaptation of Bacillus subtilis and Bacillus megaterium. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1992, 1126, 119–124. [Google Scholar] [CrossRef]
- Kaneda, T. Fatty Acids of the Genus Bacillus: An Example of Branched-Chain Preference. Bacteriol. Rev. 1977, 41, 391–418. [Google Scholar] [CrossRef]
- Mäder, U.; Hennig, S.; Hecker, M.; Homuth, G. Transcriptional Organization and Posttranscriptional Regulation of the Bacillus subtilis Branched-Chain Amino Acid Biosynthesis Genes. J. Bacteriol. 2004, 186, 2240–2252. [Google Scholar] [CrossRef]
- Coutte, F.; Niehren, J.; Dhali, D.; John, M.; Versari, C.; Jacques, P. Modeling leucine’s metabolic pathway and knockout prediction improving the production of surfactin, a biosurfactant from Bacillus subtilis. Biotechnol. J. 2015, 10, 1216–1234. [Google Scholar] [CrossRef]
- Fink, P.S. Biosynthesis of the Branched-Chain Amino Acids. In Bacillus Subtilis and Other Gram-Positive Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1993; pp. 307–317. ISBN 978-1-68367-277-7. [Google Scholar]
- Debarbouille, M.; Gardan, R.; Arnaud, M.; Rapoport, G. Role of BkdR, a Transcriptional Activator of the SigL-Dependent Isoleucine and Valine Degradation Pathway in Bacillus Subtilis. J. Bacteriol. 1999, 181, 2059–2066. [Google Scholar] [CrossRef]
- Brinsmade, S.R.; Kleijn, R.J.; Sauer, U.; Sonenshein, A.L. Regulation of CodY Activity through Modulation of Intracellular Branched-Chain Amino Acid Pools. J. Bacteriol. 2010, 192, 6357–6368. [Google Scholar] [CrossRef]
- Dhali, D.; Coutte, F.; Arias, A.A.; Auger, S.; Bidnenko, V.; Chataigné, G.; Lalk, M.; Niehren, J.; De Sousa, J.; Versari, C.; et al. Genetic engineering of the branched fatty acid metabolic pathway of Bacillus subtilis for the overproduction of surfactin C14 isoform. Biotechnol. J. 2017, 12, 1600574. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Cao, Y.; Wang, Y.; Sun, L.; Song, H. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb. Cell Factories 2019, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Besson, F.; Tenoux, I.; Hourdou, M.-L.; Michel, G. Synthesis of β-hydroxy fatty acids and β-amino fatty acids by the strains of Bacillus subtilis producing iturinic antibiotics. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1992, 1123, 51–58. [Google Scholar] [CrossRef]
- Guez, J.-S.; Vassaux, A.; Larroche, C.; Jacques, P.; Coutte, F. New Continuous Process for the Production of Lipopeptide Biosurfactants in Foam Overflowing Bioreactor. Front. Bioeng. Biotechnol. 2021, 9, 678469. [Google Scholar] [CrossRef] [PubMed]
- Leclère, V.; Béchet, M.; Adam, A.; Guez, J.-S.; Wathelet, B.; Ongena, M.; Thonart, P.; Gancel, F.; Chollet-Imbert, M.; Jacques, P. Mycosubtilin Overproduction by Bacillus subtilis BBG100 Enhances the Organism’s Antagonistic and Biocontrol Activities. Appl. Environ. Microbiol. 2005, 71, 4577–4584. [Google Scholar] [CrossRef] [PubMed]
- Guez, J.; Müller, C.; Danze, P.; Büchs, J.; Jacques, P. Respiration activity monitoring system (RAMOS), an efficient tool to study the influence of the oxygen transfer rate on the synthesis of lipopeptide by Bacillus subtilis ATCCJ. J. Biotechnol. 2008, 134, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Thimon, L.; Peypoux, F.; Maget-Dana, R.; Roux, B.; Michel, G. Interactions of bioactive lipopeptides, iturin A and surfactin from Bacillus subtilis. Biotechnol. Appl. Biochem. 1992, 16, 144–151. [Google Scholar]
- Löffler, J.; Einsele, H.; Hebart, H.; Schumacher, U.; Hrastnik, C.; Daum, G.; Löffler, J.; Daum, G. Phospholipid and sterol analysis of plasma membranes of azole-resistantCandida albicansstrains. FEMS Microbiol. Lett. 2000, 185, 59–63. [Google Scholar] [CrossRef]
- Tereshina, V.M.; Memorskay, A.S.; Kotlova, E.R.; Feofilov, E.P. Membrane lipid and cytosol carbohydrate composition in Aspergillus niger under heat shock. Microbiology 2010, 79, 40–46. [Google Scholar] [CrossRef]
- Hourdou, M.-L.; Besson, F.; Tenoux, I.; Michel, G. Fatty acid and β-amino acid syntheses in strains of Bacillus subtilis producing iturinic antibiotics. Lipids 1989, 24, 940–944. [Google Scholar] [CrossRef]
- Sonenshein, A.L. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Genet. 2007, 5, 917–927. [Google Scholar] [CrossRef]
- Brinsmade, S.R.; Alexander, E.L.; Livny, J.; Stettner, A.I.; Segrè, D.; Rhee, K.Y.; Sonenshein, A.L. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc. Natl. Acad. Sci. USA 2014, 111, 8227–8232. [Google Scholar] [CrossRef]
- Molle, V.; Nakaura, Y.; Shivers, R.P.; Yamaguchi, H.; Losick, R.; Fujita, Y.; Sonenshein, A.L. Additional Targets of the Bacillus subtilis Global Regulator CodY Identified by Chromatin Immunoprecipitation and Genome-Wide Transcript Analysis. J. Bacteriol. 2003, 185, 1911–1922. [Google Scholar] [CrossRef]
- Belitsky, B.R.; Brinsmade, S.R.; Sonenshein, A.L. Intermediate Levels of Bacillus subtilis CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB. PLoS Genet. 2015, 11, e1005600. [Google Scholar] [CrossRef] [PubMed]
- Niehren, J.; Versari, C.; John, M.; Coutte, F.; Jacques, P. Predicting changes of reaction networks with partial kinetic information. BioSystems 2016, 149, 113–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thomaides, H.B.; Davison, E.J.; Burston, L.; Johnson, H.; Brown, D.R.; Hunt, A.C.; Errington, J.; Czaplewski, L. Essential Bacterial Functions Encoded by Gene Pairs. J. Bacteriol. 2007, 189, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Berger, B.J.; English, S.; Chan, G.; Knodel, M.H. Methionine Regeneration and Aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J. Bacteriol. 2003, 185, 2418–2431. [Google Scholar] [CrossRef] [PubMed]
- Duitman, E.H.; Wyczawski, D.; Boven, L.G.; Venema, G.; Kuipers, O.P.; Hamoen, L.W. Novel Methods for Genetic Transformation of Natural Bacillus subtilis Isolates Used To Study the Regulation of the Mycosubtilin and Surfactin Synthetases. Appl. Environ. Microbiol. 2007, 73, 3490–3496. [Google Scholar] [CrossRef]
- Hamoen, L.W.; Kausche, D.; Marahiel, M.A.; van Sinderen, D.; Venema, G.; Serror, P. The Bacillus subtilis transition state regulator AbrB binds to the—35 promoter region of comK. FEMS Microbiol. Lett. 2003, 218, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Kaan, T.; Homuth, G.; Mäder, U.; Bandow, J.; Schweder, T. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 2002, 148, 3441–3455. [Google Scholar] [CrossRef] [PubMed]
- Allart, E.; Niehren, J.; Versari, C. Computing difference abstractions of linear equation systems. Theor. Comput. Sci. 2021, 893, 72–104. [Google Scholar] [CrossRef]
- Tojo, S.; Satomura, T.; Morisaki, K.; Yoshida, K.-I.; Hirooka, K.; Fujita, Y. Negative Transcriptional Regulation of the ilv-leu Operon for Biosynthesis of Branched-Chain Amino Acids through the Bacillus subtilis Global Regulator TnrA. J. Bacteriol. 2004, 186, 7971–7979. [Google Scholar] [CrossRef]
- Cai, D.; Zhu, J.; Li, Y.; Li, L.; Zhang, M.; Wang, Z.; Yang, H.; Li, J.; Yang, Z.; Chen, S. Systematic engineering of branch chain amino acid supply modules for the enhanced production of bacitracin from Bacillus licheniformis. Metab. Eng. Commun. 2020, 11, e00136. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhi, Y.; Xu, Y. Systematically engineering the biosynthesis of a green biosurfactant surfactin by Bacillus subtilis. Metab. Eng. 2019, 52, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.B. In Situ Microbial Enhancement of Oil Production. Dev. Ind. Microbiol. 1981, 22, 695–701. [Google Scholar]
- Tsuge, K.; Akiyama, T.; Shoda, M. Cloning, Sequencing, and Characterization of the Iturin A Operon. J. Bacteriol. 2001, 183, 6265–6273. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russel, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 978-087969577-4. [Google Scholar]
- Guez, J.-S.; Coutte, F.; Drucbert, A.-S.; Chihib, N.-E.; Danzé, P.-M.; Jacques, P. Resistance of the cell wall to degradation is a critical parameter for isolation of high quality RNA from natural isolates of Bacillus subtilis. Arch. Microbiol. 2009, 191, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.; Helgren, J.; Manolache, S.; Lau, A.; Lagally, M.; Denes, F. Cold-plasma modification of oxide surfaces for covalent biomolecule attachment. Biosens. Bioelectron. 2005, 21, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Peypoux, F.; Besson, F.; Michel, G.; Delcambe, L. Structure of Bacillomycin D, a New Antibiotic of the Iturin Group. J. Biol. Inorg. Chem. 2005, 118, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.; Berger, T. Bacteria Identification by Gas Chromatography of Whole Cell Fatty Acids. Hewlett Packard Appl. Note 1985, 228, 228–241. [Google Scholar]
- Chihib, N.-E.; Da Silva, M.R.; Delattre, G.; Laroche, M.; Federighi, M. Different cellular fatty acid pattern behaviours of two strains of Listeria monocytogenes Scott A and CNL 895807 under different temperature and salinity conditions. FEMS Microbiol. Lett. 2003, 218, 155–160. [Google Scholar] [CrossRef] [PubMed]


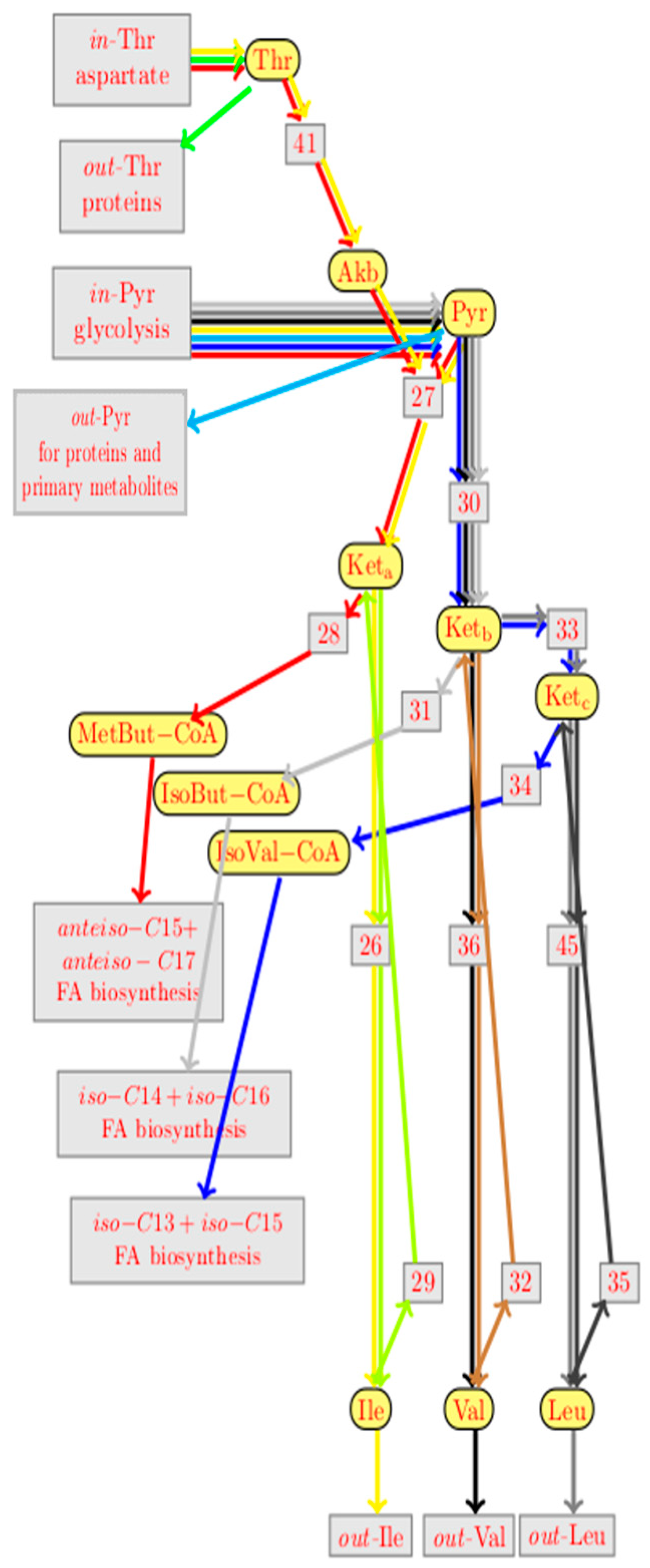
 and any reaction with name r by a boxed node
and any reaction with name r by a boxed node  . Solid edges either link a reactant to its reaction
. Solid edges either link a reactant to its reaction 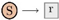 or a reaction to one of its products
or a reaction to one of its products  . There are three kinds of dashed edges, which start at the three kinds of modifiers. An accelerator edge links an accelerator to a reaction
. There are three kinds of dashed edges, which start at the three kinds of modifiers. An accelerator edge links an accelerator to a reaction  , an activator edge links an activator to a reaction
, an activator edge links an activator to a reaction  , and an inhibitor edge links an inhibitor to a reaction
, and an inhibitor edge links an inhibitor to a reaction 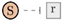 . An input edge
. An input edge 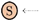 points from the context to an inflow species S, while an outflow edge
points from the context to an inflow species S, while an outflow edge  points from an outflow species S to the context. For convenience, we introduced the last kind of edges
points from an outflow species S to the context. For convenience, we introduced the last kind of edges 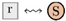 as a shortcut for a product that is degraded by a hidden reaction, i.e., as a shortcut for
as a shortcut for a product that is degraded by a hidden reaction, i.e., as a shortcut for  . Species nodes with three different colors were used, which indicate their biological roles. Yellow indicates metabolites (such as
. Species nodes with three different colors were used, which indicate their biological roles. Yellow indicates metabolites (such as  ) and blue indicates proteins (such as
) and blue indicates proteins (such as  ). There is a third color for “artificial species” that serves to modulate regulation, such as the promoter of the ilv-leu operon
). There is a third color for “artificial species” that serves to modulate regulation, such as the promoter of the ilv-leu operon  . Reactions that are potential candidates for knockouts or overexpression will be annotated in orange. Dark orange indicates candidates that were selected by our knockout prediction, while light orange indicates candidates that were not. Genes knockout predictions are represented by ⇓ and gene overexpression by ↑.
. Reactions that are potential candidates for knockouts or overexpression will be annotated in orange. Dark orange indicates candidates that were selected by our knockout prediction, while light orange indicates candidates that were not. Genes knockout predictions are represented by ⇓ and gene overexpression by ↑.
 and any reaction with name r by a boxed node
and any reaction with name r by a boxed node  . Solid edges either link a reactant to its reaction
. Solid edges either link a reactant to its reaction 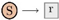 or a reaction to one of its products
or a reaction to one of its products  . There are three kinds of dashed edges, which start at the three kinds of modifiers. An accelerator edge links an accelerator to a reaction
. There are three kinds of dashed edges, which start at the three kinds of modifiers. An accelerator edge links an accelerator to a reaction  , an activator edge links an activator to a reaction
, an activator edge links an activator to a reaction  , and an inhibitor edge links an inhibitor to a reaction
, and an inhibitor edge links an inhibitor to a reaction 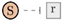 . An input edge
. An input edge  points from the context to an inflow species S, while an outflow edge
points from the context to an inflow species S, while an outflow edge  points from an outflow species S to the context. For convenience, we introduced the last kind of edges
points from an outflow species S to the context. For convenience, we introduced the last kind of edges  as a shortcut for a product that is degraded by a hidden reaction, i.e., as a shortcut for
as a shortcut for a product that is degraded by a hidden reaction, i.e., as a shortcut for  . Species nodes with three different colors were used, which indicate their biological roles. Yellow indicates metabolites (such as
. Species nodes with three different colors were used, which indicate their biological roles. Yellow indicates metabolites (such as  ) and blue indicates proteins (such as
) and blue indicates proteins (such as  ). There is a third color for “artificial species” that serves to modulate regulation, such as the promoter of the ilv-leu operon
). There is a third color for “artificial species” that serves to modulate regulation, such as the promoter of the ilv-leu operon  . Reactions that are potential candidates for knockouts or overexpression will be annotated in orange. Dark orange indicates candidates that were selected by our knockout prediction, while light orange indicates candidates that were not. Genes knockout predictions are represented by ⇓ and gene overexpression by ↑.
. Reactions that are potential candidates for knockouts or overexpression will be annotated in orange. Dark orange indicates candidates that were selected by our knockout prediction, while light orange indicates candidates that were not. Genes knockout predictions are represented by ⇓ and gene overexpression by ↑.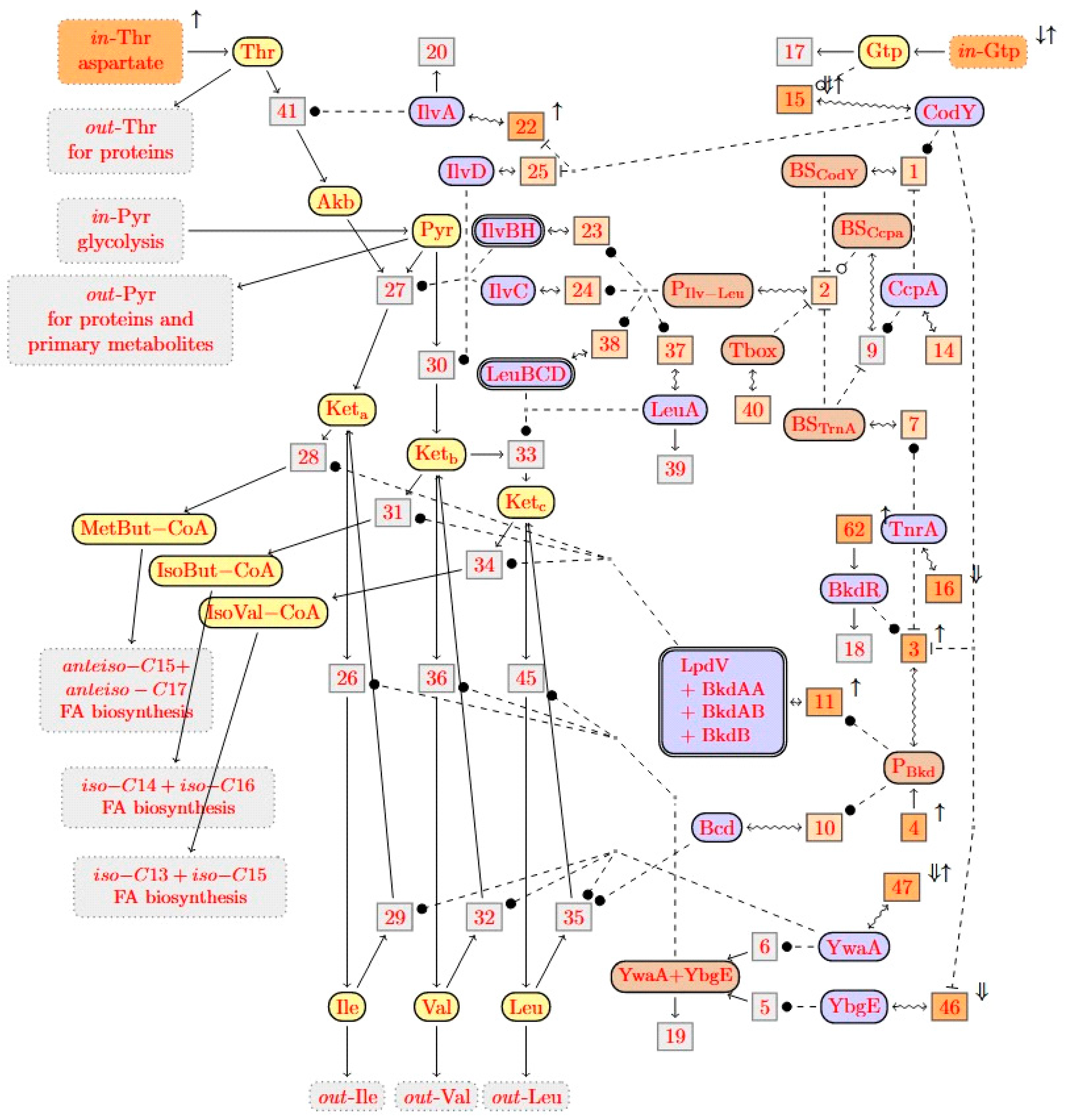
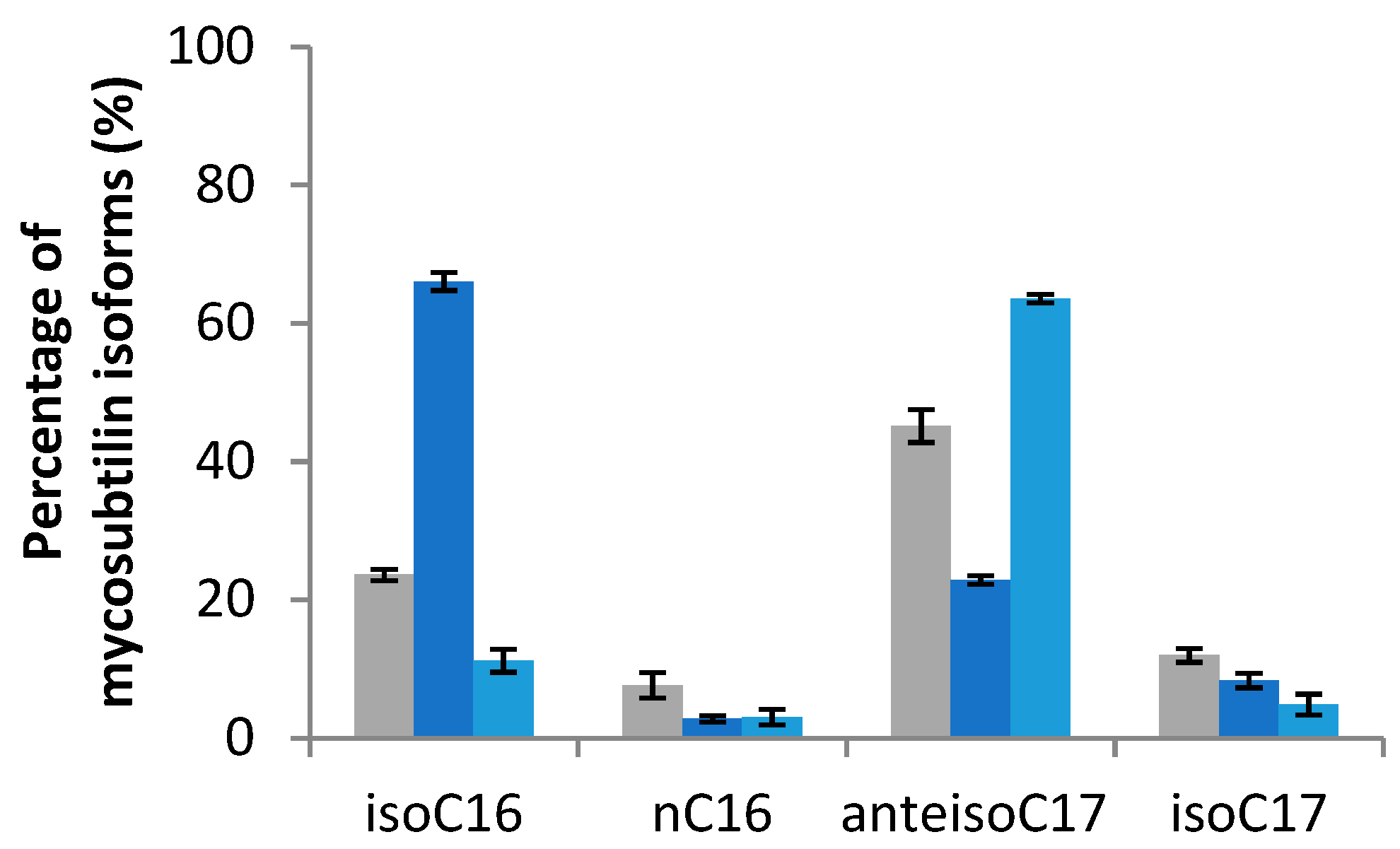
| Pathogens Strains | Mixture | iso-C16 | n-C16 | anteiso-C17 | iso-C17 | Reference |
|---|---|---|---|---|---|---|
| Botrytis cinerea | 8 | 32 | 16 | 8 | 16 | [9] |
| Aspergillus niger | 8 | 32 | 16 | 8 | 16 | This work |
| Candida albicans | 8 | >32 | 8 | 32 | 16 | [7] |
| Strains | Genotype/Phenotype | Reference or Source | |
|---|---|---|---|
| E. coli | JM 109 | recA1, endA1, gyrA96, thi, hsdR17, relA1, supE44, Δ(lac-proAB), [F′, traD36, proAB, lacI/lacZΔM15] | Promega |
| B. subtilis | ATCC 6633 | Wild-type strain/Myc+, Srf+ | Laboratory stock |
| BBG100 | ATCC 6633 PrepU neo::myc/Myc+++; Srf+, NmR | Laboratory stock [35] | |
| RFB102 | ATCC 6633 amyE::Pspac-comK-spc/SpcR | Laboratory stock [7] | |
| BBG133 | RFB102 PrepU-neo::ilvA/NmR | This work | |
| BV12I37 | ATCC 6633 ΔcodY, cat kan/CmR, KmR | [48] | |
| Plasmids | |||
| pGEM-T Easy | Cloning vector/ApR | Promega | |
| pBEST501 | pUC9:: PrepU-neo/NmR | Laboratory stock | |
| pBG142 | pGEM-T Easy lacZ::ilvA/ApR | This work | |
| pBG154 | pGEM-T Easy lacZ::PrepU-neo::ilvA/ApR, NmR | This work | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guez, J.-S.; Coucheney, F.; Guy, J.; Béchet, M.; Fontanille, P.; Chihib, N.-E.; Niehren, J.; Coutte, F.; Jacques, P. Bioinformatics Modelling and Metabolic Engineering of the Branched Chain Amino Acid Pathway for Specific Production of Mycosubtilin Isoforms in Bacillus subtilis. Metabolites 2022, 12, 107. https://doi.org/10.3390/metabo12020107
Guez J-S, Coucheney F, Guy J, Béchet M, Fontanille P, Chihib N-E, Niehren J, Coutte F, Jacques P. Bioinformatics Modelling and Metabolic Engineering of the Branched Chain Amino Acid Pathway for Specific Production of Mycosubtilin Isoforms in Bacillus subtilis. Metabolites. 2022; 12(2):107. https://doi.org/10.3390/metabo12020107
Chicago/Turabian StyleGuez, Jean-Sébastien, Françoise Coucheney, Joany Guy, Max Béchet, Pierre Fontanille, Nour-Eddine Chihib, Joachim Niehren, François Coutte, and Philippe Jacques. 2022. "Bioinformatics Modelling and Metabolic Engineering of the Branched Chain Amino Acid Pathway for Specific Production of Mycosubtilin Isoforms in Bacillus subtilis" Metabolites 12, no. 2: 107. https://doi.org/10.3390/metabo12020107
APA StyleGuez, J.-S., Coucheney, F., Guy, J., Béchet, M., Fontanille, P., Chihib, N.-E., Niehren, J., Coutte, F., & Jacques, P. (2022). Bioinformatics Modelling and Metabolic Engineering of the Branched Chain Amino Acid Pathway for Specific Production of Mycosubtilin Isoforms in Bacillus subtilis. Metabolites, 12(2), 107. https://doi.org/10.3390/metabo12020107









