Comparative Transcriptome Analysis of MeJA Responsive Enzymes Involved in Phillyrin Biosynthesis of Forsythia suspensa
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Determination of Phillyrin Content in F. suspensa Fruits and Leaves
2.3. RNA Extraction, cDNA Library Construction and Illumina Sequencing
2.4. Transcriptome Assembly and Annotation
2.5. Differentially Expressed Genes (DEGs) Analysis
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Statistical Analyses
3. Results
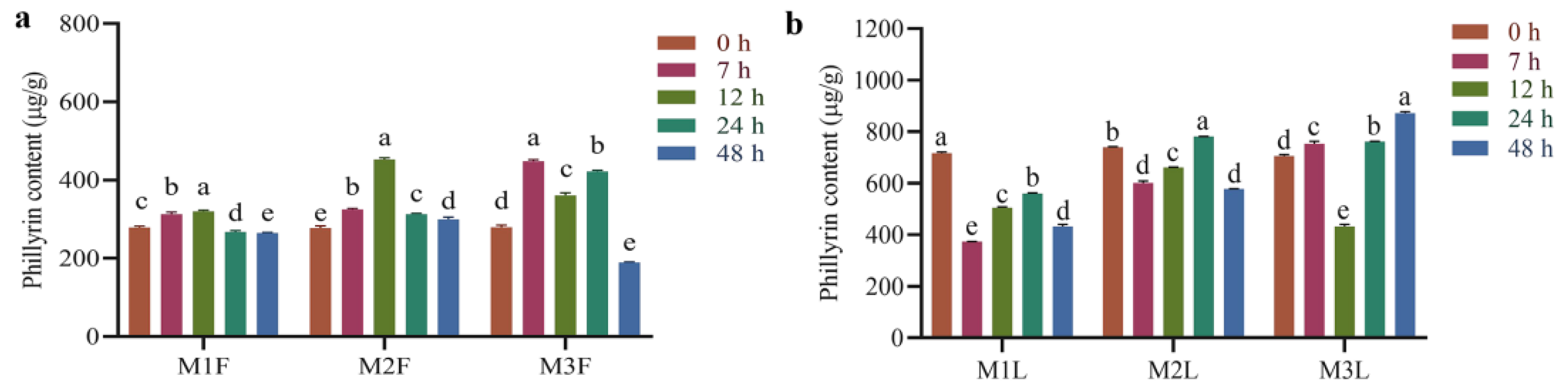
3.1. Effect of MeJA Induction on the Content of Phillyrin
3.2. Illumina Sequencing and De Novo Assembly
3.3. Functional Annotation of Unigenes
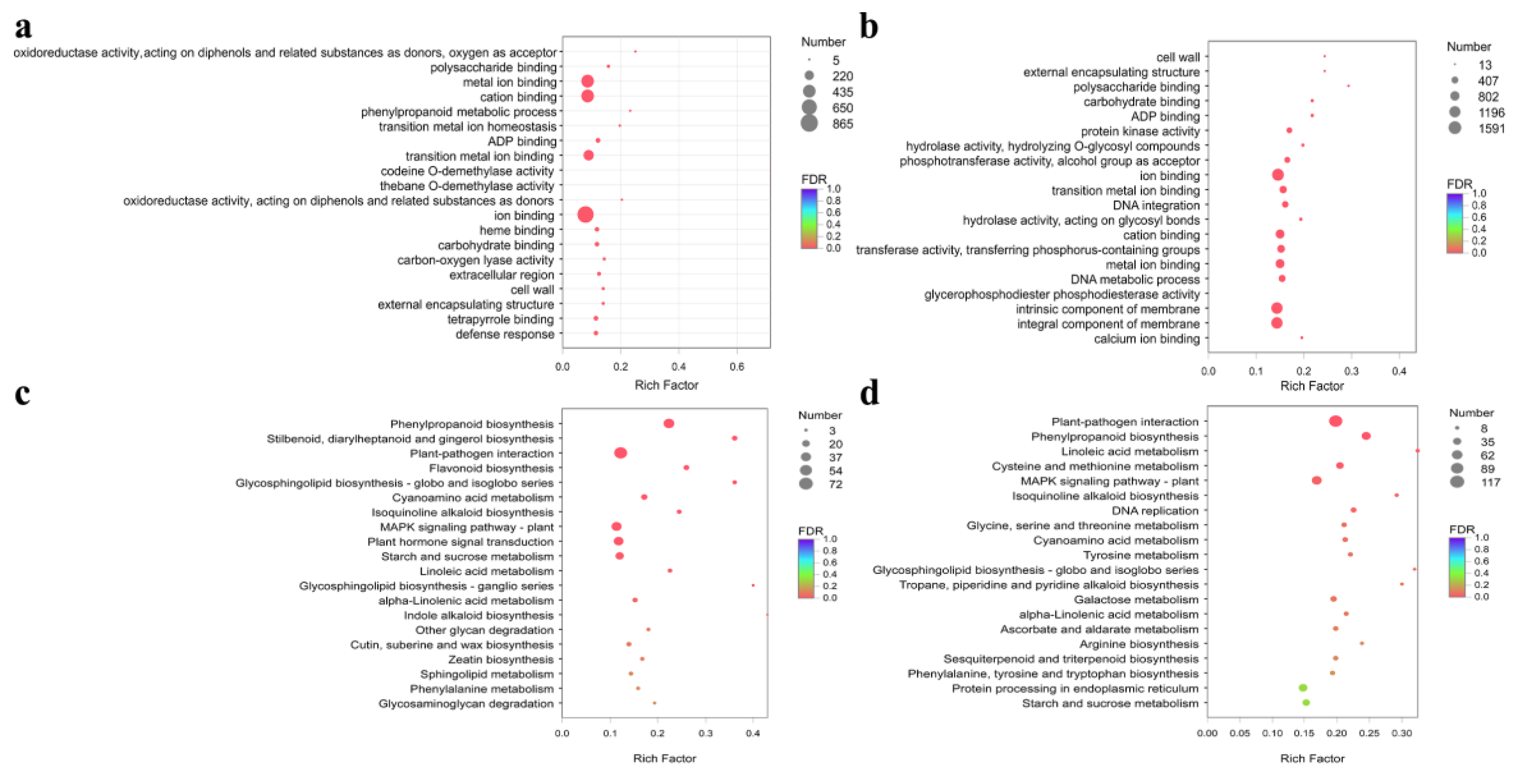
3.4. Analysis of Differentially Expressed Unigenes (DEGs)
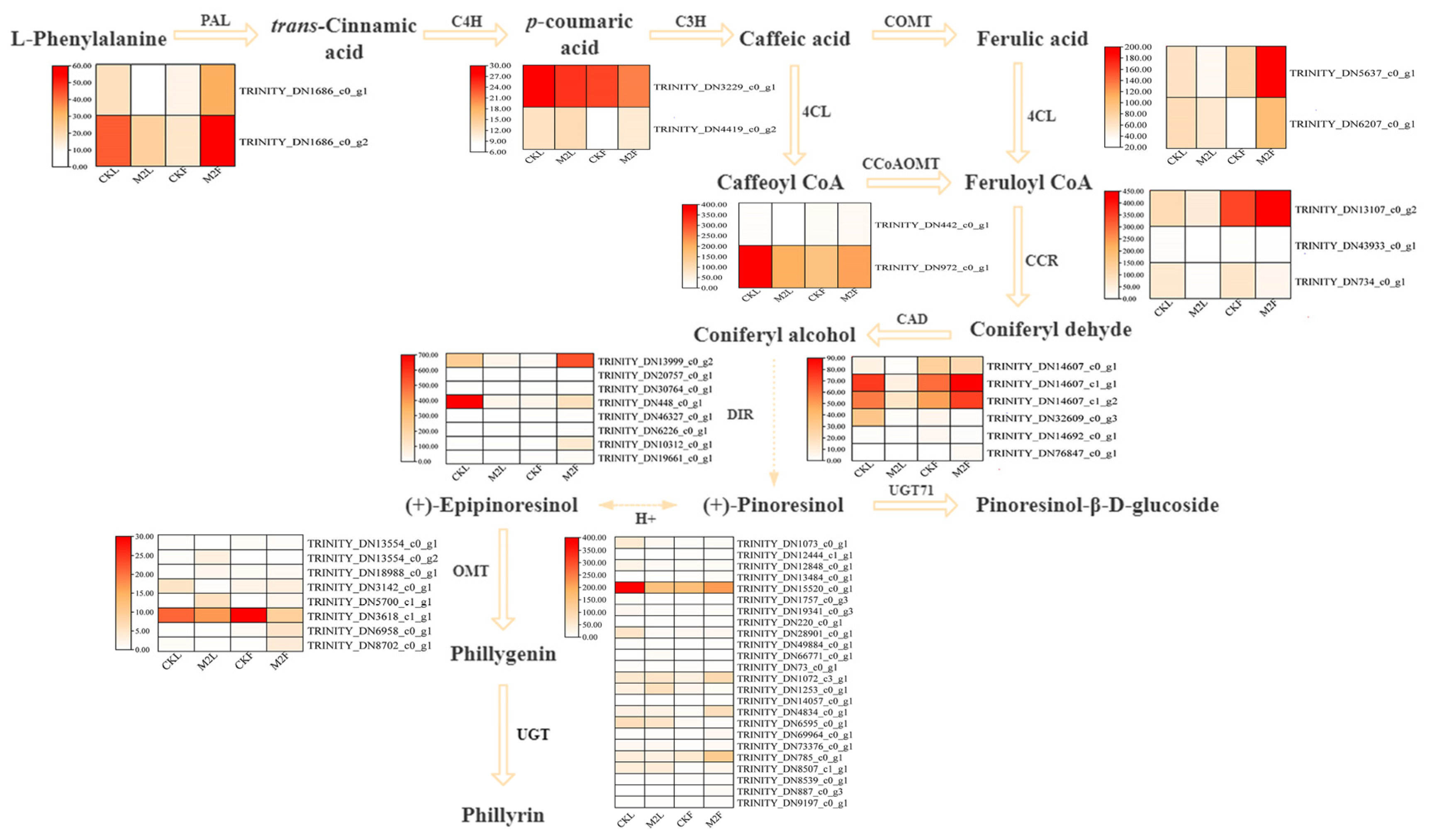
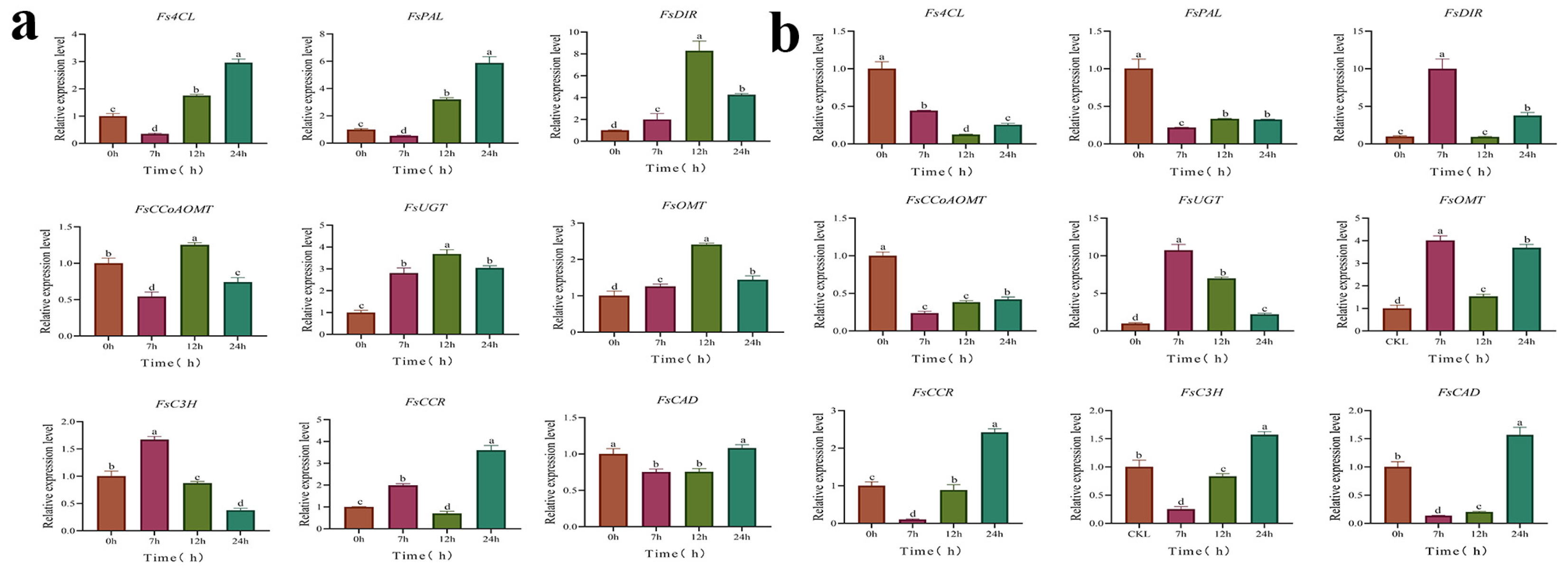
3.5. Analysis of DEGs Related to Phillyrin Biosynthetic Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, E.Q.; Ai, X.; Zang, S.; Guan, T.; Xu, X.; Li, H. Ultrasound-assisted extraction of phillyrin from Forsythia suspensa. Ultrason. Sonochem 2011, 18, 549–552. [Google Scholar] [CrossRef]
- Li, H.; Chen, F. Preparative isolation and purification of phillyrin from the medicinal plant Forsythia suspensa by high-speed counter-current chromatography. J. Chromatogr. A 2005, 1083, 102–105. [Google Scholar] [CrossRef]
- Zhou, W.; Di, L.; Shan, J.; Bi, X.; Chen, L.; Wang, L. Intestinal absorption of forsythoside A in different compositions of Shuang–Huang–Lian. Fitoterapia 2010, 82, 375–382. [Google Scholar] [CrossRef]
- Hu, K.; Guan, W.; Bi, Y.; Zhang, W.; Li, L.; Zhang, B.; Liu, Q.; Song, Y.; Li, X.; Duan, Z.; et al. Efficacy and safety of Lianhua Qingwen capsules, a repurposed Chinese herb, in patients with Coronavirus disease 2019: A multicenter, prospective, randomized controlled trial [Phytomedicine 85 (2021) 153242]. Phytomedicine 2022, 94, 153800. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gao, Y.; Yuan, Y.; Liu, M.; Gao, Y.; Yuan, Y.; Yang, K.; Shi, S.; Tian, J.; Zhang, J. Efficacy and safety of herbal medicine (Lianhuaqingwen) for treating COVID-19: A systematic review and meta-analysis. Integr. Med. Res. 2021, 10, 100644. [Google Scholar] [CrossRef] [PubMed]
- Bu, Y.; Shi, T.; Meng, M.; Bu, Y.; Shi, T.; Meng, M.; Kong, G.; Tian, Y.; Chen, Q.; Yao, X.; et al. A novel screening model for the molecular drug for diabetes and obesity based on tyrosine phosphatase Shp2. Bioorg. Med. Chem. Lett. 2011, 21, 874–878. [Google Scholar] [CrossRef]
- Xue, H.; Yuan, W. Review on Pharmacological Research of Forsythia Suspensa Leaves. Lishizhen Med. Mater. Med. Res. 2009, 20, 1149–1150. [Google Scholar]
- Yuan, J.; Qiu, Z.; Liu, J.L.; Liu, H.; Zhang, S.L. Processing techniques of Forsythia Suspensa leaves green tea and analysis of nutrient active ingredients. J. Henan Univ. Sci. Technol. 2015, 36, 78–82. [Google Scholar]
- Dong, Z.; Lu, X.; Tong, X.; Dong, Y.; Tang, L.; Liu, M. Forsythiae Fructus: A Review on its Phytochemistry, Quality Control, Pharmacology and Pharmacokinetics. Molecules 2017, 22, 1466. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Koyama, T.; Bahabadi, S.E. Essences in metabolic engineering of lignan biosynthesis. Metabolites 2015, 5, 270–290. [Google Scholar] [CrossRef]
- Du, Y.; You, L.; Ni, B.; Sai, N.; Wang, W.; Sun, M.; Xu, R.; Yao, Y.; Zhang, Z.; Qu, C.; et al. Phillyrin Mitigates Apoptosis and Oxidative Stress in Hydrogen Peroxide-Treated RPE Cells through Activation of the Nrf2 Signaling Pathway. Oxidative Med. Cell. Longev. 2020, 2020, 2684672. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, R.; Pan, W.; Huang, W.; Liu, B.; Xie, Y.; Wang, Z.; Li, C.; Jiang, H.; Huang, J.; et al. Phillyrin (KD-1) exerts anti-viral and anti-inflammatory activities against novel coronavirus (SARS-CoV-2) and human coronavirus 229E (HCoV-229E) by suppressing the nuclear factor kappa B (NF-kappaB) signaling pathway. Phytomedicine 2020, 78, 153296. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Saadeldeen, F.S.A.; Xu, L.; Zhao, Y.; Wei, J.; Wang, D.H.M.; Liu, Z.; Kang, W. The Mechanism of Phillyrin from the Leaves of Forsythia suspensa for Improving Insulin Resistance. Biomed. Res. Int. 2019, 2019, 3176483. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, S.; Ma, Y.; Zhang, T.; Zhao, G. Research progress in metabolic engineering and synthetic biology for natural lignan production. Chin. Tradit. Herb. Drugs 2016, 47, 2556–2562. [Google Scholar]
- Rahman, M.M.A.; Dewick, P.M.; Jackson, D.E.; Lucas John, A. Biosynthesis of lignans in Forsythia intermedia. Phytochemistry 1990, 29, 1841–1846. [Google Scholar] [CrossRef]
- Fukasawa, A.T.; Kung, S.D.; Watson, J.C. Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Mol. Biol. 1996, 30, 711–722. [Google Scholar] [CrossRef]
- Gertsch, J.; Tobler, R.T.; Brun, R.; Sticher, O.; Heilmann, J. Antifungal, Antiprotozoal, Cytotoxic and Piscicidal Properties of Justicidin B and a New Arylnaphthalide Lignan from Phyllanthus piscatorum. Planta Med. 2003, 69, 420–424. [Google Scholar]
- Kumar, A.; Ellis, B.E. 4-Coumarate:CoA ligase gene family in Rubus idaeus: cDNA structures, evolution, and expression. Plant Mol. Biol. 2003, 31, 327–340. [Google Scholar] [CrossRef]
- Anterola, A.; Lewis, N. Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 2002, 61, 221–294. [Google Scholar] [CrossRef]
- Guo, D.; Chen, F.; Inoue, K.; Blount, J.W.; Dixon, R.A. Downregulation of caffeic acid 3-0-methyltransferase and caffeoyl CoA 3-0-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell Online 2001, 13, 73. [Google Scholar] [CrossRef]
- Luderitz, T.; Grisebach, H. Enzymic Synthesis of Lignin Precursors Comparison of Cinnamoyl-CoA Reductase and Cinnamyl Alcohol: NADP iDehydrogenase from Spruce (Picea abies L.) and Soybean (GZycine max L.). Eur. J. Biochem. 1981, 119, 115–124. [Google Scholar] [CrossRef]
- Davin, L.B.; Wang, H.B.; Crowell, A.L.; Davin, L.B.; Wang, H.B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective bimo-lecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 1997, 275, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhang, S.; He, Z.; He, y.; He, S.; Liu, L.; Sun, X.; Li, Q. Comparative transcriptome analyses identify genes involved into the biosynthesis of forsythin and forsythoside A in Forsythia suspensa. Funct. Integr. Genom. 2022, 22, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Berim, A.; Ebel, R.; Schneider, B.; Petersen, M. UDP-glucose:(6-methoxy)podophyllotoxin 7-O-glucosyltransferase from suspension cultures of Linum nodiflorum. Phytochemistry 2008, 69, 374–381. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Verpoorte, R.; Heijden, R.V.D.; Hoge, J.H.C.; Hoopen, T. Plant cell biotechnology for the production of secondary metabolites. Pure Appl. Chem. 1994, 66, 2307–2310. [Google Scholar] [CrossRef]
- Zhao, J.; Zhu, W.H.; Hu, Q. Selection of fungal elicitors to increase indole alkaloid accumulation in Catharanthus roseus suspension cell culture. Enzym. Microb. Technol. 2001, 28, 666–672. [Google Scholar] [CrossRef]
- Diallo, A.O.; Agharbaoui, Z.; Badawi, M.A.; Ali-Benali, M.A.; Moheb, A.; Houde, M.; Sarhan, F. Transcriptome analysis of an mvp mutant reveals important changes in global gene expression and a role for methyl jasmonate in vernalization and flowering in wheat. J. Exp. Bot. 2014, 65, 2271–2286. [Google Scholar] [CrossRef]
- Misra, R.C.; Maiti, P.; Chanotiya, C.S.; Shanker, K.; Ghosh, S. Methyl jasmonate-elicited transcriptional responses and pentacyclic triterpene biosynthesis in sweet basil. Plant Physiol. 2014, 164, 1028–1044. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic. Acids Res. 2005, 33, D501–D504. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic. Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic. Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Dewey, C.N.; Bo, L. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shen, J.; Wu, Y.; Jiang, Z.; Xu, Y.; Zheng, T.; Wang, J.; Cheng, T.; Zhang, Q.; Pan, H. Selection and validation of appropriate reference genes for gene expression studies in Forsythia. Physiol. Mol. Biol. Plants 2020, 26, 173–188. [Google Scholar] [CrossRef]
- Lewis, N.G.; Davin, L.B.; Sarkanen, S. Lignin and Lignan Biosynthesis: Distinctions and Reconciliations. ACS Symp. 1998, 697, 1–27. [Google Scholar]
- Zhou, C.; Lu, M.; Cheng, J.; Rohani, E.R.; Hamezah, H.S.; Han, R.; Tong, X. Review on the Pharmacological Properties of Phillyrin. Molecules 2022, 27, 3670. [Google Scholar] [CrossRef] [PubMed]
- Moharramnejad, S.; Azam, A.T.; Panahandeh, J.; Dehghanian, Z.; Ashraf, M. Effect of Methyl Jasmonate and Salicylic Acid on In Vitro Growth, Stevioside Production, and Oxidative Defense System in Stevia rebaudiana. Sugar Tech 2019, 21, 1031–1038. [Google Scholar]
- Satake, H.; Ono, E.; Murata, J. Recent advances in the metabolic engineering of lignan biosynthesis pathways for the production of transgenic plant-based foods and supplements. J. Agric. Food Chem. 2013, 61, 11721–11729. [Google Scholar] [CrossRef]
- Malik, S.; Biba, O.; Grúz, J.; Arroo, R.R.J.; Strnad, M. Biotechnological approaches for producing aryltetralin lignans from Linum species. Phytochem. Rev. 2014, 13, 893–913. [Google Scholar] [CrossRef]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef] [PubMed]
- Berim, A.; Spring, O.; Conrad, J.; Maitrejean, M.; Boland, W.; Petersen, M. Enhancement of lignan biosynthesis in suspension cultures of Linum nodiflorum by coronalon, indanoyl-isoleucine and methyl jasmonate. Planta 2005, 222, 769–776. [Google Scholar] [CrossRef]
- Wang, J.; Fan, S.; Li, A.; Chen, T.; Sun, B.; Yang, X. Analysis of the Forsythin and Forsythiaside A in different parts of Forsythia suspensa and its medicinal use discussion. Mod. Chin. Med. 2013, 15, 556–559. [Google Scholar]
- Ye, H.; Wu, Z.; Li, F.R. Problems and thoughts of folium Forsythia used for medical purpose. Chin. J. Clin. Ration. Drug Use 2011, 4, 74–75. [Google Scholar]
- Vanholme, R.; Demedts, B.; Morreel, K.; John, R.; Wout, B. (2010) Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.; Deng, K.; He, X.; Zhan, R.; Tang, L. Methyl Jasmonate affects metabolism and gene transcription of volatile terpenoids from Amomum villosum Lour. Mod. Tradit. Chin. Med. Mater. Med.-World Sci. Technol. 2014, 16, 1528–1536. [Google Scholar]
- Sun, B.; Wang, P.; Wang, R.; Li, Y.; Xu, S. Molecular Cloning and Characterization of a meta/para-O-Methyltransferase from Lycoris aurea. Int. J. Mol. Sci. 2018, 19, 1911. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Xu, J.; Wei, J.; Sun, J.; Xu, Y.; Yang, X.; Zhang, Y.; Liu, J.; Sui, C. Expression analysis of glycosyltransferase BcUGT1 from Bupleurum chinense DC. and its expression in E. coli and the target protein purification. Yao Xue Xue Bao = Acta Pharm. Sin. 2013, 48, 1345–1352. [Google Scholar]



| Type | Unigene |
|---|---|
| Total number | 87,564 |
| Total base | 87,582,604 |
| Largest length (bp) | 15,856 |
| Smallest length (bp) | 201 |
| Average length (bp) | 1000.21 |
| GC percent (%) | 44 |
| N50 average length (bp) | 1851 |
| Q30 (%) | 94.55 |
| Q20 (%) | 98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhang, J.; Liu, H.; Shang, H.; Zhao, X.; Xu, H.; Zhang, H.; Hou, D. Comparative Transcriptome Analysis of MeJA Responsive Enzymes Involved in Phillyrin Biosynthesis of Forsythia suspensa. Metabolites 2022, 12, 1143. https://doi.org/10.3390/metabo12111143
Liu X, Zhang J, Liu H, Shang H, Zhao X, Xu H, Zhang H, Hou D. Comparative Transcriptome Analysis of MeJA Responsive Enzymes Involved in Phillyrin Biosynthesis of Forsythia suspensa. Metabolites. 2022; 12(11):1143. https://doi.org/10.3390/metabo12111143
Chicago/Turabian StyleLiu, Xiaoran, Jiaqi Zhang, Hao Liu, Huixiang Shang, Xingli Zhao, Huawei Xu, Hongxiao Zhang, and Dianyun Hou. 2022. "Comparative Transcriptome Analysis of MeJA Responsive Enzymes Involved in Phillyrin Biosynthesis of Forsythia suspensa" Metabolites 12, no. 11: 1143. https://doi.org/10.3390/metabo12111143
APA StyleLiu, X., Zhang, J., Liu, H., Shang, H., Zhao, X., Xu, H., Zhang, H., & Hou, D. (2022). Comparative Transcriptome Analysis of MeJA Responsive Enzymes Involved in Phillyrin Biosynthesis of Forsythia suspensa. Metabolites, 12(11), 1143. https://doi.org/10.3390/metabo12111143








