Urine Metabolites Enable Fast Detection of COVID-19 Using Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Subjects
2.3. Sample Preparation
2.4. Flow Injection–Tandem MS Analysis
2.5. Data Analysis and Statistical Classifiers
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pritt, B.S.; Wang, P.; Nuzzo, J.; Zimmermann, S.; Burnham, C.-A.D. Deadly Pathogens, Transformative Technologies, and Protracted Pandemics: Challenges and Opportunities in Laboratory Medicine. Clin. Chem. 2022, 68, 1–3. [Google Scholar] [CrossRef]
- Fernandes, Q.; Inchakalody, V.P.; Merhi, M.; Mestiri, S.; Taib, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Lima, N.M.; Fernandes, B.L.M.; Alves, G.F.; de Souza, J.C.Q.; Siqueira, M.M.; Patrícia do Nascimento, M.; Moreira, O.B.O.; Sussulini, A.; de Oliveira, M.A.L. Mass spectrometry applied to diagnosis, prognosis, and therapeutic targets identification for the novel coronavirus SARS-CoV-2: A review. Anal. Chim. Acta 2022, 1195, 339385. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.M. A Push for Real Normal: Mass Screening for COVID-19. Clin. Chem. 2022, 68, 4–6. [Google Scholar] [CrossRef]
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef]
- Utama, R.; Hapsari, R.; Puspitasari, I.; Sari, D.; Hendrianingtyas, M.; Nurainy, N. Self-collected gargle specimen as a patient-friendly sample collection method for COVID-19 diagnosis in a population context. Sci. Rep. 2022, 12, 3706. [Google Scholar] [CrossRef]
- Pasomsub, E.; Watcharananan, S.P.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Suksuwan, W.; Sungkanuparph, S.; Phuphuakrat, A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: A cross-sectional study. Clin. Microbiol. Infect. 2021, 27, e281–e285. [Google Scholar] [CrossRef]
- Liao, W.T.; Hsu, M.Y.; Shen, C.F.; Hung, K.F.; Cheng, C.M. Home Sample Self-Collection for COVID-19 Patients. Adv. Biosyst. 2020, 4, e2000150. [Google Scholar] [CrossRef]
- Petruzzi, G.; De Virgilio, A.; Pichi, B.; Mazzola, F.; Zocchi, J.; Mercante, G.; Spriano, G.; Pellini, R. COVID-19: Nasal and oropharyngeal swab. Head Neck 2020, 42, 1303–1304. [Google Scholar] [CrossRef]
- Bwire, G.M.; Majigo, M.V.; Njiro, B.J.; Mawazo, A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 719–725. [Google Scholar] [CrossRef]
- Yelin, I.; Aharony, N.; Tamar, E.S.; Argoetti, A.; Messer, E.; Berenbaum, D.; Shafran, E.; Kuzli, A.; Gandali, N.; Shkedi, O.; et al. Evaluation of COVID-19 RT-qPCR Test in Multi sample Pools. Clin. Infect. Dis. 2020, 71, 2073–2078. [Google Scholar] [CrossRef] [PubMed]
- Stokes, W.; Berenger, B.M.; Scott, B.; Szelewicki, J.; Singh, T.; Portnoy, D.; Turnbull, L.; Pabbaraju, K.; Shokoples, S.; Wong, A.A.; et al. One Swab Fits All: Performance of a Rapid, Antigen-Based SARS-CoV-2 Test Using a Nasal Swab, Nasopharyngeal Swab for Nasal Collection, and RT–PCR Confirmation from Residual Extraction Buffer. J. Appl. Lab. Med. 2022, 7, jfac004. [Google Scholar] [CrossRef] [PubMed]
- Gokulan, C.G.; Kiran, U.; Kuncha, S.K.; Mishra, R.K. Temporal stability and detection sensitivity of the dry swab-based diagnosis of SARS-CoV-2. J. Biosci. 2021, 46, 95. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, E.D.; Fonseca, W.T.; de Oliveira, T.R.; de Correia, C.; Faça, V.M.; de Morais, B.P.; Silvestrini, V.C.; Pott-Junior, H.; Teixeira, F.R.; Faria, R.C. COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor. Sens. Actuators B Chem. 2022, 353, 131128. [Google Scholar] [CrossRef]
- Garza, K.Y.; Silva, A.A.R.; Rosa, J.R.; Keating, M.F.; Povilaitis, S.C.; Spradlin, M.; Sanches, P.H.G.; Moura, A.V.; Gutierrez, J.M.; Lin, J.Q.; et al. Rapid Screening of COVID-19 Disease Directly from Clinical Nasopharyngeal Swabs using the MasSpec Pen Technology. Anal. Chem. 2021, 93, 12582–12593. [Google Scholar] [CrossRef] [PubMed]
- Dhar, B.C. Diagnostic assay and technology advancement for detecting SARS-CoV-2 infections causing the COVID-19 pandemic. Anal. Bioanal. Chem. 2022, 414, 2903–2934. [Google Scholar] [CrossRef]
- Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers-Diagnostic and Therapeutic Solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. [Google Scholar] [CrossRef]
- Moore, K.J.M.; Cahill, J.; Aidelberg, G.; Aronoff, R.; Bektaş, A.; Bezdan, D.; Butler, D.J.; Chittur, S.V.; Codyre, M.; Federici, F.; et al. Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications. J. Biomol. Tech. 2021, 32, 228–275. [Google Scholar] [CrossRef]
- Drobysh, M.; Ramanaviciene, A.; Viter, R.; Chen, C.F.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A. Biosensors for the Determination of SARS-CoV-2 Virus and Diagnosis of COVID-19 Infection. Int. J. Mol. Sci. 2022, 23, 666. [Google Scholar] [CrossRef]
- Li, Y.; Hou, G.; Zhou, H.; Wang, Y.; Tun, H.M.; Zhu, A.; Zhao, J.; Xiao, F.; Lin, S.; Liu, D.; et al. Multi-platform omics analysis reveals molecular signature for COVID-19 pathogenesis, prognosis and drug target discovery. Signal. Transduct. Target. Ther. 2021, 6, 155. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef] [PubMed]
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323, 2249–2251. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Aoki, K.; Nagasawa, T.; Ishii, Y.; Yagi, S.; Okuma, S.; Kashiwagi, K.; Maeda, T.; Miyazaki, T.; Yoshizawa, S.; Tateda, K. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2021, 27, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.-P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Sutjipto, S.; Lee, P.H.; Tay, J.Y.; Mendis, S.M.; Abdad, M.Y.; Marimuthu, K.; Ng, O.T.; Cui, L.; Chan, M.; Soon, M.; et al. The Effect of Sample Site, Illness Duration, and the Presence of Pneumonia on the Detection of SARS-CoV-2 by Real-time Reverse Transcription PCR. Open Forum Infect. Dis. 2020, 7, ofaa335. [Google Scholar] [CrossRef]
- Patel, R. Advances in Testing for Infectious Diseases—Looking Back and Projecting Forward. Clin. Chem. 2022, 68, 10–15. [Google Scholar] [CrossRef]
- Li, L.; Shim, T.; Zapanta, P.E. Optimization of COVID-19 testing accuracy with nasal anatomy education. Am. J. Otolaryngol. 2021, 42, 102777. [Google Scholar] [CrossRef]
- Marra, P.; Colacurcio, V.; Bisogno, A.; De Luca, P.; Calvanese, M.; Petrosino, M.; De Bonis, E.; Troisi, D.; Cassandro, C.; Cavaliere, M.; et al. Evaluation of Discomfort in Nasopharyngeal Swab Specimen Collection for SARS-CoV-2 Diagnosis. Clin. Ter. 2021, 172, 448–452. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, D.; Moon, J.W.; Chae, S.W.; Rhyu, I.J. Complications of Nasopharyngeal Swabs and Safe Procedures for COVID-19 Testing Based on Anatomical Knowledge. J. Korean Med. Sci. 2022, 37, e88. [Google Scholar] [CrossRef]
- Lin, L.; Song, Y.; Wang, Q.; Pu, J.; Sun, F.Y.; Zhang, Y.; Zhou, X.; Larson, H.J.; Hou, Z. Public Attitudes and Factors of COVID-19 Testing Hesitancy in the United Kingdom and China: Comparative Infodemiology Study. JMIR Infodemiol. 2021, 1, e26895. [Google Scholar] [CrossRef] [PubMed]
- Ehrenstein, B.; Schwarz, T.; Fleck, M.; Günther, F. Hygiene measures against COVID-19 in routine outpatient care: Acceptance by the patients? Z. Rheumatol. 2021, 80, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Liu, W.; Ding, X.; Liang, S.; Zheng, Y.; Zhu, X.; Quan, S.; Yi, X.; Xiang, N.; Du, J.; et al. Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19. Cell Rep. 2022, 38, 110271. [Google Scholar] [CrossRef]
- Dewulf, J.P.; Martin, M.; Marie, S.; Oguz, F.; Belkhir, L.; De Greef, J.; Yombi, J.C.; Wittebole, X.; Laterre, P.-F.; Jadoul, M.; et al. Urine metabolomics links dysregulation of the tryptophan-kynurenine pathway to inflammation and severity of COVID-19. Sci. Rep. 2022, 12, 9959. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e1420. [Google Scholar] [CrossRef]
- Blasco, H.; Bessy, C.; Plantier, L.; Lefevre, A.; Piver, E.; Bernard, L.; Marlet, J.; Stefic, K.; Benz-de Bretagne, I.; Cannet, P.; et al. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 2020, 10, 16824. [Google Scholar] [CrossRef]
- Song, J.-W.; Lam, S.M.; Fan, X.; Cao, W.-J.; Wang, S.-Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.-P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e185. [Google Scholar] [CrossRef]
- Danlos, F.-X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef]
- Gray, N.; Lawler, N.G.; Yang, R.; Morillon, A.-C.; Gay, M.C.L.; Bong, S.-H.; Holmes, E.; Nicholson, J.K.; Whiley, L. A simultaneous exploratory and quantitative amino acid and biogenic amine metabolic profiling platform for rapid disease phenotyping via UPLC-QToF-MS. Talanta 2021, 223, 121872. [Google Scholar] [CrossRef]
- Kimhofer, T.; Lodge, S.; Whiley, L.; Gray, N.; Loo, R.L.; Lawler, N.G.; Nitschke, P.; Bong, S.-H.; Morrison, D.L.; Begum, S.; et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19, 4442–4454. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, B.; Sharma, L.; Roberts, L.; Peng, X.; Bermejo, S.; Leighton, I.; Casanovas-Massana, A.; Minasyan, M.; Farhadian, S.; Ko, A.I.; et al. Cutting Edge: Severe SARS-CoV-2 Infection in Humans Is Defined by a Shift in the Serum Lipidome, Resulting in Dysregulation of Eicosanoid Immune Mediators. J. Immunol. 2021, 206, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Doğan, H.O.; Şenol, O.; Bolat, S.; Yıldız, Ş.N.; Büyüktuna, S.A.; Sarıismailoğlu, R.; Doğan, K.; Hasbek, M.; Hekim, S.N. Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 2021, 93, 2340–2349. [Google Scholar] [CrossRef]
- Mohammed, A.F.K.; Alghetaa, H.; Miranda, K.; Wilson, K.; P. Singh, N.; Cai, G.; Putluri, N.; Nagarkatti, P.; Nagarkatti, M. Δ9-Tetrahydrocannabinol Prevents Mortality from Acute Respiratory Distress Syndrome through the Induction of Apoptosis in Immune Cells, Leading to Cytokine Storm Suppression. Int. J. Mol. Sci. 2020, 21, 6244. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H.; et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef]
- Shi, D.; Yan, R.; Lv, L.; Jiang, H.; Lu, Y.; Sheng, J.; Xie, J.; Wu, W.; Xia, J.; Xu, K.; et al. The serum metabolome of COVID-19 patients is distinctive and predictive. Metab. Clin. Exp. 2021, 118, 154739. [Google Scholar] [CrossRef]
- Lv, L.; Jiang, H.; Chen, Y.; Gu, S.; Xia, J.; Zhang, H.; Lu, Y.; Yan, R.; Li, L. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal. Chim. Acta 2021, 1152, 338267. [Google Scholar] [CrossRef]
- Seger, C.; Salzmann, L. After another decade: LC-MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.-F. (Ed.) Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7). Chin. Med. J. 2020, 133, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G.; Voss, A.; Scheithauer, S. Inactivation of coronaviruses by heat. J. Hosp. Infect. 2020, 105, 348–349. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Durbin, B.P.; Hardin, J.S.; Hawkins, D.M.; Rocke, D.M. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 2002, 18, S105–S110. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yi, J.; Huang, C.; Zhang, J.; Fu, S.; Li, Z.; Lyu, Q.; Xu, Y.; Wang, K.; Yang, H.; et al. Rapid Detection of COVID-19 Using MALDI-TOF-Based Serum Peptidome Profiling. Anal. Chem. 2021, 93, 4782–4787. [Google Scholar] [CrossRef]
- Kwee, T.C.; Kwee, R.M. Chest CT in COVID-19: What the Radiologist Needs to Know. Radiographics 2020, 40, 1848–1865. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Berhane, S.; Taylor, M.; Adriano, A.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021, 3, CD013705. [Google Scholar] [CrossRef]
- Böger, B.; Fachi, M.M.; Vilhena, R.O.; Cobre, A.F.; Tonin, F.S.; Pontarolo, R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control 2021, 49, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Sirajuddin, A.; Zhang, X.; Liu, G.; Teng, Z.; Zhao, S.; Lu, M. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19). Eur. Radiol. 2020, 30, 4874–4882. [Google Scholar] [CrossRef] [PubMed]
- Ufuk, F.; Savaş, R. Chest CT features of the novel coronavirus disease (COVID-19). Turk. J. Med. Sci. 2020, 50, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 2020, 296, E41–E45. [Google Scholar] [CrossRef]
- Feng, H.; Liu, Y.; Lv, M.; Zhong, J. A case report of COVID-19 with false negative RT-PCR test: Necessity of chest CT. Jpn J. Radiol. 2020, 38, 409–410. [Google Scholar] [CrossRef]
- Hossein, H.; Ali, K.M.; Hosseini, M.; Sarveazad, A.; Safari, S.; Yousefifard, M. Value of chest computed tomography scan in diagnosis of COVID-19; a systematic review and meta-analysis. Clin. Transl. Imaging 2020, 8, 469–481. [Google Scholar] [CrossRef]
- Gibellini, L.; De Biasi, S.; Paolini, A.; Borella, R.; Boraldi, F.; Mattioli, M.; Lo Tartaro, D.; Fidanza, L.; Caro-Maldonado, A.; Meschiari, M.; et al. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia. EMBO Mol. Med. 2020, 12, e13001. [Google Scholar] [CrossRef]
- Surazakov, A.; Klassen, A.; Gizinger, O. The bioenergetics of COVID-19 immunopathology and the therapeutic potential of biophysical radiances. J. Photochem. Photobiol. B Biol. 2020, 213, 112083. [Google Scholar] [CrossRef]
- Gvozdjakova, A.; Klauco, F.; Kucharska, J.; Sumbalova, Z. Is mitochondrial bioenergetics and coenzyme Q10 the target of a virus causing COVID-19? Bratisl. Lek. Listy 2020, 121, 775–778. [Google Scholar] [CrossRef]
- Trombetta, A.C.; Farias, G.B.; Gomes, A.M.C.; Godinho-Santos, A.; Rosmaninho, P.; Conceição, C.M.; Laia, J.; Santos, D.F.; Almeida, A.R.M.; Mota, C.; et al. Severe COVID-19 Recovery Is Associated with Timely Acquisition of a Myeloid Cell Immune-Regulatory Phenotype. Front. Immunol. 2021, 12, 691725. [Google Scholar] [CrossRef]
- Yazdanpanah, F.; Hamblin, M.R.; Rezaei, N. The immune system and COVID-19: Friend or foe? Life Sci. 2020, 256, 117900. [Google Scholar] [CrossRef] [PubMed]
- Paces, J.; Strizova, Z.; Smrz, D.; Cerny, J. COVID-19 and the immune system. Physiol. Res. 2020, 69, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Atila, A.; Alay, H.; Yaman, M.E.; Akman, T.C.; Cadirci, E.; Bayrak, B.; Celik, S.; Atila, N.E.; Yaganoglu, A.M.; Kadioglu, Y.; et al. The serum amino acid profile in COVID-19. Amino. Acids 2021, 53, 1569–1588. [Google Scholar] [CrossRef] [PubMed]
- Ansone, L.; Briviba, M.; Silamikelis, I.; Terentjeva, A.; Perkons, I.; Birzniece, L.; Rovite, V.; Rozentale, B.; Viksna, L.; Kolesova, O.; et al. Amino Acid Metabolism is Significantly Altered at the Time of Admission in Hospital for Severe COVID-19 Patients: Findings from Longitudinal Targeted Metabolomics Analysis. Microbiol. Spectr. 2021, 9, e00338-21. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, M.; Peschka, M.; Schmiedel, S.; Haddad, M.; Frye, M.; Maas, C.; Lohse, A.; Huber, S.; Kirchhof, P.; Nofer, J.-R.; et al. Disturbed lipid and amino acid metabolisms in COVID-19 patients. J. Mol. Med. 2022, 100, 555–568. [Google Scholar] [CrossRef]
- Suhail, S.; Zajac, J.; Fossum, C.; Lowater, H.; McCracken, C.; Severson, N.; Laatsch, B.; Narkiewicz-Jodko, A.; Johnson, B.; Liebau, J.; et al. Role of Oxidative Stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) Infection: A Review. Protein J. 2020, 39, 644–656. [Google Scholar] [CrossRef]
- Chernyak, B.V.; Popova, E.N.; Prikhodko, A.S.; Grebenchikov, O.A.; Zinovkina, L.A.; Zinovkin, R.A. COVID-19 and Oxidative Stress. Biochemistry 2020, 85, 1543–1553. [Google Scholar] [CrossRef]
- Pincemail, J.; Cavalier, E.; Charlier, C.; Cheramy–Bien, J.-P.; Brevers, E.; Courtois, A.; Fadeur, M.; Meziane, S.; Goff, C.L.; Misset, B.; et al. Oxidative Stress Status in COVID-19 Patients Hospitalized in Intensive Care Unit for Severe Pneumonia. A Pilot Study. Antioxidants 2021, 10, 257. [Google Scholar] [CrossRef]
- Dasari, C.M.; Bhukya, R. Comparative analysis of protein synthesis rate in COVID-19 with other human coronaviruses. Infect. Genet. Evol. 2020, 85, 104432. [Google Scholar] [CrossRef]
- Yuan, S.; Peng, L.; Park, J.J.; Hu, Y.; Devarkar, S.C.; Dong, M.B.; Shen, Q.; Wu, S.; Chen, S.; Lomakin, I.B.; et al. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. Mol. Cell 2020, 80, 1055–1066.e1056. [Google Scholar] [CrossRef]
- Woods, J.A.; Hutchinson, N.T.; Powers, S.K.; Roberts, W.O.; Gomez-Cabrera, M.C.; Radak, Z.; Berkes, I.; Boros, A.; Boldogh, I.; Leeuwenburgh, C.; et al. The COVID-19 pandemic and physical activity. Sport. Med. Health Sci. 2020, 2, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Altay, O.; Arif, M.; Li, X.; Yang, H.; Aydın, M.; Alkurt, G.; Kim, W.; Akyol, D.; Zhang, C.; Dinler-Doganay, G.; et al. Combined Metabolic Activators Accelerates Recovery in Mild-to-Moderate COVID-19. Adv. Sci. 2021, 8, 2101222. [Google Scholar] [CrossRef] [PubMed]
- Chuan-Yuan, L. Can Glycine Mitigate COVID-19 Associated Tissue Damage and Cytokine Storm? Radiat. Res. 2020, 194, 199–201. [Google Scholar] [CrossRef]
- Silvagno, F.; Vernone, A.; Pescarmona, G.P. The Role of Glutathione in Protecting against the Severe Inflammatory Response Triggered by COVID-19. Antioxidants 2020, 9, 624. [Google Scholar] [CrossRef]
- Bak, D.W.; Bechtel, T.J.; Falco, J.A.; Weerapana, E. Cysteine reactivity across the subcellular universe. Curr. Opin. Chem. Biol. 2019, 48, 96–105. [Google Scholar] [CrossRef]
- Bonvini, A.; Coqueiro, A.Y.; Tirapegui, J.; Calder, P.C.; Rogero, M.M. Immunomodulatory role of branched-chain amino acids. Nutr. Rev. 2018, 76, 840–856. [Google Scholar] [CrossRef]
- Tsai, S.C.; Lu, C.C.; Bau, D.T.; Chiu, Y.J.; Yen, Y.T.; Hsu, Y.M.; Fu, C.W.; Kuo, S.C.; Lo, Y.S.; Chiu, H.Y.; et al. Approaches towards fighting the COVID-19 pandemic (Review). Int. J. Mol. Med. 2021, 47, 3–22. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef]
- Lawler, N.G.; Gray, N.; Kimhofer, T.; Boughton, B.; Gay, M.; Yang, R.; Morillon, A.-C.; Chin, S.-T.; Ryan, M.; Begum, S.; et al. Systemic Perturbations in Amine and Kynurenine Metabolism Associated with Acute SARS-CoV-2 Infection and Inflammatory Cytokine Responses. J. Proteome Res. 2021, 20, 2796–2811. [Google Scholar] [CrossRef]
- Oh, M.-H.; Sun, I.-H.; Zhao, L.; Leone, R.D.; Sun, I.-M.; Xu, W.; Collins, S.L.; Tam, A.J.; Blosser, R.L.; Patel, C.H.; et al. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J. Clin. Investig. 2020, 130, 3865–3884. [Google Scholar] [CrossRef] [PubMed]
- Kretzmann, N.A.; Fillmann, H.; Mauriz, J.L.; Marroni, C.A.; Marroni, N.; González-Gallego, J.; Tuñón, M.J. Effects of glutamine on proinflammatory gene expression and activation of nuclear factor kappa B and signal transducers and activators of transcription in TNBS-induced colitis. Inflamm. Bowel Dis. 2008, 14, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mesa, J.E.; Galindo-Coral, S.; Montes, M.C.; Muñoz Martin, A.J. Thrombosis and Coagulopathy in COVID-19. Curr. Probl. Cardiol. 2021, 46, 100742. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Nagao, M.; Toh, R.; Irino, Y.; Shinohara, M.; Iino, T.; Yoshikawa, S.; Tanaka, H.; Satomi-Kobayashi, S.; Ishida, T.; et al. Critical role of glutamine metabolism in cardiomyocytes under oxidative stress. Biochem. Biophys. Res. Commun. 2021, 534, 687–693. [Google Scholar] [CrossRef]
- Mohajeri, M.; Horriatkhah, E.; Mohajery, R. The effect of glutamine supplementation on serum levels of some inflammatory factors, oxidative stress, and appetite in COVID-19 patients: A case-control study. Inflammopharmacology 2021, 29, 1769–1776. [Google Scholar] [CrossRef]
- Kerner, J.; Hoppel, C.L. Carnitine and β-Oxidation. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 384–387. [Google Scholar]
- Houten, S.M.; Wanders, R.J.A.; Ranea-Robles, P. Metabolic interactions between peroxisomes and mitochondria with a special focus on acylcarnitine metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165720. [Google Scholar] [CrossRef]
- Boenzi, S.; Diodato, D. Biomarkers for mitochondrial energy metabolism diseases. Essays. Biochem. 2018, 62, 443–454. [Google Scholar] [CrossRef]
- Indiveri, C.; Iacobazzi, V.; Tonazzi, A.; Giangregorio, N.; Infantino, V.; Convertini, P.; Console, L.; Palmieri, F. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Mol. Asp. Med. 2011, 32, 223–233. [Google Scholar] [CrossRef]
- Kerner, J.; Hoppel, C. Fatty acid import into mitochondria. Biochim. Biophys. Acta 2000, 1486, 1–17. [Google Scholar] [CrossRef]
- Chen, W.S.; Liu, M.H.; Cheng, M.L.; Wang, C.H. Decreases in Circulating Concentrations of Short-Chain Acylcarnitines are Associated with Systolic Function Improvement After Decompensated Heart Failure. Int. Heart J. 2020, 61, 1014–1021. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Liang, Y.; Hu, R.; Xu, W.; Liu, Y. Plasma Targeted Metabolomics Analysis for Amino Acids and Acylcarnitines in Patients with Prediabetes, Type 2 Diabetes Mellitus, and Diabetic Vascular Complications. Diabetes Metab. J. 2021, 45, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Korobkova, E.O.; Kozhevnikova, M.V.; Ilgisonis, I.S.; Shakaryants, G.A.; Appolonova, S.A.; Kukharenko, A.V.; Larcova, E.V.; Maltseva, A.A.; Khabarova, N.V.; Belenkov, Y.N. Metabolomic profiling in patients with metabolic syndrome. Kardiologiia 2020, 60, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, H.; Alsrhani, A.; Zafar, A.; Javed, H.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.A.; Ahmed, Z.; Younas, S. COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health 2020, 13, 1833–1839. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, A.; Gheibi Hayat, S.M.; Taghizadeh, H.; Akbari, A.; Inabadi, M.; Savardashtaki, A.; Johnston, T.P.; Sahebkar, A. COVID-19 and cardiac injury: Clinical manifestations, biomarkers, mechanisms, diagnosis, treatment, and follow up. Expert Rev. Anti. Infect. 2021, 19, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Du, Z.; Zhu, F.; Cao, Z.; An, Y.; Gao, Y.; Jiang, B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet 2020, 395, e52. [Google Scholar] [CrossRef]
- Huang, C.; Huang, L.; Wang, Y.; Li, X.; Ren, L.; Gu, X.; Kang, L.; Guo, L.; Liu, M.; Zhou, X.; et al. 6-month consequences of COVID-19 in patients discharged from hospital: A cohort study. Lancet 2021, 397, 220–232. [Google Scholar] [CrossRef]
- Otsubo, C.; Bharathi, S.; Uppala, R.; Ilkayeva, O.R.; Wang, D.; McHugh, K.; Zou, Y.; Wang, J.; Alcorn, J.F.; Zuo, Y.Y.; et al. Long-chain Acylcarnitines Reduce Lung Function by Inhibiting Pulmonary Surfactant. J. Biol. Chem. 2015, 290, 23897–23904. [Google Scholar] [CrossRef]


| Classifier a (Training + Validation) | Withheld Set 1 | Withheld Set 2 | |||
|---|---|---|---|---|---|
| Neg-NH | Pos-H | Neg-H | Pos-H | Neg-H | |
| Total = 246 | 104 n (%) | 42 n (%) | 24 n (%) | 57 n (%) | 19 n (%) |
| Age—mean (min–max) b | 38.2 (20–89) | 56.2 (21–86) | 58.8 (26–81) | 56.3 (26–77) | 59.8 (30–83) |
| Female c | 61 (58.7) | 13 (31.0) | 11 (45.8) | 23 (40.4) | 10 (52.6) |
| Male c | 43 (41.3) | 29 (69.0) | 13 (54.2) | 34 (59.6) | 9 (47.4) |
| Symptoms | |||||
| Fever | 0 (0.0) | 23 (54.8) | 11 (45.8) | 36 (63.2) | 10 (52.6) |
| Cough | 0 (0.0) | 30 (71.4) | 14 (58.3) | 35 (61.4) | 11 (57.9) |
| Myalgia | 0 (0.0) | 8 (19.0) | 2 (8.3) | 13 (22.8) | 3 (15.8) |
| Sore throat | 1 (1.0) | 7 (16.7) | 8 (33.3) | 8 (14.0) | 2 (10.5) |
| Headache | 3 (2.9) | 12 (28.6) | 2 (8.3) | 7 (12.3) | 5 (26.3) |
| Coryza | 0 (0.0) | 5 (11.9) | 3 (12.5) | 6 (10.5) | 2 (10.5) |
| Dyspnea | 0 (0.0) | 29 (69.0) | 16 (66.7) | 29 (50.9) | 12 (63.2) |
| Oxygen saturation < 95% | 0 (0.0) | 17 (40.5) | 10 (41.7) | 19 (33.3) | 3 (15.8) |
| Tiredness/fatigue | 0 (0.0) | 3 (7.1) | 2 (8.3) | 7 (12.3) | 3 (15.8) |
| Loss of smell or taste | 0 (0.0) | 8 (19.0) | 9 (37.5) | 12 (21.1) | 5 (26.3) |
| Vomiting or nausea | 0 (0.0) | 2 (4.8) | 3 (12.5) | 9 (15.8) | 1 (5.3) |
| Diarrhea | 0 (0.0) | 11 (26.2) | 2 (8.3) | 12 (21.1) | 3 (15.8) |
| Comorbidity | |||||
| SAH d | 16 (15.4) | 21 (50.0) | 10 (41.7) | 29 (50.9) | 8 (42.1) |
| Cardiovascular disease | 2 (1.9) | 7 (16.7) | 4 (16.7) | 10 (17.5) | 4 (21.1) |
| Obesity | 12 (11.5) | 9 (21.4) | 1 (4.2) | 13 (22.8) | 4 (21.1) |
| Diabetes mellitus | 3 (2.9) | 17 (40.5) | 2 (8.3) | 18 (31.6) | 4 (21.1) |
| Neoplasia | 0 (0.0) | 3 (7.1) | 0 (0.0) | 1 (1.8) | 1 (5.3) |
| Lung disease | 8 (7.7) | 3 (7.1) | 6 (25.0) | 5 (8.8) | 4 (21.1) |
| COPD e | 1 (1.0) | 1 (2.4) | 2 (8.3) | 3 (5.3) | 2 (10.5) |
| Smoker or ex-smoker | 6 (5.8) | 3 (7.1) | 4 (16.7) | 3 (5.3) | 3 (15.8) |
| Asthma | 2 (1.9) | 2 (4.8) | 2 (8.3) | 2 (3.5) | 1 (5.3) |
| Kidney disease | 0 (0.0) | 2 (4.8) | 0 (0.0) | 1 (1.8) | 1 (5.3) |
| Tomography Findings | |||||
| Ground glass opacity | 0 (0.0) | 40 (95.2) | 19 (79.2) | 54 (94.7) | 16 (84.2) |
| Consolidations | 0 (0.0) | 20 (47.6) | 13 (54.2) | 32 (56.1) | 8 (42.1) |
| Crazy-paving appearance | 0 (0.0) | 19 (45.2) | 10 (41.7) | 22 (38.6) | 8 (42.1) |
| reticular pattern | 0 (0.0) | 6 (14.3) | 6 (25.0) | 16 (28.1) | 2 (10.5) |
| Pulmonary commitment degree | 0 (0.0) | 35 (83.3) | 16 (66.7) | 49 (86.0) | 12 (63.2) |
| Suggestive of viral infection | 0 (0.0) | 40 (95.2) | 19 (79.2) | 54 (94.7) | 16 (84.2) |
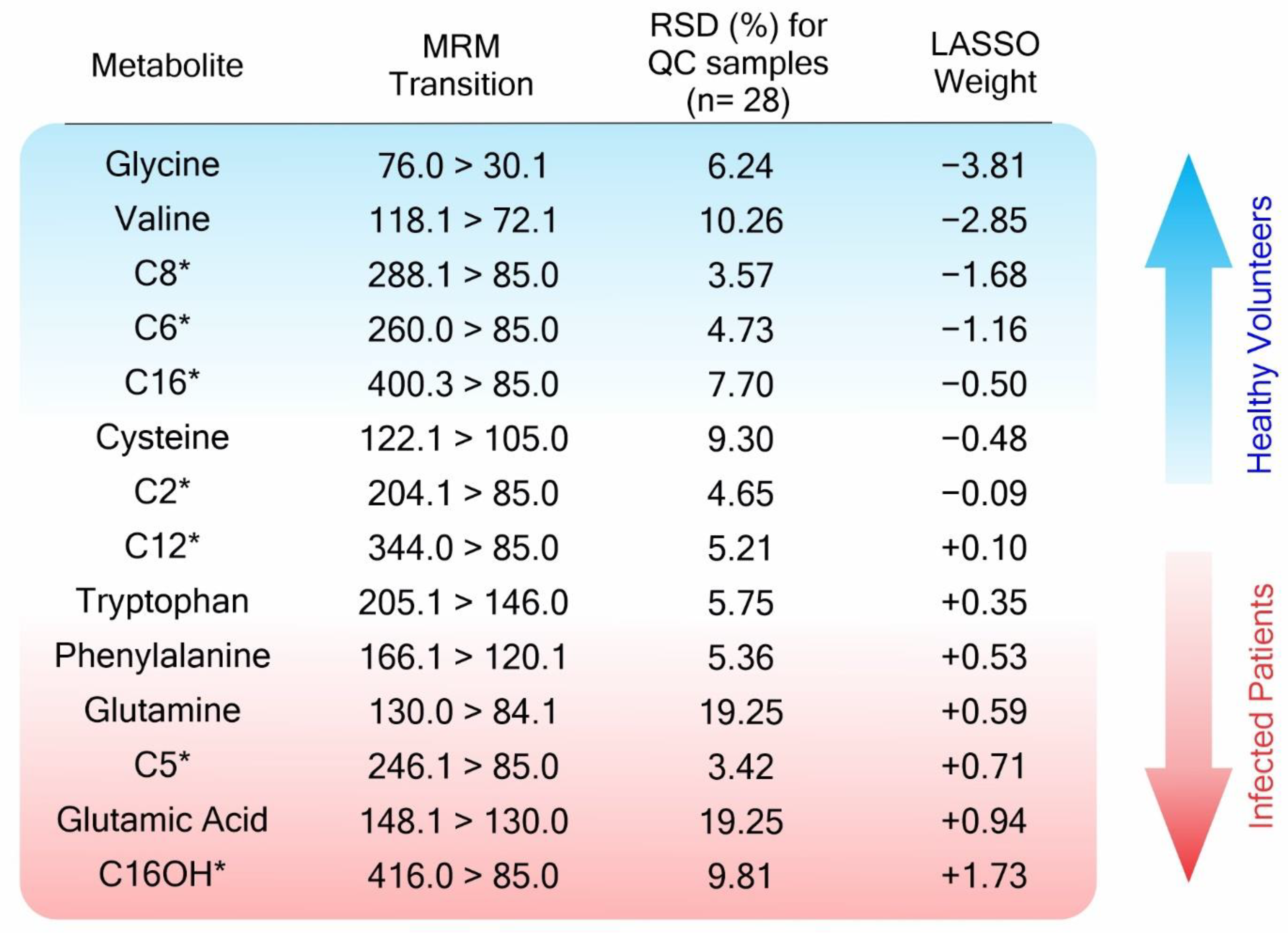
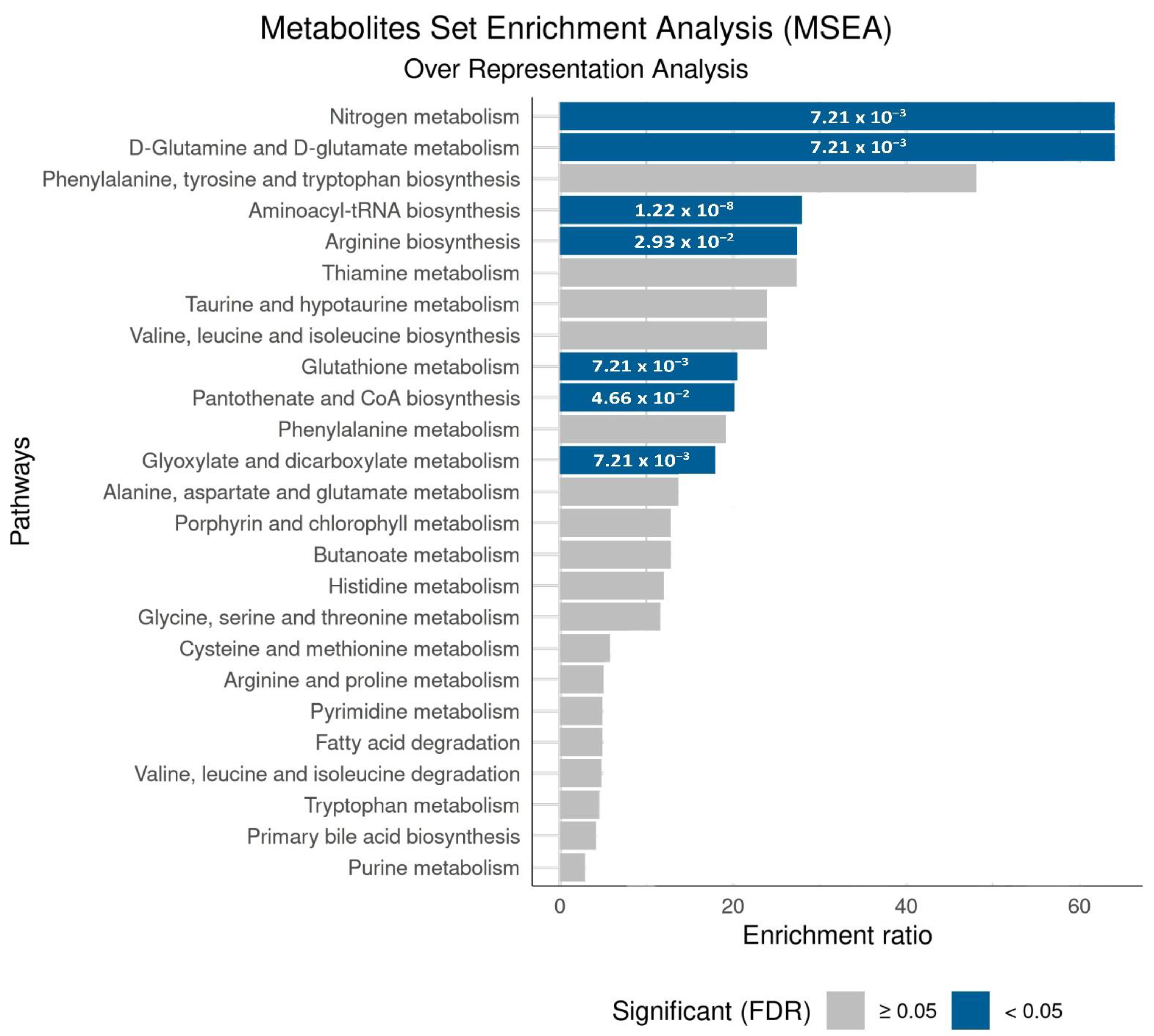
| Amino Acids | Impacted Pathway (MSEA) | Related Pathway | Impact in COVID-19 |
|---|---|---|---|
| Glycine (Gly) | Aminoacyl-tRNA biosynthesis Glyoxylate and dicarboxylate metabolism Glutathione metabolism | Immune regulation [82,83] Oxidative stress [83,84] | [70,71,72,76,77,78] |
| Valine (Val) | Aminoacyl-tRNA biosynthesis Pantothenate and CoA biosynthesis | Immune regulation [86] | [70,71,72] |
| Cysteine (Cys) | Aminoacyl-tRNA biosynthesis Pantothenate and CoA biosynthesis | Oxidative stress [82,85,87]; Protein regulation [85,88] | [76,77,78,79,80,81] |
| Tryptophan (Try) | Aminoacyl-tRNA biosynthesis | Immune regulation [35,41,46,74,89,90] | [70,71,72] |
| Phenylalanine (Phe) | Aminoacyl-tRNA biosynthesis | Bioenergetics [41,46]; Immune regulation [35,74,89,90] | [67,68,69,70,71,72] |
| Glutamine (Gln) | Aminoacyl-tRNA biosynthesis D-Glutamine and D-glutamate metabolism Nitrogen metabolism Glyoxylate and dicarboxylate metabolism Arginine biosynthesis | Immune regulation [74,91,92]; Metabolic changes [46,93]; Oxidative stress [94,95] | [70,71,72,73,74,75,76,77,78] |
| Glutamate (Glu) (glutamic acid) | Aminoacyl-tRNA biosynthesis D-Glutamine and D-glutamate metabolism Nitrogen metabolism Glyoxylate and dicarboxylate metabolism Arginine biosynthesis | Metabolic changes [39,49,93]; Oxidative stress [39,46] | [73,74,75,76,77,78] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura, A.V.; de Oliveira, D.C.; Silva, A.A.R.; da Rosa, J.R.; Garcia, P.H.D.; Sanches, P.H.G.; Garza, K.Y.; Mendes, F.M.M.; Lambert, M.; Gutierrez, J.M.; et al. Urine Metabolites Enable Fast Detection of COVID-19 Using Mass Spectrometry. Metabolites 2022, 12, 1056. https://doi.org/10.3390/metabo12111056
Moura AV, de Oliveira DC, Silva AAR, da Rosa JR, Garcia PHD, Sanches PHG, Garza KY, Mendes FMM, Lambert M, Gutierrez JM, et al. Urine Metabolites Enable Fast Detection of COVID-19 Using Mass Spectrometry. Metabolites. 2022; 12(11):1056. https://doi.org/10.3390/metabo12111056
Chicago/Turabian StyleMoura, Alexandre Varao, Danilo Cardoso de Oliveira, Alex Ap. R. Silva, Jonas Ribeiro da Rosa, Pedro Henrique Dias Garcia, Pedro Henrique Godoy Sanches, Kyana Y. Garza, Flavio Marcio Macedo Mendes, Mayara Lambert, Junier Marrero Gutierrez, and et al. 2022. "Urine Metabolites Enable Fast Detection of COVID-19 Using Mass Spectrometry" Metabolites 12, no. 11: 1056. https://doi.org/10.3390/metabo12111056
APA StyleMoura, A. V., de Oliveira, D. C., Silva, A. A. R., da Rosa, J. R., Garcia, P. H. D., Sanches, P. H. G., Garza, K. Y., Mendes, F. M. M., Lambert, M., Gutierrez, J. M., Granado, N. M., dos Santos, A. C., de Lima, I. L., Negrini, L. D. d. O., Antonio, M. A., Eberlin, M. N., Eberlin, L. S., & Porcari, A. M. (2022). Urine Metabolites Enable Fast Detection of COVID-19 Using Mass Spectrometry. Metabolites, 12(11), 1056. https://doi.org/10.3390/metabo12111056








