Metabolomics Signatures and Subsequent Maternal Health among Mothers with a Congenital Heart Defect-Affected Pregnancy
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Study Participants
2.2. Targeted Metabolite Profiling
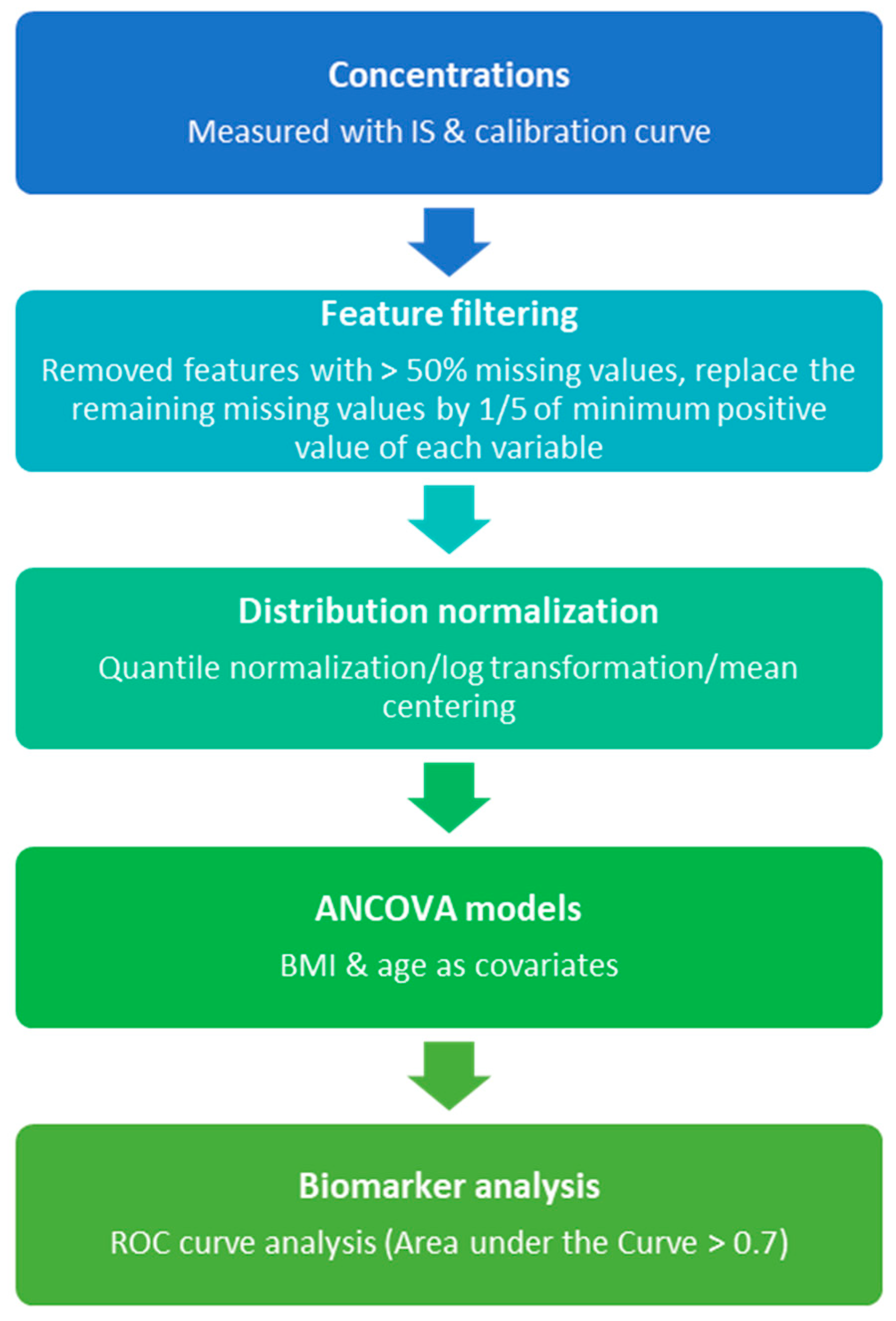
2.3. Metabolomics Data Analysis
2.4. Pathway Analysis
3. Results
3.1. Characteristics of Study Participants
3.2. Targeted Metabolomics Profiling
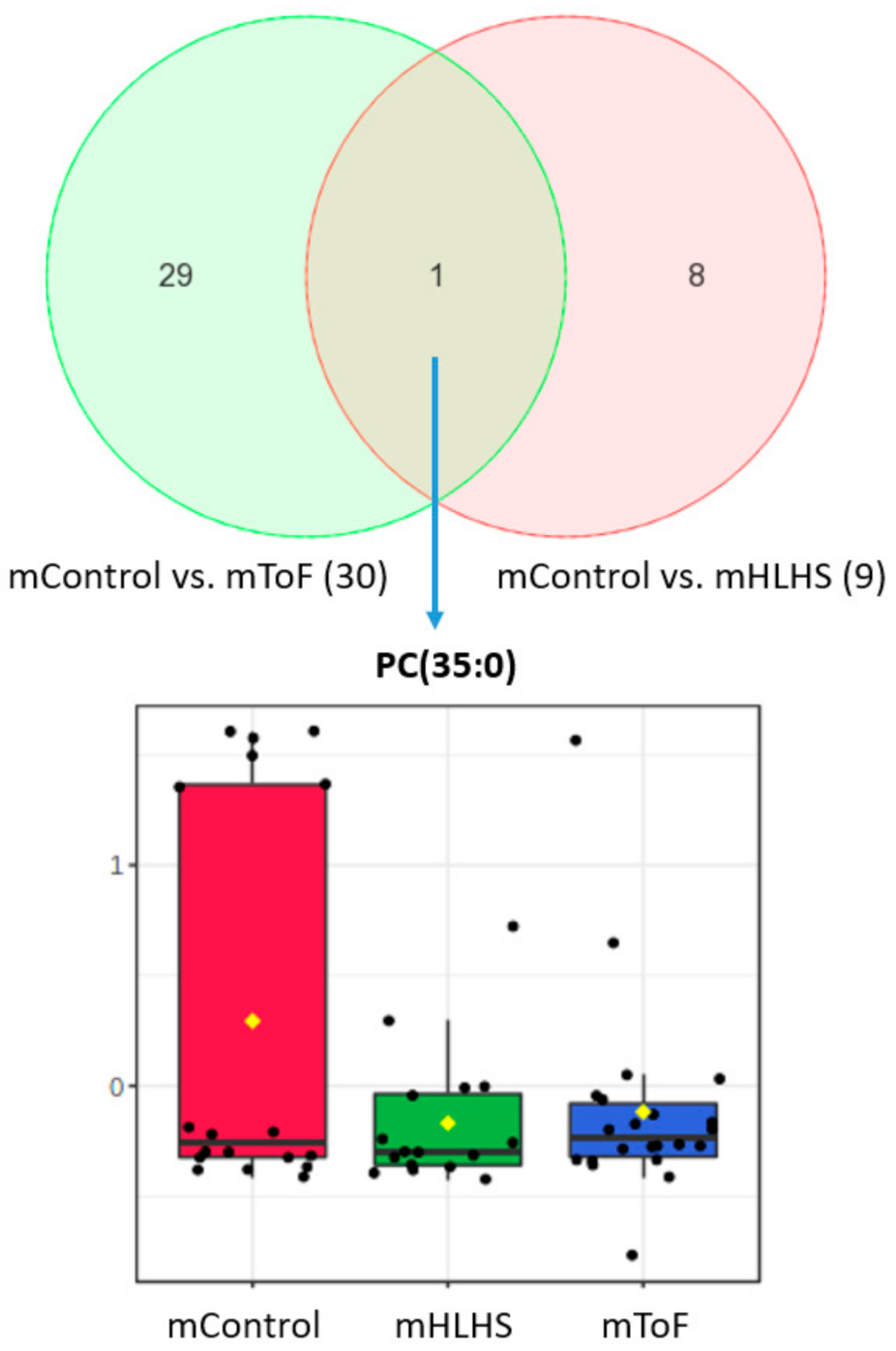
3.3. Maternal Metabolites Associated with CHD-Affected Pregnancies
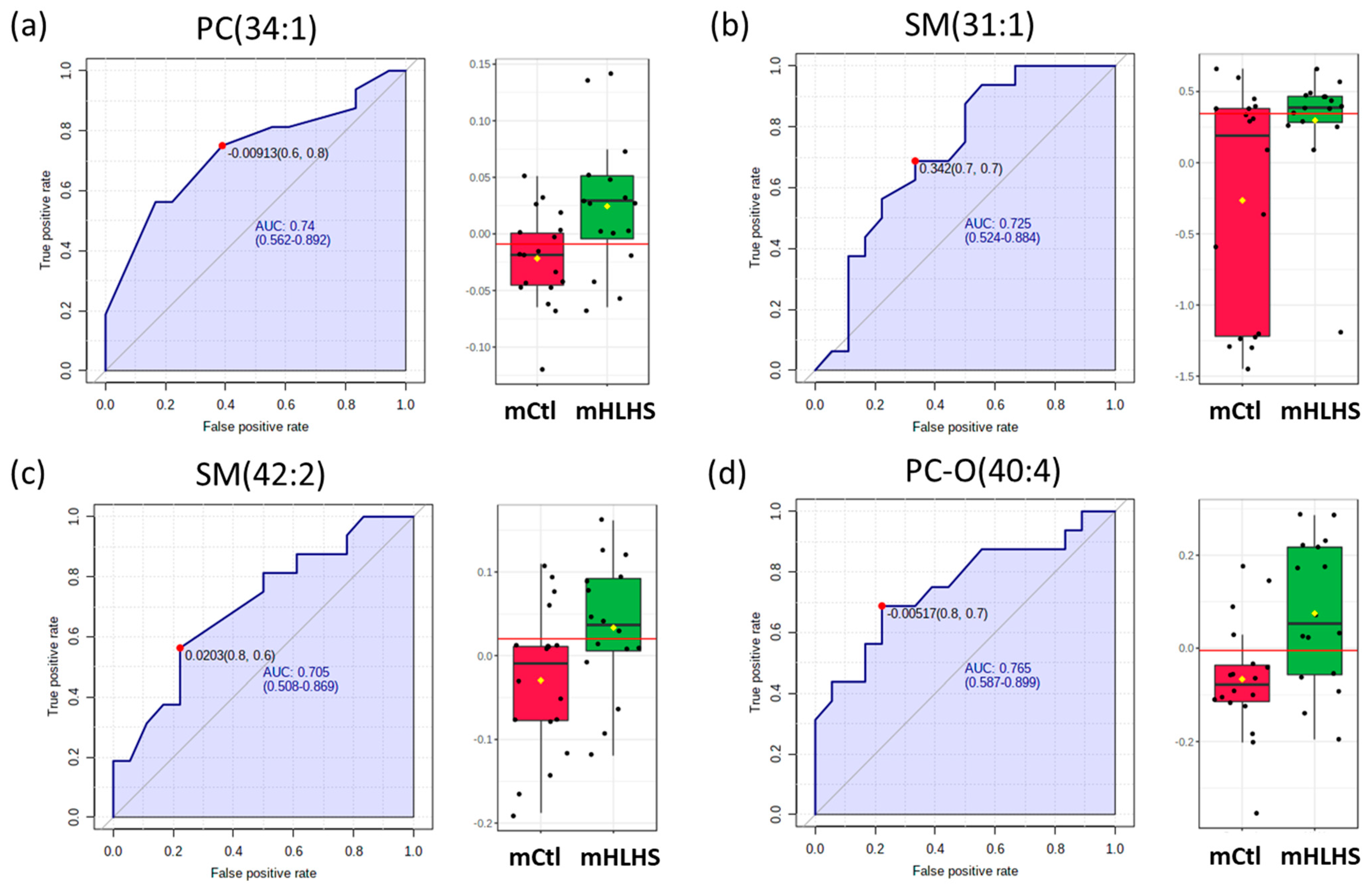
3.4. Biomarker Analysis
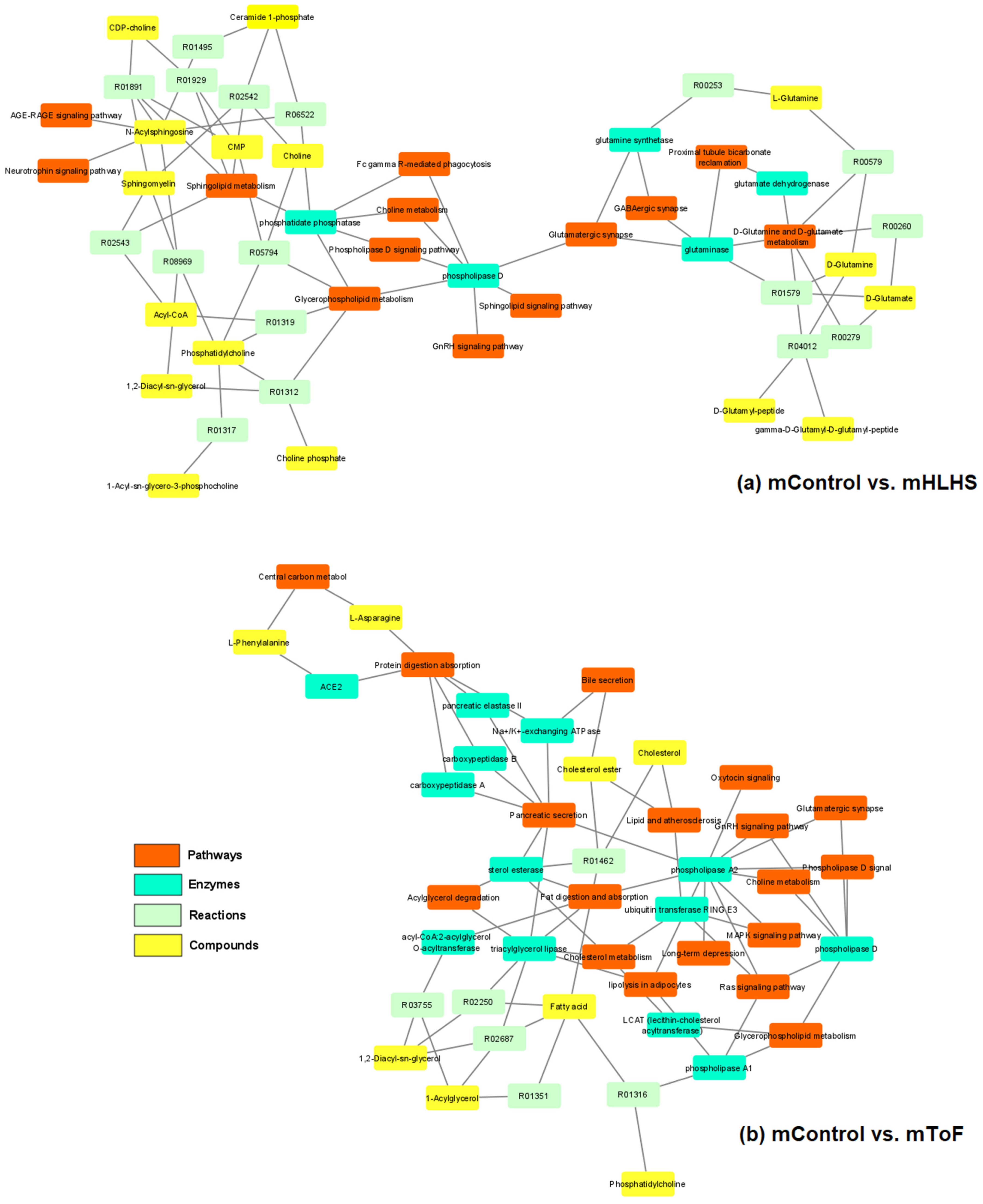
3.5. Pathway Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Botto, L.D.; Mulinare, J.; Erickson, J.D. Occurrence of congenital heart defects in relation to maternal mulitivitamin use. Am. J. Epidemiol. 2000, 151, 878–884. [Google Scholar] [CrossRef]
- Gilboa, S.M.; Devine, O.J.; Kucik, J.E.; Oster, M.E.; Riehle-Colarusso, T.; Nembhard, W.N.; Xu, P.; Correa, A.; Jenkins, K.; Marelli, A.J. Congenital Heart Defects in the United States: Estimating the Magnitude of the Affected Population in 2010. Circulation 2016, 134, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Nembhard, W.N.; Tang, X.; Hu, Z.; MacLeod, S.; Stowe, Z.; Webber, D. Maternal and infant genetic variants, maternal periconceptional use of selective serotonin reuptake inhibitors, and risk of congenital heart defects in offspring: Population based study. BMJ 2017, 356, j832. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cleves, M.A.; Ghaffar, S.; Zhao, W.; Mosley, B.S.; Hobbs, C.A. First-year survival of infants born with congenital heart defects in Arkansas (1993–1998): A survival analysis using registry data. Birth Defects Res. Part A Clin. Mol. Teratol. 2003, 67, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, S.M.; Salemi, J.L.; Nembhard, W.N.; Fixler, D.E.; Correa, A. Mortality resulting from congenital heart disease among children and adults in the United States, 1999 to 2006. Circulation 2010, 122, 2254–2263. [Google Scholar] [CrossRef]
- Nembhard, W.N.; Waller, D.K.; Sever, L.E.; Canfield, M.A. Patterns of first-year survival among infants with selected congenital anomalies in Texas, 1995–1997. Teratology 2001, 64, 267–275. [Google Scholar] [CrossRef]
- Friedman, P.; Yilmaz, A.; Ugur, Z.; Jafar, F.; Whitten, A.; Ustun, I.; Turkoglu, O.; Graham, S.; Singh, R.B. Urine metabolomic biomarkers for prediction of isolated fetal congenital heart defect. J. Matern.-Fetal Neonatal Med. 2021, 2021, 1–8. [Google Scholar] [CrossRef]
- Troisi, J.; Cavallo, P.; Richards, S.; Symes, S.; Colucci, A.; Sarno, L.; Landolfi, A.; Scala, G.; Adair, D.; Ciccone, C.; et al. Noninvasive screening for congenital heart defects using a serum metabolomics approach. Prenat. Diagn. 2021, 41, 743–753. [Google Scholar] [CrossRef]
- Xie, D.; Luo, Y.; Xiong, X.; Lou, M.; Liu, Z.; Wang, A.; Xiong, L.; Kong, F.; Wang, Y.; Wang, H. Study on the Potential Biomarkers of Maternal Urine Metabolomics for Fetus with Congenital Heart Diseases Based on Modified Gas Chromatograph-Mass Spectrometer. BioMed Res. Int. 2019, 2019, 1905416. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Ertl, R.; Mandal, R.; Bjorndahl, T.C.; Syngelaki, A.; Han, B.; Dong, E.; Liu, P.B.; Alpay-Savasan, Z.; Wishart, D.S.; et al. Metabolomic prediction of fetal congenital heart defect in the first trimester. Am. J. Obs. Gynecol. 2014, 211, e1–e204. [Google Scholar] [CrossRef]
- Rochfort, S. Metabolomics Reviewed: A New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Chrysayi-Tokousbalides, M. Metabolomics in pesticide research and development: Review and future perspectives. Metabolomics 2011, 7, 35–53. [Google Scholar] [CrossRef]
- Beger, R.D. A Review of Applications of Metabolomics in Cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef]
- De Raad, M.; Fischer, C.R.; Northen, T.R. High-throughput platforms for metabolomics. Curr. Opin. Chem. Biol. 2016, 30, 7–13. [Google Scholar] [CrossRef]
- Macel, M.; Van Dam, N.M.; Keurentjes, J.J.B. Metabolomics: The chemistry between ecology and genetics. Mol. Ecol. Resour. 2010, 10, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Maity, S.; Jannasch, A.; Adamec, J.; Nalepa, T.; Höök, T.O.; Sepúlveda, M.S. Starvation causes disturbance in amino acid and fatty acid metabolism in Diporeia. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2012, 161, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lu, Y.; Huang, S.; Gao, L.; Liang, X.; Wu, Y.; Wang, J.; Huang, Q.; Tang, L.; Wang, G.; et al. Identifying Early Urinary Metabolic Changes with Long-Term Environmental Exposure to Cadmium by Mass-Spectrometry-Based Metabolomics. Environ. Sci. Technol. 2014, 48, 6409–6418. [Google Scholar] [CrossRef]
- Grison, S.; Favé, G.; Maillot, M.; Manens, L.; Delissen, O.; Blanchardon, E.; Dublineau, I.; Aigueperse, J.; Bohand, S.; Martin, J.C.; et al. Metabolomics reveals dose effects of low-dose chronic exposure to uranium in rats: Identification of candidate biomarkers in urine samples. Metabolomics. 2016, 12, 154. [Google Scholar] [CrossRef]
- Xu, M.-Y.; Sun, Y.-J.; Wang, P.; Xu, H.-Y.; Chen, L.-P.; Zhu, L.; Wu, Y.-J. Metabolomics Analysis and Biomarker Identification for Brains of Rats Exposed Subchronically to the Mixtures of Low-Dose Cadmium and Chlorpyrifos. Chem. Res. Toxicol. 2015, 28, 1216–1223. [Google Scholar] [CrossRef]
- Gyawali, P.; Karpe, A.V.; Hillyer, K.E.; Nguyen, T.V.; Hewitt, J.; Beale, D.J. A multi-platform metabolomics approach to identify possible biomarkers for human faecal contamination in Greenshell™ mussels (Perna canaliculus). Sci. Total Environ. 2021, 771, 145363. [Google Scholar] [CrossRef]
- Sulek, K.; Han, T.-L.; Villas-Boas, S.; Wishart, D.S.; Soh, S.E.; Kwek, K.; Gluckman, P.D.; Chong, Y.S.; Kenny, L.; Baker, P. Hair metabolomics: Identification of fetal compromise provides proof of concept for biomarker discovery. Theranostics 2014, 4, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, L.; Zhang, W.; Zhang, J.; Du, X.; Huang, Q.; Tian, M.; Shen, H. Serum metabolome biomarkers associate low-level environmental perfluorinated compound exposure with oxidative/nitrosative stress in humans. Environ. Pollut. 2017, 229, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Maity, S.; Jannasch, A.; Adamec, J.; Watkins, J.M.; Nalepa, T.; Höök, T.O.; Sepúlveda, M.S. Elucidating causes of Diporeia decline in the Great Lakes via metabolomics: Physiological responses after exposure to different stressors. Physiol. Biochem. Zool. PBZ 2013, 86, 213–223. [Google Scholar] [CrossRef]
- Klein, M.S.; Shearer, J. Metabolomics and Type 2 Diabetes: Translating Basic Research into Clinical Application. J. Diabetes Res. 2016, 2016, 3898502. [Google Scholar] [CrossRef]
- Urpi-Sarda, M.; Almanza-Aguilera, E.; Tulipani, S.; Tinahones, F.J.; Salas-Salvadó, J.; Andres-Lacueva, C. Metabolomics for biomarkers of type 2 diabetes mellitus: Advances and nutritional intervention trends. Curr. Cardiovasc. Risk Rep. 2015, 9, 12. [Google Scholar] [CrossRef]
- Milburn, M.V.; Lawton, K.A. Application of Metabolomics to Diagnosis of Insulin Resistance. Annu. Rev. Med. 2013, 64, 291–305. [Google Scholar] [CrossRef]
- Moco, S.; Collino, S.; Rezzi, S.; Martin, F.-P.J. Metabolomics perspectives in pediatric research. Pediatr. Res. 2013, 73, 570–576. [Google Scholar] [CrossRef]
- Papamichael, M.; Katsardis, C.; Sarandi, E.; Georgaki, S.; Frima, E.-S.; Varvarigou, A.; Tsoukalas, D. Application of Metabolomics in Pediatric Asthma: Prediction, Diagnosis and Personalized Treatment. Metabolites 2021, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Dona, A.C.; Coffey, S.; Figtree, G. Translational and emerging clinical applications of metabolomics in cardiovascular disease diagnosis and treatment. Eur. J. Prev. Cardiol. 2020, 23, 1578–1589. [Google Scholar] [CrossRef]
- Senn, T.; Hazen, S.L.; Tang, W.H.W. Translating Metabolomics to Cardiovascular Biomarkers. Prog. Cardiovasc. Dis. 2012, 55, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.D.; Asnani, A.; Gerszten, R.E. Application of Metabolomics to Cardiovascular Biomarker and Pathway Discovery. J. Am. Coll. Cardiol. 2008, 52, 117–123. [Google Scholar] [CrossRef]
- Jasbi, P.; Wang, D.; Cheng, S.L.; Fei, Q.; Cui, J.Y.; Liu, L.; Wei, Y.; Raftery, D.; Gu, H. Breast cancer detection using targeted plasma metabolomics. J. Chromatogr. B 2019, 1105, 26–37. [Google Scholar] [CrossRef]
- Putluri, N.; Maity, S.; Kommagani, R.; Creighton, C.J.; Putluri, V.; Chen, F.; Nanda, S.; Bhowmik, S.K.; Terunuma, A.; Dorsey, T.; et al. Pathway-Centric Integrative Analysis Identifies RRM2 as a Prognostic Marker in Breast Cancer Associated with Poor Survival and Tamoxifen Resistance. Neoplasia 2014, 16, 390–402. [Google Scholar] [CrossRef]
- Shajahan-Haq, A.N.; Cheema, M.S.; Clarke, R. Application of Metabolomics in Drug Resistant Breast Cancer Research. Metabolites 2015, 5, 100–118. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Maity, S.; Coarfa, C.; Rajapakshe, K.; Chen, L.; Jin, F.; Putluri, V.; Tinker, L.F.; Mo, Q.; Chen, F.; et al. A Prospective Targeted Serum Metabolomics Study of Pancreatic Cancer in Postmenopausal Women. Cancer Prev. Res. 2019, 12, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, A.; Heuchel, R.; Forshed, J.; Lehtiö, J.; Löhr, M.; Nordström, A. Discrimination of pancreatic cancer and pancreatitis by LC-MS metabolomics. Metabolomics 2017, 13, 61. [Google Scholar] [CrossRef]
- Sakai, A.; Suzuki, M.; Kobayashi, T.; Nishiumi, S.; Yamanaka, K.; Hirata, Y.; Nakagawa, T.; Azuma, T.; Yoshida, M. Pancreatic cancer screening using a multiplatform human serum metabolomics system. Biomark. Med. 2016, 10, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Simpson, B.N.; Kim, M.; Chuang, Y.-F.; Beason-Held, L.; Kitner-Triolo, M.; Kraut, M.; Lirette, S.T.; Windham, B.G.; Griswold, M.E.; Legido-Quigley, C.; et al. Blood metabolite markers of cognitive performance and brain function in aging. J. Cereb. Blood Flow Metab. 2016, 36, 1212–1223. [Google Scholar] [CrossRef]
- Srivastava, S. Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics. Metabolites 2019, 9, 301. [Google Scholar] [CrossRef]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward personalized medicine. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Tuyiringire, N.; Tusubira, D.; Munyampundu, J.-P.; Tolo, C.U.; Muvunyi, C.M.; Ogwang, P.E. Application of metabolomics to drug discovery and understanding the mechanisms of action of medicinal plants with anti-tuberculosis activity. Clin. Transl. Med. 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- York, B.; Li, F.; Lin, F.; Marcelo, K.L.; Mao, J.; Dean, A.; Gonzales, N.; Gooden, D.; Maity, S.; Coarfa, C.; et al. Pharmacological inhibition of CaMKK2 with the selective antagonist STO-609 regresses NAFLD. Sci. Rep. 2017, 7, 11793. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Anthony, D.C. Metabolomics in multiple sclerosis disease course and progression. Mult. Scler. J. 2020, 26, 591–598. [Google Scholar] [CrossRef]
- Varma, V.R.; Oommen, A.M.; Varma, S.; Casanova, R.; An, Y.; Andrews, R.M.; O’Brien, R.; Pletnikova, O.; Troncoso, J.C.; Toledo, J.; et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: A targeted metabolomics study. PLoS Med. 2018, 15, e1002482. [Google Scholar] [CrossRef]
- Villoslada, P.; Alonso, C.; Agirrezabal, I.; Kotelnikova, E.; Zubizarreta, I.; Pulido-Valdeolivas, I.; Saiz, A.; Comabella, M.; Montalban, X.; Villar, L.; et al. Metabolomic signatures associated with disease severity in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e321. [Google Scholar] [CrossRef]
- Huang, J.; Mo, J.; Zhao, G.; Lin, Q.; Wei, G.; Deng, W.; Chen, D.; Yu, B. Application of the amniotic fluid metabolome to the study of fetal malformations, using Down syndrome as a specific model. Mol. Med. Rep. 2017, 16, 7405–7415. [Google Scholar] [CrossRef]
- Parfieniuk, E.; Zbucka-Kretowska, M.; Ciborowski, M.; Kretowski, A.; Barbas, C. Untargeted metabolomics: An overview of its usefulness and future potential in prenatal diagnosis. Expert Rev. Proteom. 2018, 15, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.T.; Mayrink, J.; Leite, D.F.; Costa, M.L.; Calderon, I.M.; Rocha, E.A.; Vettorazzi, J.; Feitosa, F.E.; Cecatti, J.G. Metabolomics applied to maternal and perinatal health: A review of new frontiers with a translation potential. Clinics 2019, 74, e894. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Mi, K.; Jiang, Y.; Chen, J.; Lv, D.; Qian, Z.; Sun, H.; Shang, D. Construction and Analysis of Human Diseases and Metabolites Network. Front. Bioeng. Biotechnol. 2020, 8, 398. [Google Scholar] [CrossRef]
- Tang, X.; Cleves, M.A.; Nick, T.G.; Li, M.; MacLeod, S.L.; Erickson, S.W.; Li, J.; Shaw, G.M.; Mosley, B.S.; Hobbs, C.A.; et al. Obstructive heart defects associated with candidate genes, maternal obesity, and folic acid supplementation. Am. J. Med. Genet. Part A 2015, 167, 1231–1242. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Olshan, A.F.; Hobbs, C.A.; Shaw, G.M. Discovery of genetic susceptibility factors for human birth defects: An opportunity for a National Agenda. Am. J. Med. Genet. Part A 2011, 155, 1794–1797. [Google Scholar] [CrossRef] [PubMed]
- Boneva, R.S.; Botto, L.D.; Moore, C.A.; Yang, Q.; Correa, A.; Erickson, J.D. Mortality associated with congenital heart defects in the United States: Trends and racial disparities, 1979–1997. Circulation 2001, 103, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Narchi, H.; Kulaylat, N. Heart disease in infants of diabetic mothers. Images Paediatr. Cardiol. 2000, 2, 17–23. [Google Scholar] [PubMed]
- Levy, H.L.; Guldberg, P.; Güttler, F.; Hanley, W.B.; Matalon, R.; Rouse, B.M.; Trefz, F.; Azen, C.; Allred, E.N.; De La Cruz, F.; et al. Congenital heart disease in maternal phenylketonuria: Report from the Maternal PKU Collaborative Study. Pediatr. Res. 2001, 49, 636–642. [Google Scholar] [CrossRef]
- Hedermann, G.; Hedley, P.L.; Thagaard, I.N.; Krebs, L.; Ekelund, C.K.; Sørensen, T.I.A.; Christiansen, M. Maternal obesity and metabolic disorders associate with congenital heart defects in the offspring: A systematic review. PLoS ONE 2021, 16, e0252343. [Google Scholar] [CrossRef] [PubMed]
- Testerink, N.; van der Sanden, M.H.; Houweling, M.; Helms, J.B.; Vaandrager, A.B. Depletion of phosphatidylcholine affects endoplasmic reticulum morphology and protein traffic at the Golgi complex. J. Lipid Res. 2009, 50, 2182–2192. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Furse, S.; Billing, G.; Snowden, S.G.; Smith, J.; Goldberg, G.; Koulman, A. Relationship between the lipid composition of maternal plasma and infant plasma through breast milk. Metabolomics 2019, 15, 129. [Google Scholar] [CrossRef]
- Mourad, A.M.; de Carvalho Pincinato, E.; Mazzola, P.G.; Sabha, M.; Moriel, P. Influence of soy lecithin administration on hypercholesterolemia. Cholesterol 2010, 2010, 824813. [Google Scholar] [CrossRef]
- Gauster, M.; Rechberger, G.; Sovic, A.; Hörl, G.; Steyrer, E.; Sattler, W.; Frank, S. Endothelial lipase releases saturated and unsaturated fatty acids of high density lipoprotein phosphatidylcholine. J. Lipid Res. 2005, 46, 1517–1525. [Google Scholar] [CrossRef]
- Dullaart, R.P.; Gansevoort, R.T.; Dikkeschei, B.D.; de Zeeuw, D.; de Jong, P.E.; Van Tol, A. Role of elevated lecithin: Cholesterol acyltransferase and cholesteryl ester transfer protein activities in abnormal lipoproteins from proteinuric patients. Kidney Int. 1993, 44, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, C.; Pechlaner, R.; Willeit, P.; Langley, S.R.; Mangino, M.; Mayr, U.; Menni, C.; Moayyeri, A.; Santer, P.; Rungger, G.; et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014, 129, 1821–1831. [Google Scholar] [CrossRef]
- Ganna, A.; Salihovic, S.; Sundström, J.; Broeckling, C.D.; Hedman, Å.K.; Magnusson, P.K.; Pedersen, N.L.; Larsson, A.; Siegbahn, A.; Zilmer, M.; et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014, 10, e1004801. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-K.; Lee, D.H.; Kim, J.K.; Park, M.-J.; Yan, J.-J.; Song, D.-K.; Vaziri, N.D.; Noh, J.-W. Lysophosphatidylcholine, oxidized low-density lipoprotein and cardiovascular disease in Korean hemodialysis patients: Analysis at 5 years of follow-up. J. Korean Med. Sci. 2013, 28, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Grimes, S.B.; Wild, R. Effect of Pregnancy on Lipid Metabolism and Lipoprotein Levels; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., de Herder, W.W., Dhatariya, K., Dungan, K., Hershman, J.M., Hofland, J., Kalra, S., et al., Eds.; Endotext, MDText.com, Inc.: South Dartmouth, MA, USA, 2018. [Google Scholar]
- Xi, P.; Jiang, Z.; Zheng, C.; Lin, Y.; Wu, G. Regulation of protein metabolism by glutamine: Implications for nutrition and health. Front. Biosci. (Landmark Ed.) 2011, 16, 578–597. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef]
- Durante, W. The Emerging Role of l-Glutamine in Cardiovascular Health and Disease. Nutrients 2019, 11, 2092. [Google Scholar] [CrossRef]
- Cedars, A.; Manlhiot, C.; Ko, J.-M.; Bottiglieri, T.; Arning, E.; Weingarten, A.; Opotowsky, A.; Kutty, S. Metabolomic Profiling of Adults with Congenital Heart Disease. Metabolites 2021, 11, 525. [Google Scholar] [CrossRef]
- Shao, M.; Huang, C.; Li, Z.; Yang, H.; Feng, Q. Effects of glutamine and valsartan on the brain natriuretic peptide and N-terminal pro-B-type natriuretic peptide of patients with chronic heart failure. Pak. J. Med. Sci. 2015, 31, 82–86. [Google Scholar] [CrossRef]
- Gong, W.; Liang, Q.; Zheng, D.; Zhong, R.; Wen, Y.; Wang, X. Congenital heart defects of fetus after maternal exposure to organic and inorganic environmental factors: A cohort study. Oncotarget 2017, 8, 100717–100723. [Google Scholar] [CrossRef] [PubMed]
- Peyvandi, S.; Baer, R.J.; Chambers, C.D.; Norton, M.E.; Rajagopal, S.; Ryckman, K.K.; Moon-Grady, A.; Jelliffe-Pawlowski, L.L.; Steurer, M.A. Environmental and Socioeconomic Factors Influence the Live-Born Incidence of Congenital Heart Disease: A Population-Based Study in California. J. Am. Heart Assoc. 2020, 9, e015255. [Google Scholar] [CrossRef] [PubMed]
- Helle, E.; Priest, J.R. Maternal Obesity and Diabetes Mellitus as Risk Factors for Congenital Heart Disease in the Offspring. J. Am. Heart Assoc. 2020, 9, e011541. [Google Scholar] [CrossRef]
- Øyen, N.; Diaz, L.J.; Leirgul, E.; Boyd, H.A.; Priest, J.; Mathiesen, E.R.; Quertermous, T.; Wohlfahrt, J.; Melbye, M. Prepregnancy Diabetes and Offspring Risk of Congenital Heart Disease: A Nationwide Cohort Study. Circulation 2016, 133, 2243–2253. [Google Scholar] [CrossRef]
- Feng, Y.; Yu, D.; Yang, L.; Da, M.; Wang, Z.; Lin, Y.; Ni, B.; Wang, S.; Mo, X. Maternal lifestyle factors in pregnancy and congenital heart defects in offspring: Review of the current evidence. Ital. J. Pediatr. 2014, 40, 85. [Google Scholar] [CrossRef] [PubMed]
- Clausen, H.; Noren, E.; Valtonen, S.; Koivu, A.; Sairanen, M.; Liuba, P. Evaluation of Circulating Cardiovascular Biomarker Levels for Early Detection of Congenital Heart Disease in Newborns in Sweden. JAMA Netw. Open. 2020, 3, e2027561. [Google Scholar] [CrossRef]
- Neves, A.L.; Cabral, M.; Leite-Moreira, A.; Monterroso, J.; Ramalho, C.; Guimarães, H.; Barros, H.; Guimarães, J.T.; Henriques-Coelho, T.; Areias, J.C. Myocardial Injury Biomarkers in Newborns with Congenital Heart Disease. Pediatr. Neonatol. 2016, 57, 488–495. [Google Scholar] [CrossRef]



| mControl 1 (18) | mHLHS 2 (16) | mToF 3 (22) | p * | |
|---|---|---|---|---|
| Mean ± Std/Counts | ||||
| Pre-pregnancy BMI 4 | 25.9 ± 6.6 | 24.5 ± 4.6 | 26.6 ± 7.6 | 0.6 |
| Post-partum BMI | 31.6 ± 6.0 | 28.6 ± 3.7 | 31.0 ± 6.8 | 0.3 |
| BMI change | 5.8 ± 2.7 | 4.2 ± 2.4 | 4.4 ± 2.2 | 0.1 |
| Weight change (kg) | 15.4 ± 8.0 | 11.3 ± 6.8 | 11.6 ± 5.9 | 0.1 |
| Maternal age at delivery | 25.8 ± 5.6 | 27.3 ± 5.5 | 26.6 ± 5.6 | 0.7 |
| Maternal age at EDD 5 | 25.9 ± 5.7 | 27.3 ± 5.5 | 26.6 ± 5.6 | 0.8 |
| Maternal age at conception | 25.2 ± 5.7 | 26.8 ± 5.5 | 26.0 ± 5.7 | 0.7 |
| Maternal age at delivery | 25.8 ± 5.6 | 27.3 ± 5.5 | 26.6 ± 5.6 | 0.7 |
| Race | 0.6 | |||
| Non-Hispanic white | 17 | 14 | 20 | |
| Non-Hispanic black | 1 | 1 | 2 | |
| Asian/Pacific Islander | 0 | 1 | 0 | |
| Alcohol consumption | 0.2 | |||
| Yes (B1-M3) ** | 10 | 6 | 6 | |
| No | 8 | 10 | 16 | |
| Maternal smoking | 0.9 | |||
| Heavy smokers or 15+ cigs per day | 2 | 1 | 2 | |
| Medium smokers or 5–14 cigs per day | 3 | 3 | 2 | |
| Light smokers or ≤1–4 cigs per day | 1 | 2 | 3 | |
| No smoking | 12 | 10 | 15 | |
| High blood pressure during pregnancy | 0.7 | |||
| Yes | 1 | 3 | 5 | |
| No | 1 | 1 | 1 | |
| No answer | 16 | 12 | 16 | |
| High blood pressure medicine use | 0.3 | |||
| Yes | 0 | 1 | 0 | |
| No | 1 | 3 | 6 | |
| No answer | 17 | 12 | 16 | |
| Type II Diabetes | 0.9 | |||
| Yes | 2 | 2 | 2 | |
| No | 16 | 14 | 20 | |
| Metabolites | mControl | mHLHS | mControl vs. mHLHS (mControl/mHLHS) | ||
|---|---|---|---|---|---|
| Mean ± Std (μM) | p * | FC * | Trend | ||
| PC (34:1) | 149.56 ± 29.13 | 157 ± 36.54 | 0.014 | −1.03 | Controls down |
| TG (52:6) | 2.98 ± 1.44 | 2.81 ± 2.03 | 0.016 | 2.02 | Controls up |
| PC (41:4) | 1.65 ± 0.67 | 1.90 ± 0.62 | 0.020 | −1.27 | Controls down |
| SM (31:1) | 0.49 ± 0.26 | 0.48 ± 0.25 | 0.023 | −1.43 | Controls down |
| SM (42:2) | 39.98 ± 7.62 | 41.38 ± 6.63 | 0.027 | −1.07 | Controls down |
| PC (41:1) | 0.66 ± 0.36 | 0.40 ± 0.36 | 0.029 | 1.38 | Controls up |
| Glutamine | 548 ± 73.95 | 484.10 ± 63.81 | 0.046 | 1.04 | Controls up |
| PC (35:0) | 0.12 ± 0.18 | 0.008 ± 0.009 | 0.046 | 1.39 | Controls up |
| PC-O (40:4) | 0.95 ± 0.28 | 1.20 ± 0.49 | 0.049 | −1.10 | Controls down |
| Metabolites | mControl | mToF | mControl vs. mToF (mControl/mToF) | ||
|---|---|---|---|---|---|
| Mean ± Std (μM) | p * | FC * | Trend | ||
| LPC (18:2) | 14.62 ± 7.17 | 9.87 ± 3.76 | 0.004 | 1.15 | Controls up |
| Asparagine | 45.87 ± 7.47 | 39.85 ± 10.07 | 0.006 | 1.08 | Controls up |
| PC (31:0) | 0.54 ± 0.22 | 0.37 ± 0.18 | 0.010 | 1.16 | Controls up |
| LPC (24:0) | 0.105 ± 0.11 | 0.111 ± 0.10 | 0.014 | −2.11 | Controls down |
| PC (40:8) | 6.96 ± 5.94 | 3.71 ± 4.23 | 0.014 | 1.36 | Controls up |
| PC-O (40:0) | 0.006 ± 0.003 | 0.004 ± 0.003 | 0.017 | −2.54 | Controls down |
| TG (52:7) | 1.20 ± 0.82 | 1.86 ± 0.95 | 0.017 | −2.33 | Controls down |
| TG (44:1) | 1.86 ± 1.59 | 0.78 ± 1.39 | 0.018 | 1.55 | Controls up |
| LPC (20:3) | 2.42 ± 1.11 | 1.54 ± 0.73 | 0.020 | 1.20 | Controls up |
| PC (38:1) | 0.59 ± 0.22 | 0.42 ± 0.23 | 0.021 | 1.17 | Controls up |
| LPC (15:0) | 0.59 ± 0.19 | 0.44 ± 0.13 | 0.025 | 1.08 | Controls up |
| PC-O (30:2) | 0.008 ± 0.001 | 0.007 ± 0.001 | 0.025 | 1.07 | Controls up |
| PC (38:0) | 0.66 ± 0.31 | 0.46 ± 0.31 | 0.026 | 1.20 | Controls up |
| PC-O (33:2) | 0.005 ± 0.001 | 0.004 ± 0.001 | 0.028 | 1.08 | Controls up |
| PC-O (35:4) | 0.29 ± 0.35 | 0.15 ± 0.05 | 0.032 | −1.82 | Controls down |
| Phenylalanine | 65.61 ± 13.5 | 52.38 ± 8.63 | 0.032 | 1.04 | Controls up |
| PC (33:0) | 0.85 ± 0.25 | 0.69 ± 0.17 | 0.034 | 1.05 | Controls up |
| TG (48:3) | 5.78 ± 6.72 | 2.60 ± 5.63 | 0.037 | 1.59 | Controls up |
| PC (44:3) | 1.65 ± 2.7 | 4.89 ± 5.42 | 0.038 | −1.82 | Controls down |
| LPC (18:0) | 22.04 ± 8.58 | 17.62 ± 6.67 | 0.040 | 1.09 | Controls up |
| LPC (17:0) | 1.43 ± 0.45 | 1.144 ± 0.44 | 0.041 | 1.08 | Controls up |
| PC (35:2) | 8.02 ± 2.15 | 6.43 ± 1.57 | 0.041 | 1.06 | Controls up |
| CE (22:6) | 65.41 ± 23.51 | 75.38 ± 21.32 | 0.041 | −1.08 | Controls down |
| PC (35:0) | 0.12 ± 0.18 | 0.03 ± 0.09 | 0.042 | 1.36 | Controls up |
| PC (30:1) | 0.009 ± 0.002 | 0.008 ± 0.001 | 0.045 | 1.07 | Controls up |
| TG (48:2) | 13.74 ± 8.14 | 9.71 ± 8.08 | 0.047 | 1.13 | Controls up |
| TG (46:2) | 3.72 ± 2.61 | 2.30 ± 3.28 | 0.047 | 1.36 | Controls up |
| TG (48:1) | 15.35 ± 9.04 | 10.49 ± 8.54 | 0.048 | 1.14 | Controls up |
| DG (32:2) | 0.77 ± 0.40 | 0.55 ± 0.37 | 0.048 | 1.12 | Controls up |
| PC (46:2) | 8.91 ± 4.99 | 6.26 ± 4.14 | 0.049 | 1.12 | Controls up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, P.-C.; Maity, S.; Patel, J.; Lupo, P.J.; Nembhard, W.N. Metabolomics Signatures and Subsequent Maternal Health among Mothers with a Congenital Heart Defect-Affected Pregnancy. Metabolites 2022, 12, 100. https://doi.org/10.3390/metabo12020100
Hsu P-C, Maity S, Patel J, Lupo PJ, Nembhard WN. Metabolomics Signatures and Subsequent Maternal Health among Mothers with a Congenital Heart Defect-Affected Pregnancy. Metabolites. 2022; 12(2):100. https://doi.org/10.3390/metabo12020100
Chicago/Turabian StyleHsu, Ping-Ching, Suman Maity, Jenil Patel, Philip J. Lupo, and Wendy N. Nembhard. 2022. "Metabolomics Signatures and Subsequent Maternal Health among Mothers with a Congenital Heart Defect-Affected Pregnancy" Metabolites 12, no. 2: 100. https://doi.org/10.3390/metabo12020100
APA StyleHsu, P.-C., Maity, S., Patel, J., Lupo, P. J., & Nembhard, W. N. (2022). Metabolomics Signatures and Subsequent Maternal Health among Mothers with a Congenital Heart Defect-Affected Pregnancy. Metabolites, 12(2), 100. https://doi.org/10.3390/metabo12020100







