Flavor Profiling Using Comprehensive Mass Spectrometry Analysis of Metabolites in Tomato Soups
Abstract
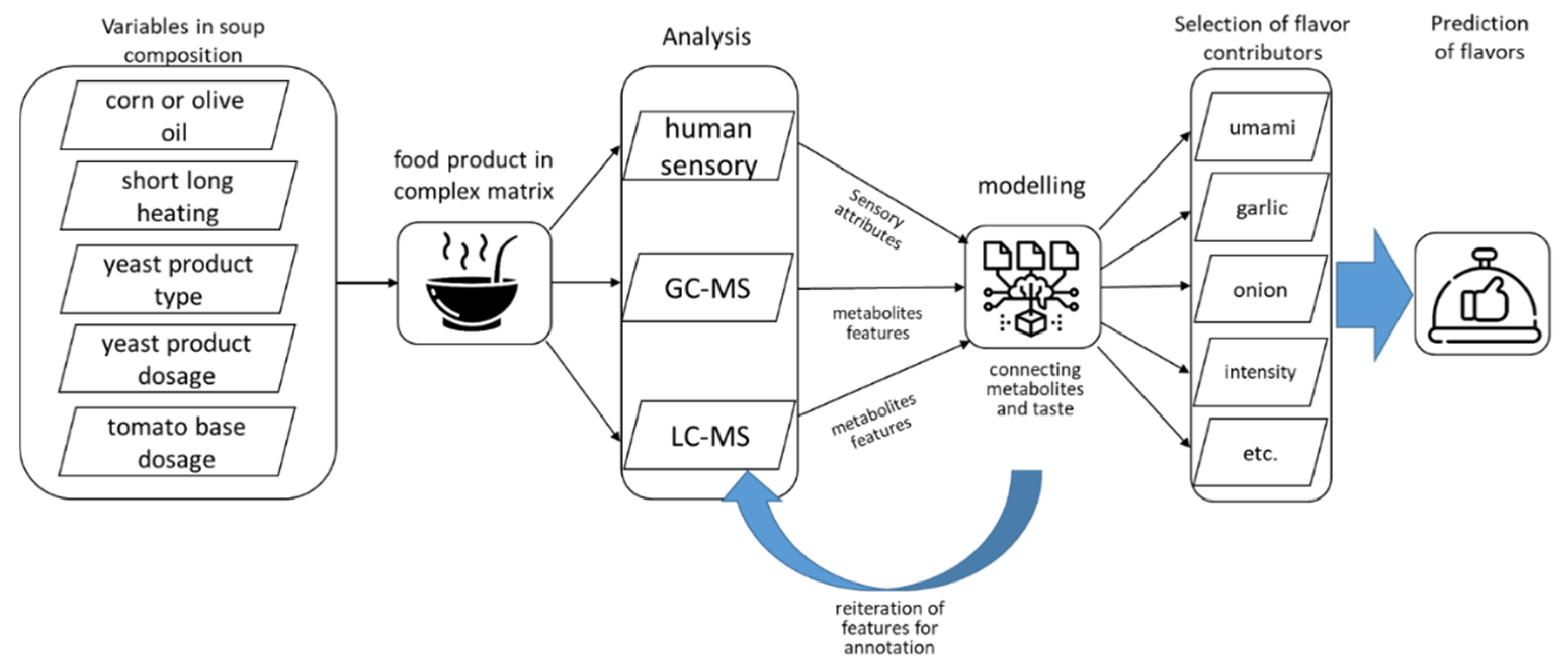
1. Introduction
2. Materials and Methods
2.1. Soup Composition
2.2. Sensory Analysis Description
2.3. Liquid Chromatography–Mass Spectrometry
2.4. Gas Chromatography–Mass Spectrometry
2.5. Sensory Data Processing
2.6. Statistical Analysis
3. Results
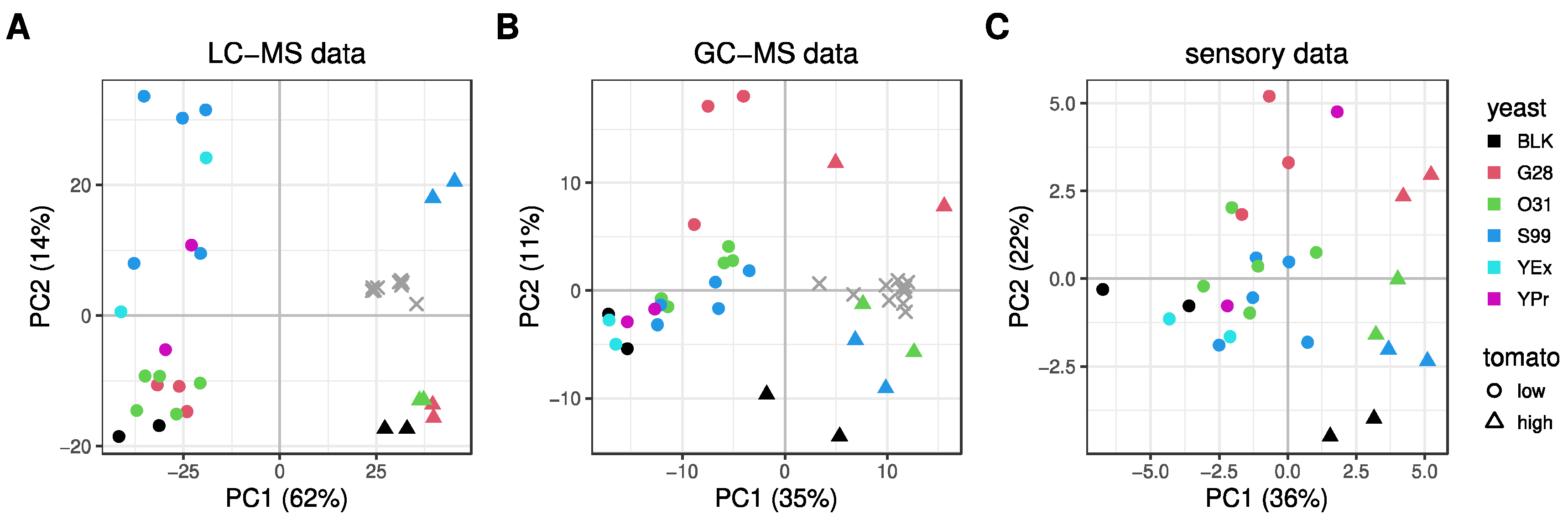
3.1. Explorative Analysis
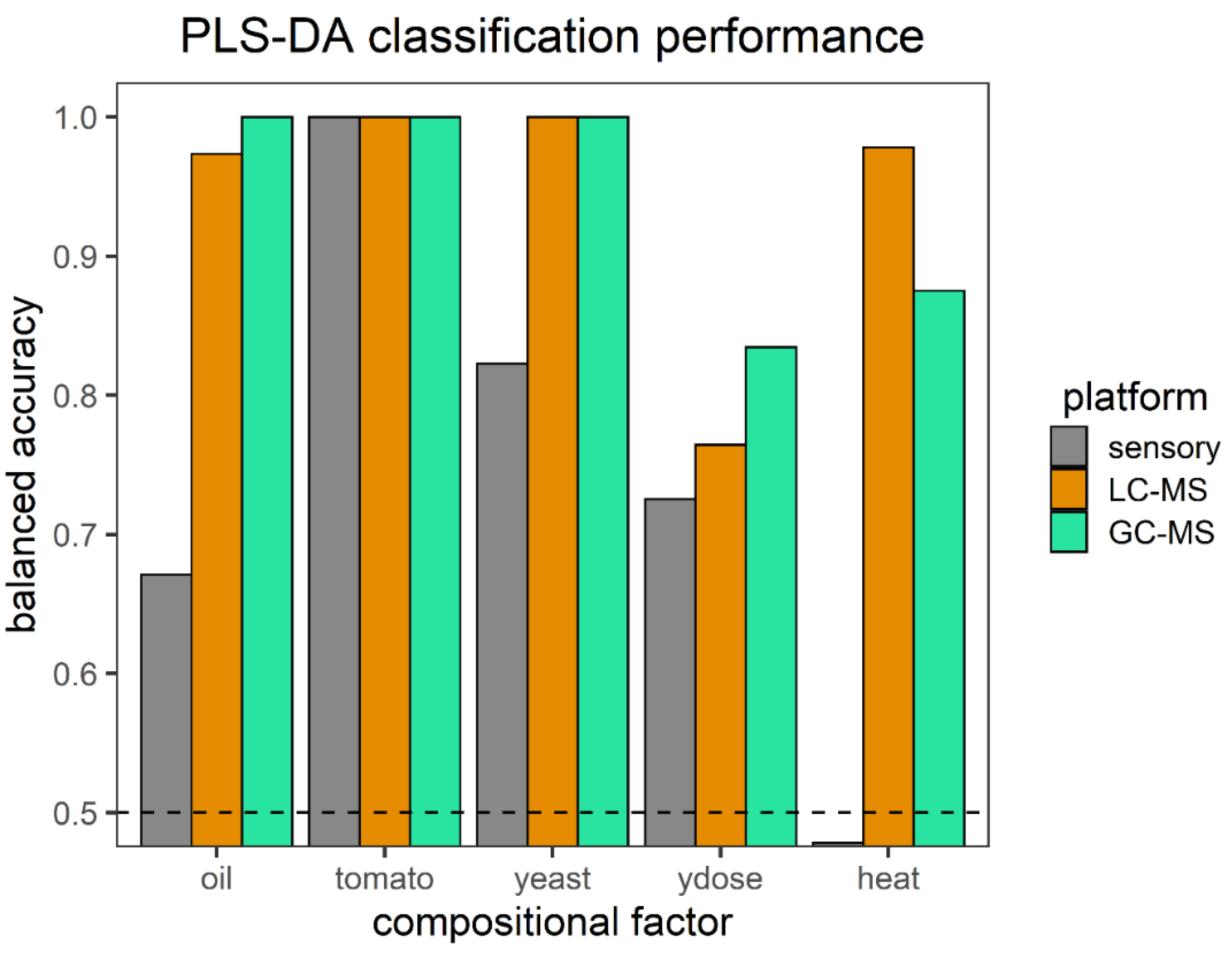
3.2. Classification Performance of the Soup Compositions on All Platforms
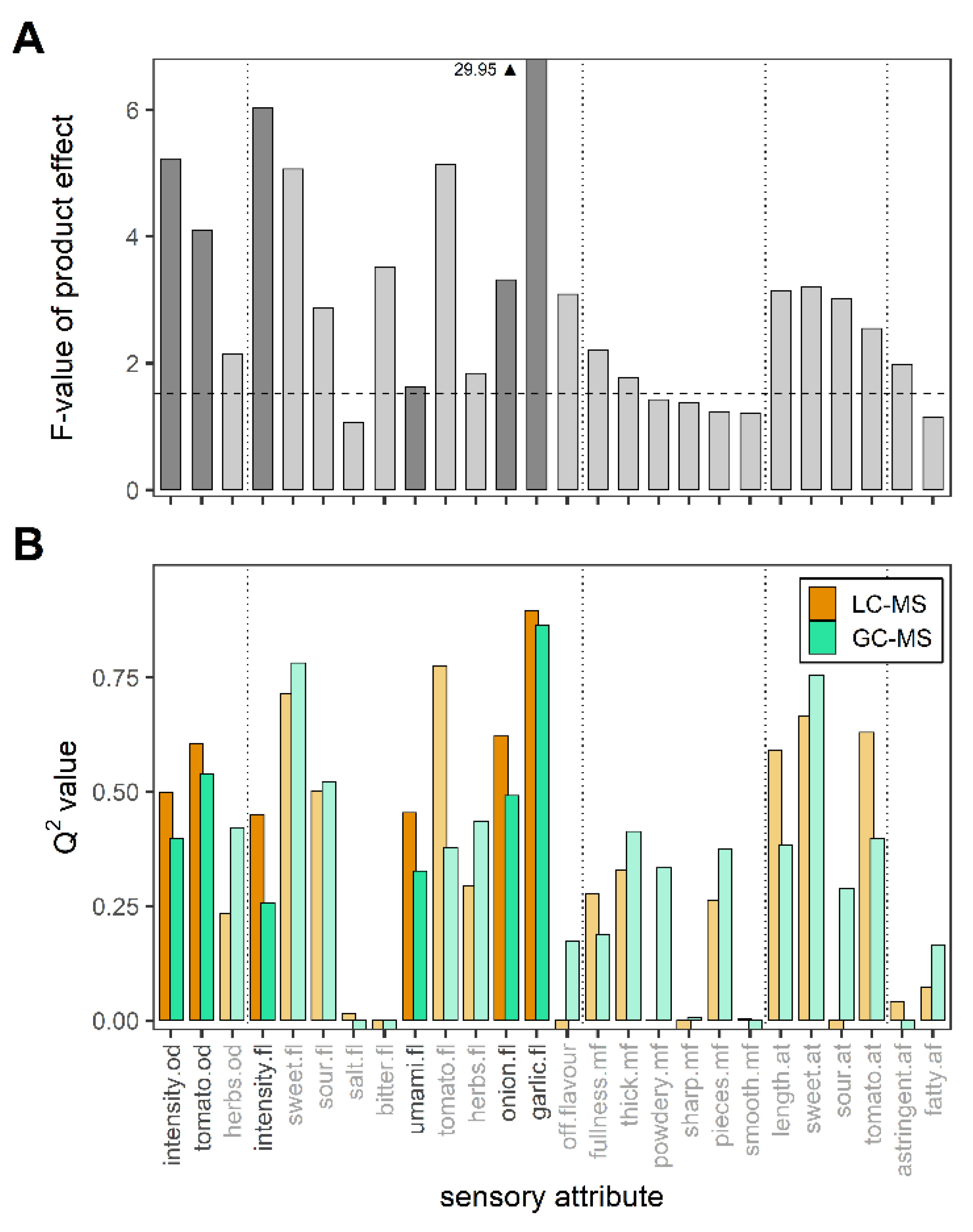
3.3. Discriminative Performance of the Sensory Panel
3.4. Relationship between Metabolomics Platforms and Sensory Attributes
3.5. Annotation of Features in LC-MS and GC-MS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clausen, M.R.; Pedersen, B.H.; Bertram, H.C.; Kidmose, U. Quality of sour cherry juice of different clones and cultivars (Prunus cerasus L.) determined by a combined sensory and NMR spectroscopic approach. J. Agric. Food Chem. 2011, 59, 12124–12130. [Google Scholar] [CrossRef]
- Pavagadhi, S.; Swarup, S. Metabolomics for Evaluating Flavor-Associated Metabolites in Plant-Based Products. Metabolites 2020, 10, 197. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, F.; Bordoni, A. Foodomics: A new comprehensive approach to food and nutrition. Genes Nutr. 2013, 8, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I. Metabolomics in food science. Adv. Food Nutr. Res. 2012, 67, 1–24. [Google Scholar] [PubMed]
- Naviglio, D.; Gallo, M. Application of Analytical Chemistry to Foods and Food Technology. Foods 2020, 9, 1296. [Google Scholar] [CrossRef] [PubMed]
- Farres, M.; Pina, B.; Tauler, R. LC-MS based metabolomics and chemometrics study of the toxic effects of copper on Saccharomyces cerevisiae. Metallomics 2016, 8, 790–798. [Google Scholar] [CrossRef]
- Montesano, D.; Rocchetti, G.; Cossignani, L.; Senizza, B.; Pollini, L.; Lucini, L.; Blasi, F. Untargeted Metabolomics to Evaluate the Stability of Extra-Virgin Olive Oil with Added Lycium barbarum Carotenoids during Storage. Foods 2019, 8, 179. [Google Scholar] [CrossRef]
- Baldina, S.; Picarella, M.E.; Troise, A.D.; Pucci, A.; Ruggieri, V.; Ferracane, R.; Barone, A.; Fogliano, V.; Mazzucato, A. Metabolite Profiling of Italian Tomato Landraces with Different Fruit Types. Front. Plant Sci. 2016, 7, 664. [Google Scholar] [CrossRef]
- Diez-Simon, C.; Eichelsheim, C.; Mumm, R.; Hall, R.D. Chemical and Sensory Characteristics of Soy Sauce: A Review. J. Agric. Food Chem. 2020, 68, 11612–11630. [Google Scholar] [CrossRef]
- Lopez-Sanchez, P.; de Vos, R.C.; Jonker, H.H.; Mumm, R.; Hall, R.D.; Bialek, L.; Leenman, R.; Strassburg, K.; Vreeken, R.; Hankemeier, T.; et al. Comprehensive metabolomics to evaluate the impact of industrial processing on the phytochemical composition of vegetable purees. Food Chem. 2015, 168, 348–355. [Google Scholar] [CrossRef]
- Sherman, E.; Coe, M.; Grose, C.; Martin, D.; Greenwood, D.R. Metabolomics Approach to Assess the Relative Contributions of the Volatile and Non-volatile Composition to Expert Quality Ratings of Pinot Noir Wine Quality. J. Agric. Food Chem. 2020, 68, 13380–13396. [Google Scholar] [CrossRef]
- Amino, Y.; Wakabayashi, H.; Akashi, S.; Ishiwatari, Y. Structural analysis and taste evaluation of gamma-glutamyl peptides comprising sulfur-containing amino acids. Biosci. Biotechnol. Biochem. 2018, 82, 383–394. [Google Scholar] [CrossRef]
- Kong, Y.; Yang, X.; Ding, Q.; Zhang, Y.Y.; Sun, B.G.; Chen, H.T.; Sun, Y. Comparison of non-volatile umami components in chicken soup and chicken enzymatic hydrolysate. Food Res. Int. 2017, 102, 559–566. [Google Scholar] [CrossRef]
- Schmidtke, L.M.; Blackman, J.W.; Clark, A.C.; Grant-Preece, P. Wine metabolomics: Objective measures of sensory properties of semillon from GC-MS profiles. J. Agric. Food Chem. 2013, 61, 11957–11967. [Google Scholar] [CrossRef] [PubMed]
- Davarzani, N.; Diez-Simon, C.; Grossmann, J.L.; Jacobs, D.M.; van Doorn, R.; van den Berg, M.A.; Smilde, A.K.; Mumm, R.; Hall, R.D.; Westerhuis, J.A. Systematic selection of competing metabolomics methods in a metabolite-sensory relationship study. Metabolomics 2021, 17, 77. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Ramirez-Gaona, M.; Marcu, A.; Pon, A.; Guo, A.C.; Sajed, T.; Wishart, N.A.; Karu, N.; Djoumbou Feunang, Y.; Arndt, D.; Wishart, D.S. YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 2017, 45, D440–D445. [Google Scholar] [CrossRef]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef]
- Diez-Simon, C.; Ammerlaan, B.; van den Berg, M.; van Duynhoven, J.; Jacobs, D.; Mumm, R.; Hall, R.D. Comparison of volatile trapping techniques for the comprehensive analysis of food flavourings by Gas Chromatography-Mass Spectrometry. J. Chromatogr. A 2020, 1624, 461191. [Google Scholar] [CrossRef]
- Lommen, A. MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef]
- Tikunov, Y.M.; Laptenok, S.; Hall, R.D.; Bovy, A.; de Vos, R.C. MSClust: A tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 2012, 8, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Romano, P.; Bartocci, E.; Bertolini, G.; De Paoli, F.; Marra, D.; Mauri, G.; Merelli, E.; Milanesi, L. Biowep: A workflow enactment portal for bioinformatics applications. BMC Bioinform. 2007, 8 (Suppl. 1), S19. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Shi, L.; Westerhuis, J.A.; Rosen, J.; Landberg, R.; Brunius, C. Variable selection and validation in multivariate modelling. Bioinformatics 2019, 35, 972–980. [Google Scholar] [CrossRef]
- van Velzen, E.J.; Westerhuis, J.A.; van Duynhoven, J.P.; van Dorsten, F.A.; Hoefsloot, H.C.; Jacobs, D.M.; Smit, S.; Draijer, R.; Kroner, C.I.; Smilde, A.K. Multilevel data analysis of a crossover designed human nutritional intervention study. J. Proteome Res. 2008, 7, 4483–4491. [Google Scholar] [CrossRef]
- Maruyama, Y.; Yasuda, R.; Kuroda, M.; Eto, Y. Kokumi substances, enhancers of basic tastes, induce responses in calcium-sensing receptor expressing taste cells. PLoS ONE 2012, 7, e34489. [Google Scholar] [CrossRef]
- Kino Haruka, K.M. Hattori Koichi, Kino Kuniki Salty Taste Enhancer. US-2016374380-A1, 26 June 2015. [Google Scholar]
- Hemmler, D.; Roullier-Gall, C.; Marshall, J.W.; Rychlik, M.; Taylor, A.J.; Schmitt-Kopplin, P. Insights into the Chemistry of Non-Enzymatic Browning Reactions in Different Ribose-Amino Acid Model Systems. Sci. Rep. 2018, 8, 16879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cui, H.; Zhang, Q.; Hayat, K.; Yu, J.; Hussain, S.; Tahir, M.U.; Zhang, X.; Ho, C.T. Taste improvement of Maillard reaction intermediates derived from enzymatic hydrolysates of pea protein. Food Res. Int. 2021, 140, 109985. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, S.; Ishibashi, N.; Tamura, M.; Nishizaki, H.; Okai, H. Synthesis of Bitter Peptides Composed of Aspartic Acid and Glutamic Acid. Agric. Biol. Chem. 1988, 52, 871–872. [Google Scholar]
- Diez-Simon, C.; Mumm, R.; Hall, R.D. Mass spectrometry-based metabolomics of volatiles as a new tool for understanding aroma and flavour chemistry in processed food products. Metabolomics 2019, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Keles, D.; Taskin, H.; Baktemur, G.; Kafkas, E.; Buyukalaca, S. Comparitive study on volatile aroma compounds of two different garlic types (Kastamonu and Chinese) using gas chromatography mass spectrometry (HS-GC/MS) technique. Afr. J. Tradit. Complementary Altern. Med. 2014, 11, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Puccinelli, M.T.; Stan, S.D. Dietary Bioactive Diallyl Trisulfide in Cancer Prevention and Treatment. Int. J. Mol. Sci. 2017, 18, 1645. [Google Scholar] [CrossRef]
- Xu, J.J.; Elkaddi, N.; Garcia-Blanco, A.; Spielman, A.I.; Bachmanov, A.A.; Chung, H.Y.; Ozdener, M.H. Arginyl dipeptides increase the frequency of NaCl-elicited responses via epithelial sodium channel alpha and delta subunits in cultured human fungiform taste papillae cells. Sci. Rep. 2017, 7, 7483. [Google Scholar] [CrossRef]
- van der Laan, T.; Elfrink, H.; Azadi-Chegeni, F.; Dubbelman, A.C.; Harms, A.C.; Jacobs, D.M.; Braumann, U.; Velders, A.H.; van Duynhoven, J.; Hankemeier, T. Fractionation platform for target identification using off-line directed two-dimensional chromatography, mass spectrometry and nuclear magnetic resonance. Anal. Chim. Acta 2021, 1142, 28–37. [Google Scholar] [CrossRef]
- Zhang, Y.; Venkitasamy, C.; Pan, Z.; Liu, W.; Zhao, L. Novel Umami Ingredients: Umami Peptides and Their Taste. J. Food Sci. 2017, 82, 16–23. [Google Scholar] [CrossRef]
- Harth, L.; Krah, U.; Linke, D.; Dunkel, A.; Hofmann, T.; Berger, R.G. Salt Taste Enhancing l-Arginyl Dipeptides from Casein and Lysozyme Released by Peptidases of Basidiomycota. J. Agric. Food Chem. 2018, 66, 2344–2353. [Google Scholar] [CrossRef]
- Okuno, T.; Morimoto, S.; Nishikawa, H.; Haraguchi, T.; Kojima, H.; Tsujino, H.; Arisawa, M.; Yamashita, T.; Nishikawa, J.; Yoshida, M.; et al. Bitterness-Suppressing Effect of Umami Dipeptides and Their Constituent Amino Acids on Diphenhydramine: Evaluation by Gustatory Sensation and Taste Sensor Testing. Chem. Pharm. Bull. 2020, 68, 234–243. [Google Scholar] [CrossRef] [PubMed]
| Onion Flavor | Garlic Flavor | Umami Flavor | Intensity Flavor | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Feature | Putative Identification 1 | Id. Level 2 | Feature | Putative Identification 1 | Id. Level 2 | Feature | Putative Identification 1 | Id. Level 2 | Feature | Putative Identification 1 | Id. Level 2 |
| 1 | X1313 X71 | γ-Glu-Met 3 | 4 | X177 | Met-Pro | 1 4 | X1414 | 5’-S-Acetyl-2’-deoxy-5’-thiouridine 3 | 4 | X1163 | PC(16:1/2:0) or an isomer | 4 |
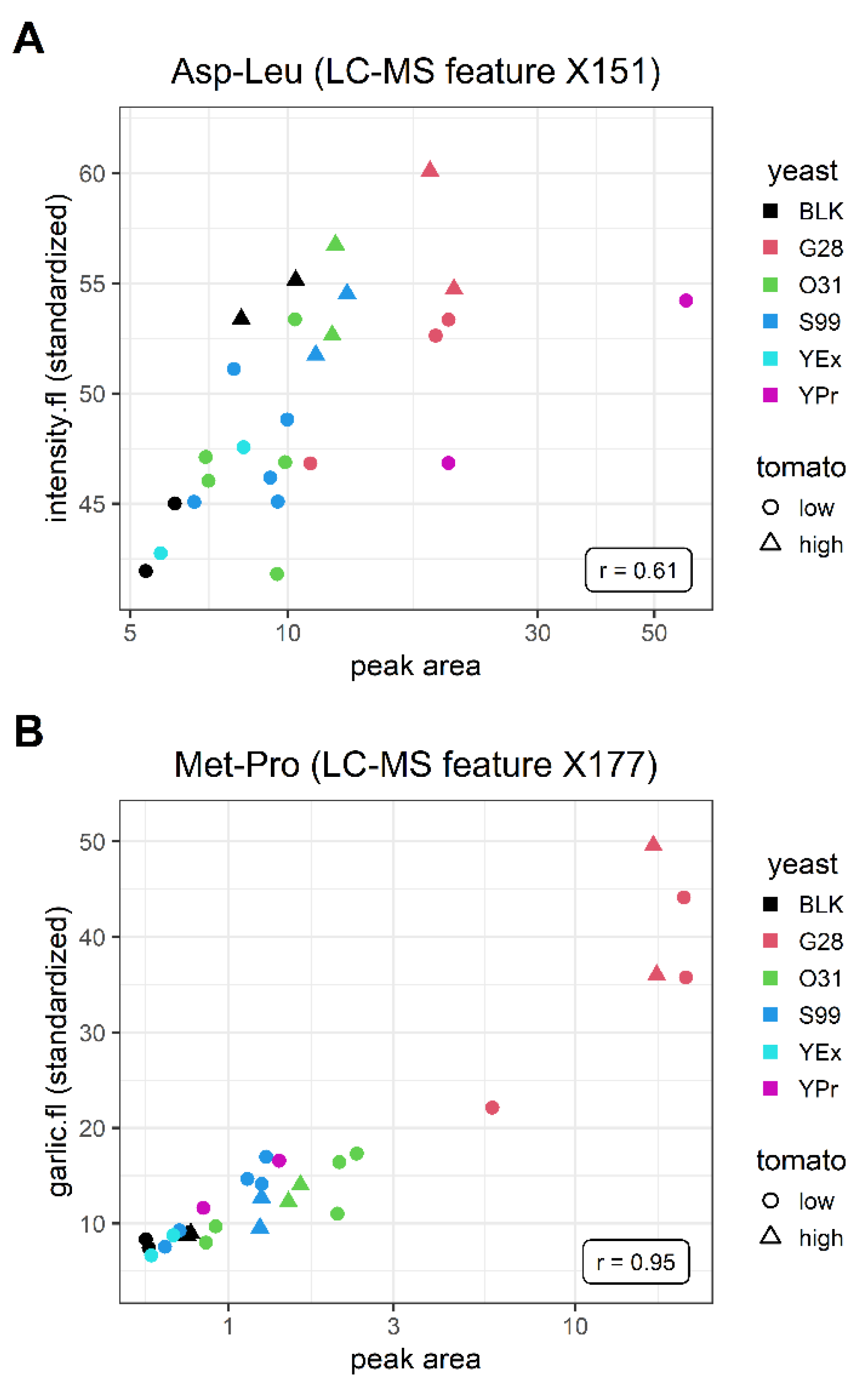
| 2 | X1317 | γ-Glu-Val | 4 | X141 | N-monopropionyl-cystine | 4 | X1650 | Ribose-isoleucine product | 4 | X151 | Asp-Leu | 1 4 |
| 3 | X1544 X280 | γ-Glu-Leu or -Ile 3 | 3 | X36 | Isomer of S-allyl-Cys | 3 5 | X153 | N2,N2-Dimethylguanosine | 4 | X1787 | Gln-Gln-His-His | 4 |
| 4 | X28 | γ-Glu-aminopropiononitrile | 4 | X152 | S-(allylthio)-Cys | 4 | X2033 | Gln-Val-Lys-Glu-Leu | 5 | X2223 | Feruloyltyramine | 4 |
| 5 | X216 | methyl xanthine derivative | 4 | X1908 X663 | Modified Ala-Ala peptide 3 | 4 | X893 | Ala-Ala-Pro-Val-Ala-Ala-Lys | 5 | X418 | Tetrahydro-1-methyl-beta-carboline-3-carboxylic acid (MTCA) | 4 |
| Onion Flavor | Garlic Flavor | Umami Flavor | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Feature | Putative Identification/Annotated Formula | Id. Level 1 | Feature | Putative Identification/Annotated Formula | Id. Level 1 | Feature | Putative Identification/Annotated Formula | Id. Level 1 |
| 1 | SPME362 | 3-Methylthiophene | 2 | SPME5189 | C8H12N2 | 3 | SPME3741 | 3-Carene | 2 |
| 2 | SPME3540 | Trimethylpyrazine | 2 | SPME2617 | C6H16N2 | 3 | SPME8427 | Δ-Elemene | 2 |
| 3 | SPME2617 | C6H16N2 | 3 | SPME6357 | C8H10N2O | 3 | SPME8624 | α-Cubebene | 2 |
| 4 | SPME4906 | C8H8O2 | 3 | SPME6926 | C6H8S2 | 4 | SPME4062 | D-Limonene | 1 |
| 5 | SPME2106 | C4H6O | 4 | SPME8203 | Di-allyl-trisulfide | 2 | SPME4134 | β-Phellandrene | 2 |
| Intensity Flavor | Intensity Odor | Tomato Odor | |||||||
| No. | Feature | Putative Identification/Annotated Formula | Id. Level 1 | Feature | Putative Identification/Annotated Formula | Id. Level 1 | Feature | Putative Identification/Annotated Formula | Id. Level 1 |
| 1 | SPME8112 | C13H28 | 4 | SPME8112 | 3-Methylthiophene | 2 | SPME5189 | C8H12N2 | 3 |
| 2 | SPME7856 | α-Ethylidene-benzeneacetaldehyde | 2 | SPME7856 | C4H6O | 4 | SPME1092 | C7H12O | 3 |
| 3 | SPME4788 | 4-Methyl-benzaldehyde, | 2 | SPME4788 | C6H16N2 | 3 | SPME7223 | C5H7BrO | 4 |
| 4 | SPME5834 | C11H20 | 4 | SPME5834 | 2-Methyl-2-butenal | 2 | SPME7181 | C3H4S3 | 4 |
| 5 | SPME3115 | C8H14O2 | 3 | SPME3115 | Dimethyl-disulfide | 1 | SPME4823 | 3-Ethyl-2,5-dimethylpyrazine | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leygeber, S.; Grossmann, J.L.; Diez-Simon, C.; Karu, N.; Dubbelman, A.-C.; Harms, A.C.; Westerhuis, J.A.; Jacobs, D.M.; Lindenburg, P.W.; Hendriks, M.M.W.B.; et al. Flavor Profiling Using Comprehensive Mass Spectrometry Analysis of Metabolites in Tomato Soups. Metabolites 2022, 12, 1194. https://doi.org/10.3390/metabo12121194
Leygeber S, Grossmann JL, Diez-Simon C, Karu N, Dubbelman A-C, Harms AC, Westerhuis JA, Jacobs DM, Lindenburg PW, Hendriks MMWB, et al. Flavor Profiling Using Comprehensive Mass Spectrometry Analysis of Metabolites in Tomato Soups. Metabolites. 2022; 12(12):1194. https://doi.org/10.3390/metabo12121194
Chicago/Turabian StyleLeygeber, Simon, Justus L. Grossmann, Carmen Diez-Simon, Naama Karu, Anne-Charlotte Dubbelman, Amy C. Harms, Johan A. Westerhuis, Doris M. Jacobs, Peter W. Lindenburg, Margriet M. W. B. Hendriks, and et al. 2022. "Flavor Profiling Using Comprehensive Mass Spectrometry Analysis of Metabolites in Tomato Soups" Metabolites 12, no. 12: 1194. https://doi.org/10.3390/metabo12121194
APA StyleLeygeber, S., Grossmann, J. L., Diez-Simon, C., Karu, N., Dubbelman, A.-C., Harms, A. C., Westerhuis, J. A., Jacobs, D. M., Lindenburg, P. W., Hendriks, M. M. W. B., Ammerlaan, B. C. H., van den Berg, M. A., van Doorn, R., Mumm, R., Hall, R. D., Smilde, A. K., & Hankemeier, T. (2022). Flavor Profiling Using Comprehensive Mass Spectrometry Analysis of Metabolites in Tomato Soups. Metabolites, 12(12), 1194. https://doi.org/10.3390/metabo12121194








