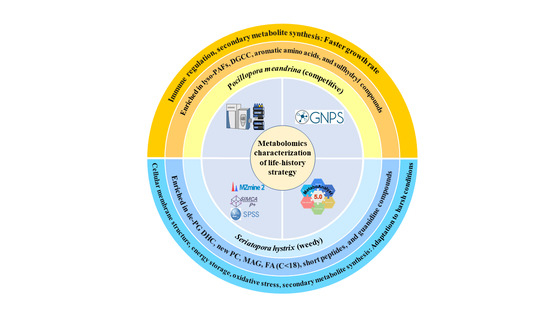
Metabolomics Characterization of Scleractinia Corals with Different Life-History Strategies: A Case Study about Pocillopora meandrina and Seriatopora hystrix in the South China Sea
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Collection
2.2. Metabolites Extraction from Coral Fragments
2.3. Mass Spectrometry Data Collection and Pre-Processing
2.4. Statistical Analysis
2.5. Molecular Network Analysis and Metabolic Pathway Analysis
2.6. Data Quality Control
3. Results
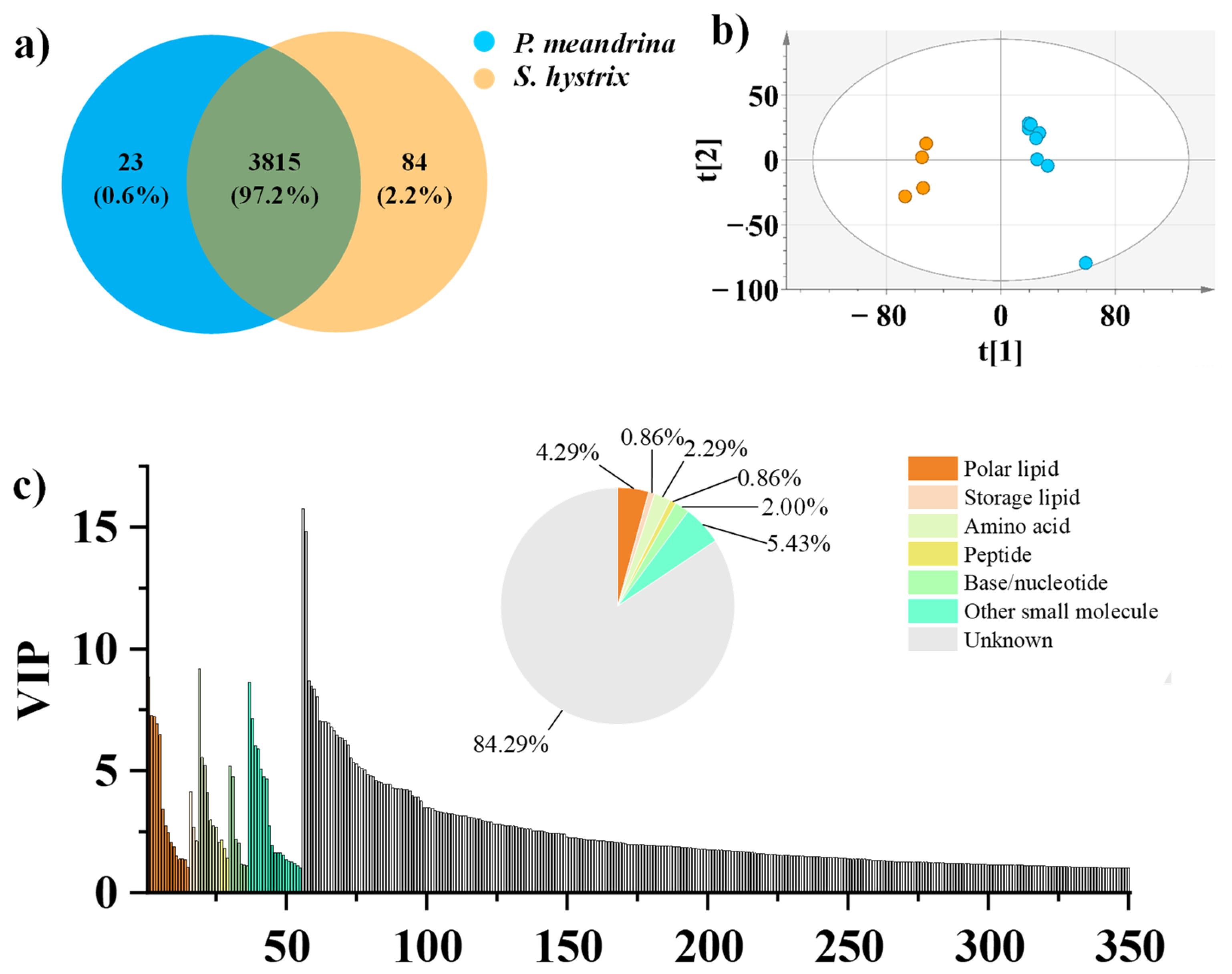
3.1. Chemical Diversity and Multivariate Statistical Analysis of the Metabolites of P. meandrina and S. hystrix
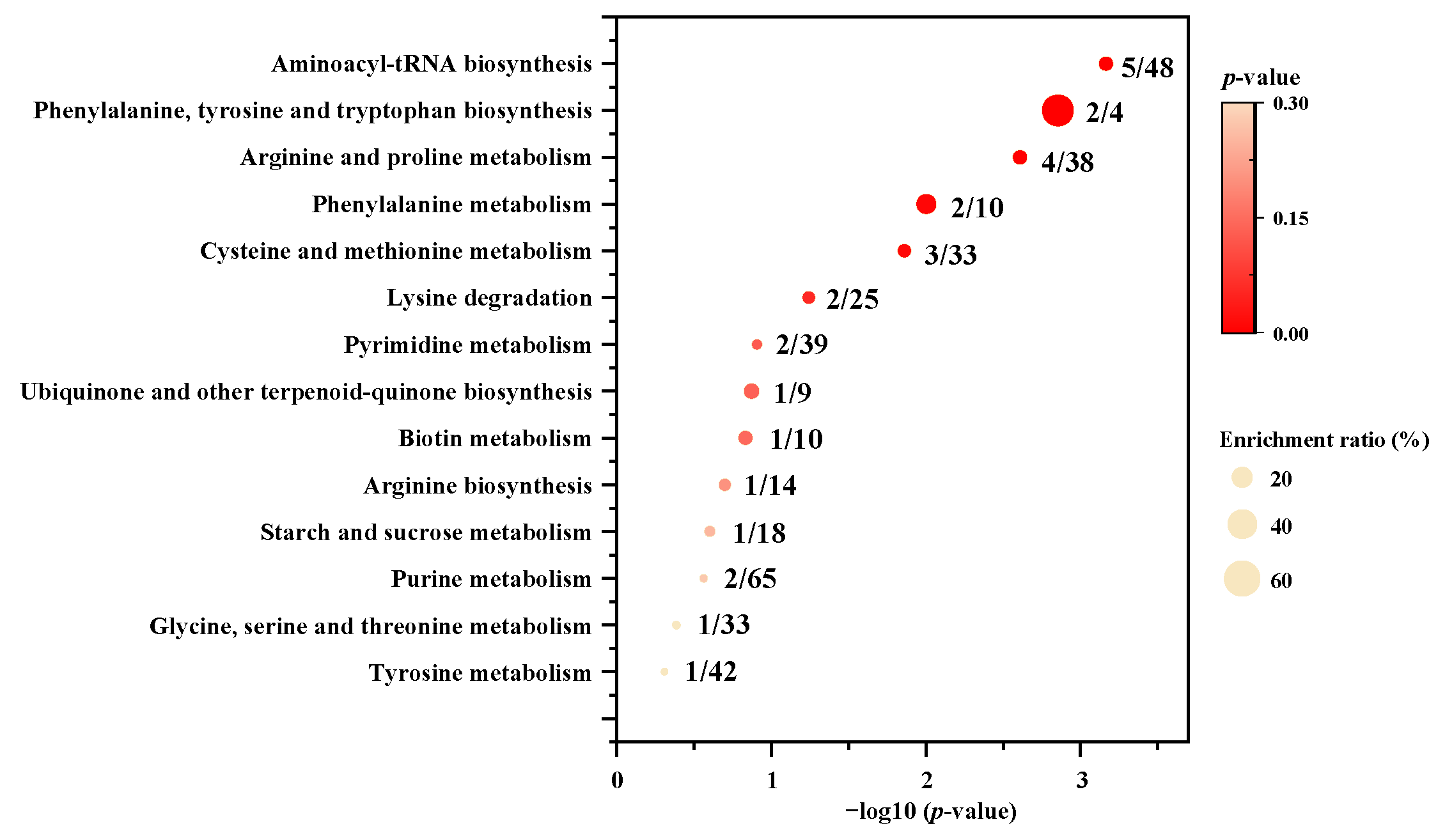
3.2. Identification of the Differential Metabolites between P. meandrina and S. hystrix
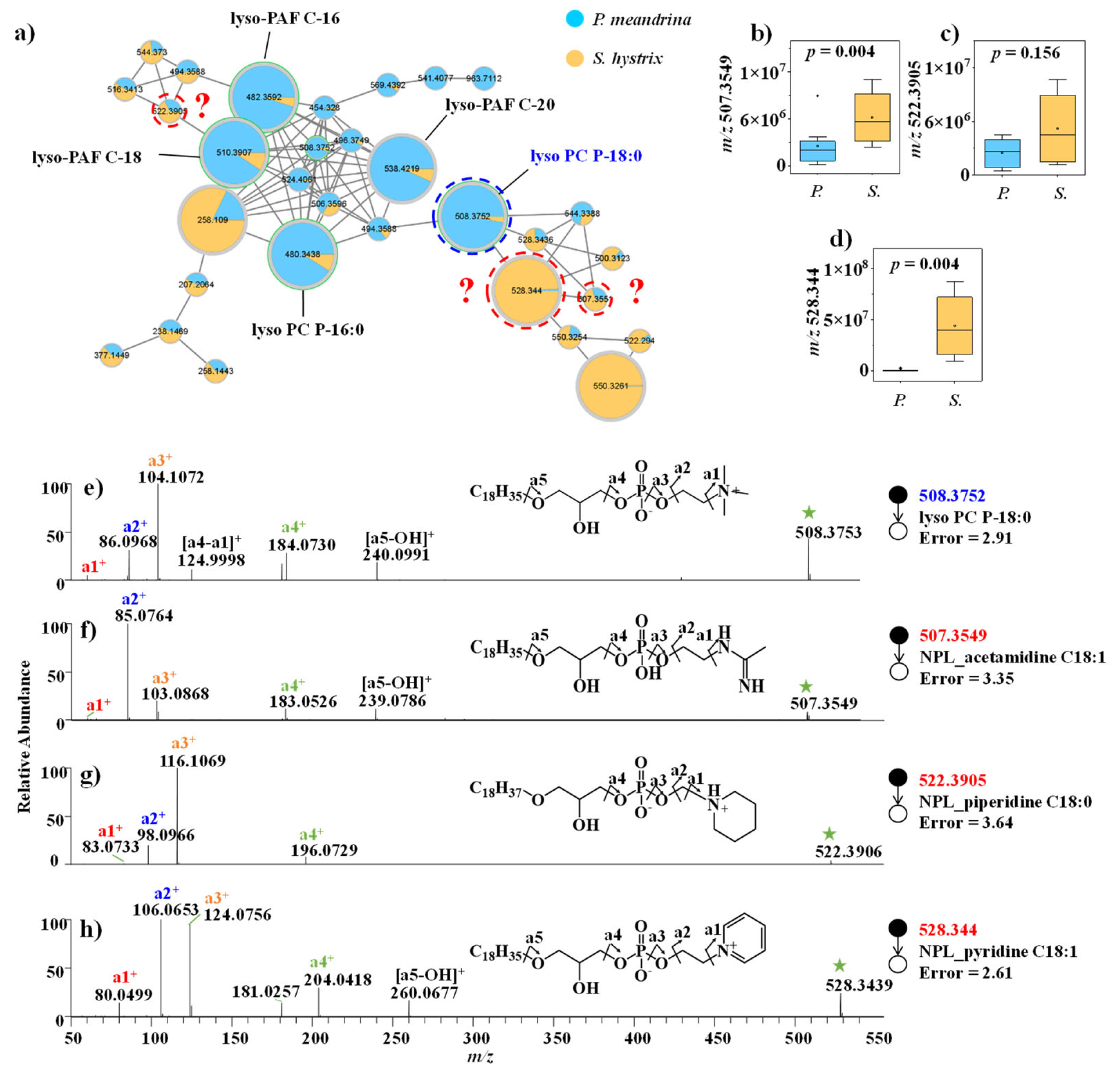
3.2.1. Lipid and FA
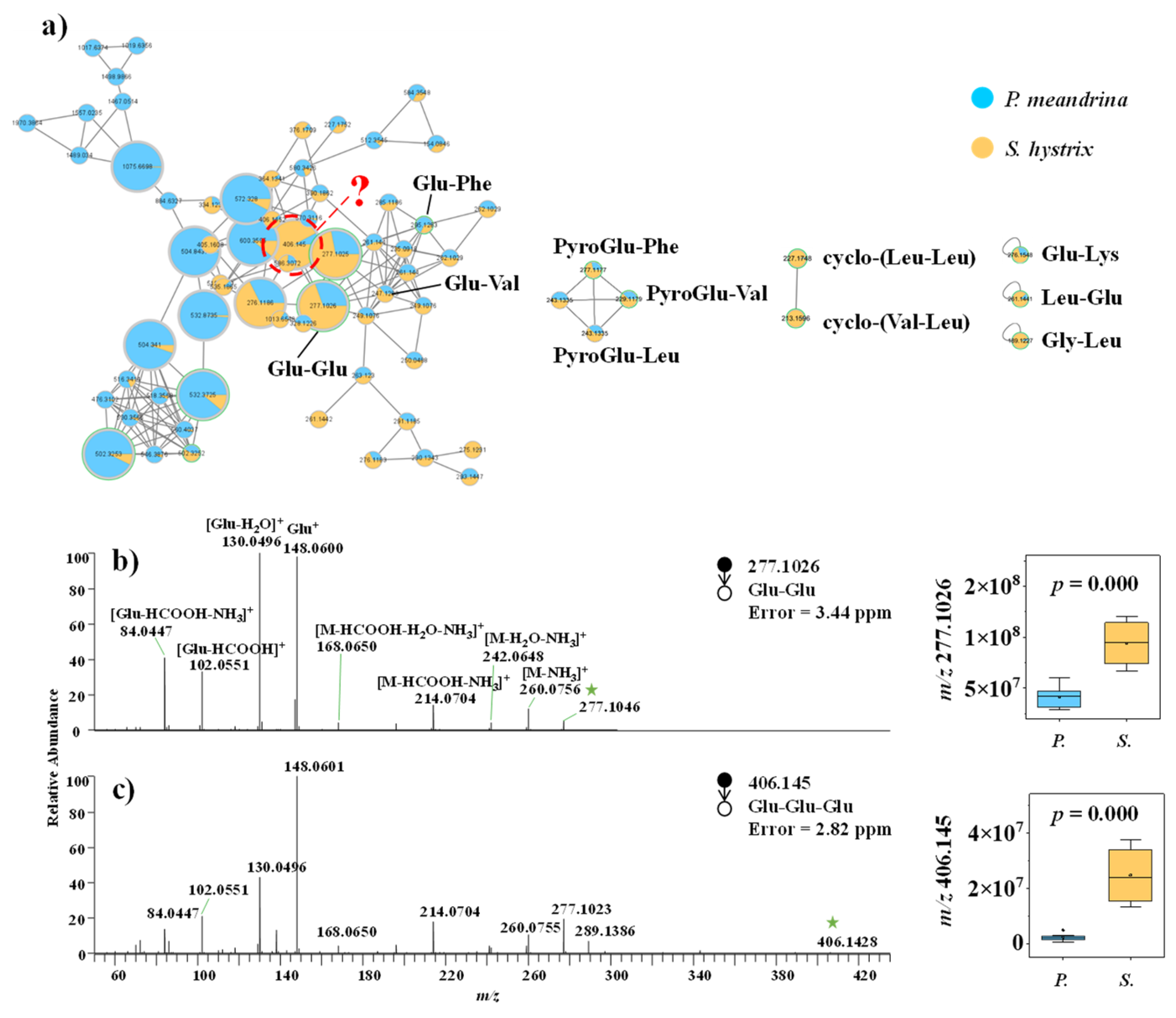
3.2.2. Small Peptide
3.2.3. Other Small Molecules
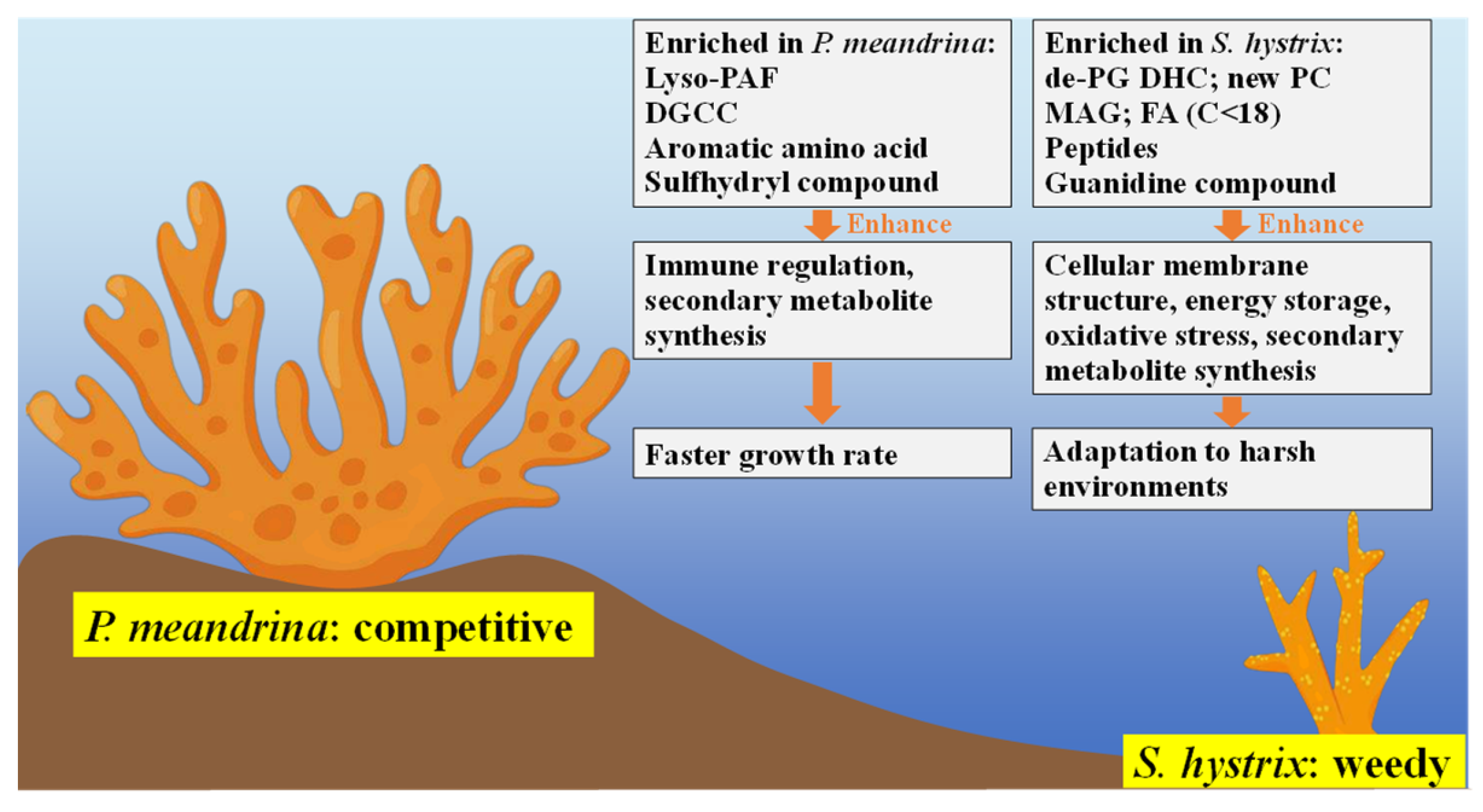
4. Discussion
4.1. Regulation of Immune Response, Cellular Membrane Structure and Energy Metabolism by Lipids and FAs
4.2. Resistance to Various Environmental and Biological Stresses by Small Peptides
4.3. Contributions of Other Small Molecule Metabolites on the Formation of Coral’s Life-History Traits
5. Conclusions and Implications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Darling, E.S.; McClanahan, T.R.; Cote, I.M. Life histories predict coral community disassembly under multiple stressors. Global Chang. Biol. 2013, 19, 1930–1940. [Google Scholar] [CrossRef] [PubMed]
- Darling, E.S.; Alvarez-Filip, L.; Oliver, T.A.; McClanahan, T.R.; Cote, I.M. Evaluating life-history strategies of reef corals from species traits. Ecol. Lett. 2012, 15, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.J.; Kuffner, I.B.; Walters, L.J.; Ritson-Williams, R.; Beach, K.S.; Becerro, M.A. Chemically mediated interactions between macroalgae Dictyota spp. and multiple life-history stages of the coral Porites astreoides. Mar. Ecol. Prog. Ser. 2011, 426, 161–170. [Google Scholar] [CrossRef]
- Pinzon, C.J.H.; Dornberger, L.; Beach-Letendre, J.; Weil, E.; Mydlarz, L.D. The link between immunity and life history traits in scleractinian corals. PeerJ 2014, 2, e628. [Google Scholar] [CrossRef]
- Leong, R.C.; Marzinelli, E.M.; Low, J.; Bauman, A.G.; Lim, E.W.X.; Lim, C.Y.; Steinberg, P.D.; Guest, J.R. Effect of coral-algal interactions on early life history processes in Pocillopora acuta in a highly disturbed coral reef system. Front. Mar. Sci. 2018, 5, 1–11. [Google Scholar] [CrossRef]
- Berry, K.L.E.; Hoogenboom, M.O.; Brinkman, D.L.; Burns, K.A.; Negri, A.P. Effects of coal contamination on early life history processes of a reef-building coral, Acropora tenuis. Mar. Pollut. Bull. 2017, 114, 505–514. [Google Scholar] [CrossRef]
- Fox, M.D.; Nelson, C.E.; Oliver, T.A.; Quinlan, Z.A.; Remple, K.; Glanz, J.; Smith, J.E.; Putnam, H.M. Differential resistance and acclimation of two coral species to chronic nutrient enrichment reflect life-history traits. Funct. Ecol. 2021, 35, 1081–1093. [Google Scholar] [CrossRef]
- McClanahan, T.R. Coral community life histories and population dynamics driven by seascape bathymetry and temperature variability. Adv. Mar. Biol. 2020, 87, 291–330. [Google Scholar]
- Roth, M.S.; Fan, T.Y.; Deheyn, D.D. Life history changes in coral fluorescence and the effects of light intensity on larval physiology and settlement in seriatopora hystrix. PLoS ONE 2013, 8, e59476. [Google Scholar]
- Hughes, T.P.; Huang, H.; Young, M.A.L. The wicked problem of china’s disappearing coral reefs. Conserv. Biol. 2013, 27, 261–269. [Google Scholar] [CrossRef]
- Wang, L.R.; Yu, K.F.; Zhao, H.T.; Zhang, Q.M. Economic valuation of the coral reefs in South China Sea. Tropical Geography 2014, 34, 44–49. [Google Scholar]
- Chen, X.Y.; Yu, K.F.; Huang, X.Y.; Wang, Y.H.; Liao, Z.H.; Zhang, R.J.; Yao, Q.C.; Wang, J.K.; Wang, W.H.; Tao, S.C.; et al. Atmospheric nitrogen deposition increases the possibility of macroalgal dominance on remote coral reefs. J. Geophys. Res-Biogeo. 2019, 124, 1355–1369. [Google Scholar] [CrossRef]
- Zhao, M.X.; Yu, K.F.; Shi, Q.; Yang, H.; Riegl, B.; Zhang, Q.; Yan, H.; Chen, T.; Liu, G.; Lin, Z. The coral communities of Yongle atoll: Status, threats and conservation significance for coral reefs in South China Sea. Mar. Freshwater Res. 2016, 67, 1888–1896. [Google Scholar] [CrossRef]
- Zhao, M.X.; Yu, K.F.; Shi, Q.; Chen, T.R.; Zhang, H.L.; Chen, T.G. Coral communities of the remote atoll reefs in the Nansha Islands, southern South China Sea. Environ. Monit. Assess. 2013, 185, 7381–7392. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N.; Brush, E.G.; Dilley, E.R.; Hixon, M.A. Autumn coral bleaching in Hawai’i. Mar. Ecol. Prog. Ser. 2021, 675, 199–205. [Google Scholar] [CrossRef]
- Sinniger, F.; Morita, M.; Harii, S. “Locally extinct” coral species Seriatopora hystrix found at upper mesophotic depths in Okinawa. Coral Reefs 2013, 32, 153. [Google Scholar] [CrossRef][Green Version]
- Baird, A.H.; Guest, J.R.; Willis, B.L. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol. Evol. Systemat. 2009, 40, 551–571. [Google Scholar] [CrossRef]
- Dongen-Vogels, V.V.; Mallefet, J. Fragment growth-rates of six cultivated coral species: A reference framework for coral transplantation. La mer 2006, 44, 99–107. [Google Scholar]
- Roach, T.N.F.; Dilworth, J.; Martin, C.H.; Jones, A.D.; Quinn, R.A.; Drury, C. Metabolomic signatures of coral bleaching history. Nat. Ecol. Evol. 2021, 5, 495–503. [Google Scholar] [CrossRef]
- Williams, A.; Chiles, E.N.; Conetta, D.; Pathmanathan, J.S.; Cleves, P.A.; Putnam, H.M.; Su, X.Y.; Bhattacharya, D. Metabolomic shifts associated with heat stress in coral holobionts. Sci. Adv. 2021, 7, eabd4210. [Google Scholar] [CrossRef]
- Shen, X.T.; Wang, R.H.; Xiong, X.; Yin, Y.D.; Cai, Y.P.; Ma, Z.J.; Liu, N.; Zhu, Z.J. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 2019, 10, 1516. [Google Scholar] [CrossRef]
- Wang, M.X.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Duhrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods. 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Melnik, A.V.; Koyama, N.; Lu, X.W.; Schorn, M.; Fang, J.S.; Aguinaldo, K.; Lincecum, T.L.; Ghequire, M.G.K.; Carrion, V.J.; et al. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat. Microbiol. 2017, 2, 17010. [Google Scholar] [CrossRef]
- Qin, Z.J.; Yu, K.F.; Chen, S.C.; Chen, B.; Liang, J.Y.; Yao, Q.C.; Yu, X.F.; Liao, Z.H.; Deng, C.Q.; Liang, Y.T. Microbiome of juvenile corals in the outer reef slope and lagoon of the South China Sea: Insight into coral acclimatization to extreme thermal environments. Environ. Microbiol. 2021, 23, 4389–4404. [Google Scholar] [CrossRef]
- Xia, J.G.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- da Silva, R.R.; Wang, M.X.; Nothias, L.F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Farag, M.A.; Porzel, A.; Al-Hanimady, M.A.; Hegazy, M.E.F.; Meyer, A.; Mohamed, T.A.; Westphal, H.; Wessjohann, L.A. Soft corals biodiversity in the Egyptian red sea: A comparative MS and NMR metabolomics approach of wild and aquarium grown species. J. Proteome Res. 2016, 15, 1274–1287. [Google Scholar] [CrossRef]
- Quinn, R.A.; Vermeij, M.J.A.; Hartmann, A.C.; d’Auriac, I.G.; Benler, S.; Haas, A.; Quistad, S.D.; Lim, Y.W.; Little, M.; Sandin, S.; et al. Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. P. Roy. Soc. B-Biol. Sci. 2016, 283, 20160469. [Google Scholar] [CrossRef]
- Vance, J.E.; Vance, D.E. Metabolic insights into phospholipid function using gene-targeted mice. J. Biol. Chem. 2005, 280, 10877–10880. [Google Scholar] [CrossRef] [PubMed]
- d’Auriac, I.G.; Quinn, R.A.; Maughan, H.; Nothias, L.F.; Little, M.; Kapono, C.A.; Cobian, A.; Reyes, B.T.; Green, K.; Quistad, S.D.; et al. Before platelets: The production of platelet-activating factor during growth and stress in a basal marine organism. P. Roy. Soc. B-Biol. Sci. 2018, 285, 20181307. [Google Scholar]
- Quinn, P.J.; Joo, F.; Vigh, L. The role of unsaturated lipids in membrane structure and stability. Prog. Biophys. Mol. Biol. 1989, 53, 71–103. [Google Scholar] [CrossRef]
- Rosset, S.; Koster, G.; Brandsma, J.; Hunt, A.N.; Postle, A.D.; D'Angelo, C. Lipidome analysis of Symbiodiniaceae reveals possible mechanisms of heat stress tolerance in reef coral symbionts. Coral Reefs 2019, 38, 1241–1253. [Google Scholar] [CrossRef]
- Anderson, K.D.; Heron, S.F.; Pratchett, M.S. Species-specific declines in the linear extension of branching corals at a subtropical reef, Lord Howe Island. Coral Reefs 2015, 34, 479–490. [Google Scholar] [CrossRef]
- Tang, C.H.; Fang, L.S.; Fan, T.Y.; Wang, L.H.; Lin, C.Y.; Lee, S.H.; Wang, W.H. Cellular membrane accommodation to thermal oscillations in the coral seriatopora caliendrum. PLoS ONE 2014, 9, e105345. [Google Scholar] [CrossRef]
- Rivest, E.B.; Chen, C.S.; Fan, T.Y.; Li, H.H.; Hofmann, G.E. Lipid consumption in coral larvae differs among sites: A consideration of environmental history in a global ocean change scenario. P. Roy. Soc. B-Biol. Sci. 2017, 284, 20162825. [Google Scholar] [CrossRef]
- Isomura, N.; Nishihira, M. Size variation of planulae and its effect on the lifetime of planulae in three pocilloporid corals. Coral Reefs 2001, 20, 309–315. [Google Scholar] [CrossRef]
- Abu Risha, M.; Ali, A.; Siengdee, P.; Trakooljul, N.; Dannenberger, D.; Wimmers, K.; Ponsuksili, S. Insights into molecular pathways and fatty acid membrane composition during the temperature stress response in the murine C2C12 cell model. Sci. Total Environ. 2022, 807, 151019. [Google Scholar] [CrossRef]
- Yamashiro, H.; Oku, H.; Onaga, K.; Iwasaki, H.; Takara, K. Coral tumors store reduced level of lipids. J. Exp. Mar. Biol. Ecol. 2001, 265, 171–179. [Google Scholar] [CrossRef]
- Rotmann, S.; Thomas, S. Coral tissue thickness as a bioindicator of mine-related turbidity stress on coral reefs at Lihir Island, Papua New Guinea. Oceanography 2012, 25, 52–63. [Google Scholar]
- Renner, M.K.; Shen, Y.C.; Cheng, X.C.; Jensen, P.R.; Frankmoelle, W.; Kauffman, C.A.; Fenical, W.; Lobkovsky, E.; Clardy, J. Cyclomarins A-C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J. Am. Chem. Soc. 1999, 121, 11273–11276. [Google Scholar] [CrossRef]
- Dai, Y.; Lin, Y.; Pang, X.Y.; Luo, X.W.; Salendra, L.; Wang, J.F.; Zhou, X.F.; Lu, Y.J.; Yang, B.; Liu, Y.H. Peptides from the soft coral-associated fungus Simplicillium sp SCSIO41209. Phytochemistry 2018, 154, 56–62. [Google Scholar] [CrossRef]
- Schmidt, C.A.; Daly, N.L.; Wilson, D.T. Coral venom toxins. Front. Ecol. Evol. 2019, 7, 1–7. [Google Scholar] [CrossRef]
- Sjogren, M.; Goransson, U.; Johnson, A.L.; Dahlstrom, M.; Andersson, R.; Bergman, J.; Jonsson, P.R.; Bohlin, L. Antifouling activity of brominated cyclopeptides from the marine sponge Geodia barretti. J. Nat. Prod. 2004, 67, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Roggatz, C.C.; Lorch, M.; Hardege, J.D.; Benoit, D.M. Ocean acidification affects marine chemical communication by changing structure and function of peptide signalling molecules. Global Chang. Biol. 2016, 22, 3914–3926. [Google Scholar] [CrossRef]
- Xie, J.; Zhao, C.F.; Han, Q.; Zhou, H.L.; Li, Q.X.; Diao, X.P. Effects of pyrene exposure on immune response and oxidative stress in the pearl oyster, Pinctada martensii. Fish Shellfish Immun. 2017, 63, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cheng, Y.S.; Xie, X.X.; Xu, Q.Y.; Chen, N. Modification of tryptophan transport system and its impact on production of L-tryptophan in Escherichia coli. Bioresour Technol. 2012, 114, 549–554. [Google Scholar] [CrossRef]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef]
- Hartmann, A.; Albert, A.; Ganzera, M. Effects of elevated ultraviolet radiation on primary metabolites in selected alpine algae and cyanobacteria. J. Photoch. Photobio. B. 2015, 149, 149–155. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parkinson, J.E.; Baker, A.C.; Baums, I.B.; Davies, S.W.; Grottoli, A.G.; Kitchen, S.A.; Matz, M.V.; Miller, M.W.; Shantz, A.A.; Kenkel, C.D. Molecular tools for coral reef restoration: Beyond biomarker discovery. Conserv. Lett. 2020, 13, 1–12. [Google Scholar] [CrossRef]
- Li, J.H.; Tian, C.F.; Xia, Y.H.; Mutanda, I.; Wang, K.B.; Wang, Y. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab. Eng. 2019, 52, 124–133. [Google Scholar] [CrossRef]
- Tang, J.; Cai, W.Q.; Yan, Z.C.; Zhang, K.D.; Zhou, Z.; Zhao, J.M.; Lin, S.J. Interactive effects of acidification and copper exposure on the reproduction and metabolism of coral endosymbiont Cladocopium goreaui. Mar. Pollut. Bull. 2022, 177, 113508. [Google Scholar] [CrossRef]
- Yang, S.H.; Lee, S.T.M.; Huang, C.R.; Tseng, C.H.; Chiang, P.W.; Chen, C.P.; Chen, H.J.; Tang, S.L. Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol. Oceanogr. 2016, 61, 1078–1086. [Google Scholar] [CrossRef]
- Lesser, M.P.; Mazel, C.H.; Gorbunov, M.Y.; Falkowski, P.G. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 2004, 305, 997–1000. [Google Scholar] [CrossRef]
- Flores-Tinoco, C.E.; Tschan, F.; Fuhrer, T.; Margot, C.; Sauer, U.; Christen, M.; Christen, B. Co-catabolism of arginine and succinate drives symbiotic nitrogen fixation. Mol. Syst. Biol. 2020, 16, 1–10. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, J.; Chen, S.; Yu, K.; Hu, J.; Wang, Y.; Zhang, J.; Qin, Z.; Zhang, R.; Kuo, T.-H.; Chung, H.-H.; et al. Metabolomics Characterization of Scleractinia Corals with Different Life-History Strategies: A Case Study about Pocillopora meandrina and Seriatopora hystrix in the South China Sea. Metabolites 2022, 12, 1079. https://doi.org/10.3390/metabo12111079
Pei J, Chen S, Yu K, Hu J, Wang Y, Zhang J, Qin Z, Zhang R, Kuo T-H, Chung H-H, et al. Metabolomics Characterization of Scleractinia Corals with Different Life-History Strategies: A Case Study about Pocillopora meandrina and Seriatopora hystrix in the South China Sea. Metabolites. 2022; 12(11):1079. https://doi.org/10.3390/metabo12111079
Chicago/Turabian StylePei, Jiying, Shiguo Chen, Kefu Yu, Junjie Hu, Yitong Wang, Jingjing Zhang, Zhenjun Qin, Ruijie Zhang, Ting-Hao Kuo, Hsin-Hsiang Chung, and et al. 2022. "Metabolomics Characterization of Scleractinia Corals with Different Life-History Strategies: A Case Study about Pocillopora meandrina and Seriatopora hystrix in the South China Sea" Metabolites 12, no. 11: 1079. https://doi.org/10.3390/metabo12111079
APA StylePei, J., Chen, S., Yu, K., Hu, J., Wang, Y., Zhang, J., Qin, Z., Zhang, R., Kuo, T.-H., Chung, H.-H., & Hsu, C.-C. (2022). Metabolomics Characterization of Scleractinia Corals with Different Life-History Strategies: A Case Study about Pocillopora meandrina and Seriatopora hystrix in the South China Sea. Metabolites, 12(11), 1079. https://doi.org/10.3390/metabo12111079