Quantitative Serum NMR Spectroscopy Stratifies COVID-19 Patients and Sheds Light on Interfaces of Host Metabolism and the Immune Response with Cytokines and Clinical Parameters
Abstract
:1. Introduction
1.1. Metabolomics in COVID-19 Research
1.2. Usage of Novel NMR Markers
1.3. Immunological Aspects of COVID-19 and Their Link to Metabolomics
1.4. Aims of the Study
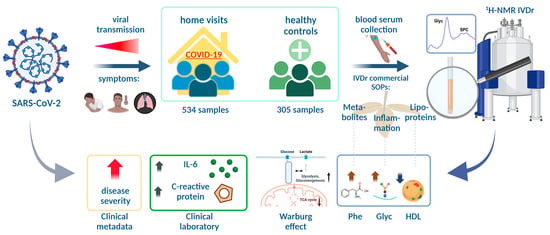
2. Materials and Methods
2.1. Patient Recruitment and Sample Collection
- (1)
- Only patients with a biobanked serum sample were included in the IVDr cohort;
- (2)
- In the IVDr cohort, 3 patients were included who had been hospitalized prior to study inclusion due to COVID-19 (inclusion of these patients was performed at the release timepoint for subsequent monitoring in an ambulatory setting after hospital release; the first sampling was performed 1, 2 and 6 days after discharge from the hospital and 2, 6 and 7 days after symptom onset);
- (3)
- The IVDr cohort consisted both of patients who entered data into the application(which was used for reports of symptoms and vital parameters) and of patients that didn’t.This allowed patients who were unable to use the app to be included (e.g., due to high age, no mobile device, etc.). Therefore, our IVDr cohort differs to a relevant extent from the published analysis [46], in which only patients with ambulant parameters before hospitalization were included.
2.2. Quantitative NMR Spectroscopy
2.3. Quantification of Cytokines
2.4. Statistics and Data Illustration
3. Results
3.1. Comparison of COVID-19 vs. Healthy Controls
3.2. Comparison of the Outpatient Cohort vs. Healthy Controls
3.3. Comparison of Hospitalized Sub-Cohort with the Sub-Cohort That Stayed in Outpatient Care
3.4. Biomarker Analysis
3.5. Cytokine Correlation Analysis
3.6. Investigation of Novel NMR-Based Inflammation Parameters in the Context of COVID-19
4. Discussion
4.1. Abnormalities in Energy and Amino Acid Metabolism Might Indicate Viral Intervention and Host Response and Might Have Immunological Implications
4.2. Profound Lipoprotein Alterations Might Be Attributable to Effects of the Virus, and Pro-Inflammatory Effects of IL-6 Are Reflected in Specific HDL Profiles
4.3. Several Clinical Parameters and Cytokines Indicate More Severe Disease Courses
4.4. Pre-Existing Conditions and Obesity Complicate the Identification of Possible Biomarkers
4.5. NMR Biomarkers Perform Well in Stratifying COVID-19 Patients
4.6. Glyc and SPC Reliably Indicate COVID-19 Severity and Offer Exciting Possibilities for the Assessment of Inflammatory Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L. COVID-19 will continue but the end of the pandemic is near. Lancet 2022, 399, 417–419. [Google Scholar] [CrossRef]
- Mussap, M.; Fanos, V. Could metabolomics drive the fate of COVID-19 pandemic? A narrative review on lights and shadows. Clin. Chem. Lab. Med. 2021, 59, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, X.; Jia, W.; Li, H. Novel Applications of Metabolomics in Personalized Medicine: A Mini-Review. Molecules 2017, 22, 1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic Phenotyping in Health and Disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letertre, M.P.M.; Giraudeau, P.; de Tullio, P. Nuclear Magnetic Resonance Spectroscopy in Clinical Metabolomics and Personalized Medicine: Current Challenges and Perspectives. Front. Mol. Biosci. 2021, 8, 698337. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, F.; Föh, B.; Mallagaray, A.; Rahmöller, J.; Ehlers, M.; Lehrian, S.; Von Kopylow, V.; Künsting, I.; Lixenfeld, A.S.; Martin, E.; et al. Metabolic and lipidomic markers differentiate COVID-19 from non-hospitalised and other intensive care patients. Front. Mol. Biosci. 2021, 8, 1091. [Google Scholar] [CrossRef]
- Lodge, S.; Nitschke, P.; Kimhofer, T.; Coudert, J.D.; Begum, S.; Bong, S.H.; Richards, T.; Edgar, D.; Raby, E.; Spraul, M.; et al. NMR Spectroscopic Windows on the Systemic Effects of SARS-CoV-2 Infection on Plasma Lipoproteins and Metabolites in Relation to Circulating Cytokines. J. Proteome Res. 2021, 20, 1382–1396. [Google Scholar] [CrossRef]
- Masuda, R.; Lodge, S.; Nitschke, P.; Spraul, M.; Schaefer, H.; Bong, S.H.; Kimhofer, T.; Hall, D.; Loo, R.L.; Bizkarguenaga, M.; et al. Integrative Modeling of Plasma Metabolic and Lipoprotein Biomarkers of SARS-CoV-2 Infection in Spanish and Australian COVID-19 Patient Cohorts. J. Proteome Res. 2021, 20, 4139–4152. [Google Scholar] [CrossRef]
- Páez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Pérez-Fragoso, A.; Meza-Sánchez, D.E.; Germán-Acacio, J.M.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; et al. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 2021, 11, 6350. [Google Scholar] [CrossRef]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; Garcia de Vicuna, A.; Seco, M.; Bosch, A.; Palazon, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef]
- Correia, B.S.B.; Ferreira, V.G.; Piagge, P.M.F.D.; Almeida, M.B.; Assunção, N.A.; Raimundo, J.R.S.; Fonseca, F.L.A.; Carrilho, E.; Cardoso, D.R. 1H qNMR-Based Metabolomics Discrimination of Covid-19 Severity. J. Proteome Res. 2022, 21, 1640–1653. [Google Scholar] [CrossRef]
- Baranovicova, E.; Bobcakova, A.; Vysehradsky, R.; Dankova, Z.; Halasova, E.; Nosal, V.; Lehotsky, J. The Ability to Normalise Energy Metabolism in Advanced COVID-19 Disease Seems to Be One of the Key Factors Determining the Disease Progression—A Metabolomic NMR Study on Blood Plasma. Appl. Sci. 2021, 11, 4231. [Google Scholar] [CrossRef]
- Ballout, R.A.; Kong, H.; Sampson, M.; Otvos, J.D.; Cox, A.L.; Agbor-Enoh, S.; Remaley, A.T. The NIH Lipo-COVID Study: A Pilot NMR Investigation of Lipoprotein Subfractions and Other Metabolites in Patients with Severe COVID-19. Biomedicines 2021, 9, 1090. [Google Scholar] [CrossRef]
- Loo, R.L.; Lodge, S.; Kimhofer, T.; Bong, S.-H.; Begum, S.; Whiley, L.; Gray, N.; Lindon, J.C.; Nitschke, P.; Lawler, N.G.; et al. Quantitative In-Vitro Diagnostic NMR Spectroscopy for Lipoprotein and Metabolite Measurements in Plasma and Serum: Recommendations for Analytical Artifact Minimization with Special Reference to COVID-19/SARS-CoV-2 Samples. J. Proteome Res. 2020, 19, 4428–4441. [Google Scholar] [CrossRef]
- Bizkarguenaga, M.; Bruzzone, C.; Gil-Redondo, R.; Sanjuan, I.; Martin-Ruiz, I.; Barriales, D.; Palacios, A.; Pasco, S.T.; González-Valle, B.; Laín, A.; et al. Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics. NMR Biomed. 2022, 35, e4637. [Google Scholar] [CrossRef]
- Ceriello, A. Hyperglycemia and the worse prognosis of COVID-19. Why a fast blood glucose control should be mandatory. Diabetes Res. Clin. Pract. 2020, 163, 108186. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Chong, J.; Xia, J. Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets. Metabolites 2021, 11, 44. [Google Scholar] [CrossRef]
- Ghini, V.; Meoni, G.; Pelagatti, L.; Celli, T.; Veneziani, F.; Petrucci, F.; Vannucchi, V.; Bertini, L.; Luchinat, C.; Landini, G.; et al. Profiling metabolites and lipoproteins in COMETA, an Italian cohort of COVID-19 patients. PLoS Pathog. 2022, 18, e1010443. [Google Scholar] [CrossRef] [PubMed]
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 2021, 17, e1009243. [Google Scholar] [CrossRef]
- Fuertes, M.; Correig, X.; Vallvé, J.C.; Amigó, N. Human Serum/Plasma Glycoprotein Analysis by 1H-NMR, an Emerging Method of Inflammatory Assessment. J. Clin. Med. 2020, 9, 354. [Google Scholar] [CrossRef]
- Otvos, J.D.; Shalaurova, I.; Wolak-Dinsmore, J.; Connelly, M.A.; Mackey, R.H.; Stein, J.H.; Tracy, R.P. GlycA: A Composite Nuclear Magnetic Resonance Biomarker of Systemic Inflammation. Clin. Chem. 2015, 61, 714–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochepied, T.; Berger, F.G.; Baumann, H.; Libert, C. α1-Acid glycoprotein: An acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003, 14, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Raziani, F.; Ebrahimi, P.; Engelsen, S.B.; Astrup, A.; Raben, A.; Tholstrup, T. Consumption of regular-fat vs reduced-fat cheese reveals gender-specific changes in LDL particle size—A randomized controlled trial. Nutr. Metab. 2018, 15, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giskeødegård, G.F.; Andreassen, T.; Bertilsson, H.; Tessem, M.-B.; Bathen, T.F. The effect of sampling procedures and day-to-day variations in metabolomics studies of biofluids. Anal. Chim. Acta 2019, 1081, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Gafson, A.R.; Thorne, T.; McKechnie, C.I.J.; Jimenez, B.; Nicholas, R.; Matthews, P.M. Lipoprotein markers associated with disability from multiple sclerosis. Sci. Rep. 2018, 8, 17026. [Google Scholar] [CrossRef]
- Lodge, S.; Nitschke, P.; Kimhofer, T.; Wist, J.; Bong, S.H.; Loo, R.L.; Masuda, R.; Begum, S.; Richards, T.; Lindon, J.C.; et al. Diffusion and Relaxation Edited Proton NMR Spectroscopy of Plasma Reveals a High-Fidelity Supramolecular Biomarker Signature of SARS-CoV-2 Infection. Anal. Chem. 2021, 93, 3976–3986. [Google Scholar] [CrossRef]
- Masuda, R.; Lodge, S.; Whiley, L.; Gray, N.; Lawler, N.; Nitschke, P.; Bong, S.-H.; Kimhofer, T.; Loo, R.L.; Boughton, B.; et al. Exploration of Human Serum Lipoprotein Supramolecular Phospholipids Using Statistical Heterospectroscopy in n-Dimensions (SHY-n): Identification of Potential Cardiovascular Risk Biomarkers Related to SARS-CoV-2 Infection. Anal. Chem. 2022, 94, 4426–4436. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255. [Google Scholar] [CrossRef]
- Mulchandani, R.; Lyngdoh, T.; Kakkar, A.K. Deciphering the COVID-19 cytokine storm: Systematic review and meta-analysis. Eur. J. Clin. Investig. 2021, 51, e13429. [Google Scholar] [CrossRef]
- Kox, M.; Waalders, N.J.B.; Kooistra, E.J.; Gerretsen, J.; Pickkers, P. Cytokine Levels in Critically Ill Patients With COVID-19 and Other Conditions. JAMA 2020, 324, 1565. [Google Scholar] [CrossRef]
- Leisman, D.E.; Ronner, L.; Pinotti, R.; Taylor, M.D.; Sinha, P.; Calfee, C.S.; Hirayama, A.V.; Mastroiani, F.; Turtle, C.J.; Harhay, M.O.; et al. Cytokine elevation in severe and critical COVID-19: A rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020, 8, 1233–1244. [Google Scholar] [CrossRef]
- Bergamaschi, L.; Mescia, F.; Turner, L.; Hanson, A.L.; Kotagiri, P.; Dunmore, B.J.; Ruffieux, H.; De Sa, A.; Huhn, O.; Morgan, M.D.; et al. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity 2021, 54, 1257–1275.e8. [Google Scholar] [CrossRef]
- Singh, Y.; Trautwein, C.; Fendel, R.; Krickeberg, N.; Berezhnoy, G.; Bissinger, R.; Ossowski, S.; Salker, M.S.; Casadei, N.; Riess, O. SARS-CoV-2 infection paralyzes cytotoxic and metabolic functions of the immune cells. Heliyon 2021, 7, e07147. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Luo, M.; Liu, J.; Jiang, W.; Yue, S.; Liu, H.; Wei, S. IL-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight 2020, 5, e139024. [Google Scholar] [CrossRef]
- Idiz, U.O.; Yurttas, T.T.; Degirmencioglu, S.; Orhan, B.; Erdogan, E.; Sevik, H.; Sevinc, M.M. Immunophenotyping of lymphocytes and monocytes and the status of cytokines in the clinical course of Covid-19 patients. J. Med. Virol. 2022, 94, 4744–4753. [Google Scholar] [CrossRef]
- Biron, C.A. Role of early cytokines, including alpha and beta interferons (IFN-α\β), in innate and adaptive immune responses to viral infections. Semin. Immunol. 1998, 10, 383–390. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Chen, Y.M.; Zheng, Y.; Yu, Y.; Wang, Y.; Huang, Q.; Qian, F.; Sun, L.; Song, Z.G.; Chen, Z.; Feng, J.; et al. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J. 2020, 39, e105896. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Galani, I.-E.; Rovina, N.; Lampropoulou, V.; Triantafyllia, V.; Manioudaki, M.; Pavlos, E.; Koukaki, E.; Fragkou, P.C.; Panou, V.; Rapti, V.; et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021, 22, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; Sowell, R.T.; Kaech, S.M.; Pearce, E.L. Metabolic Instruction of Immunity. Cell 2017, 169, 570–586. [Google Scholar] [CrossRef] [PubMed]
- Batabyal, R.; Freishtat, N.; Hill, E.; Rehman, M.; Freishtat, R.; Koutroulis, I. Metabolic dysfunction and immunometabolism in COVID-19 pathophysiology and therapeutics. Int. J. Obes. 2021, 45, 1163–1169. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Bhupathiraju, S.N.; Hu, F.B. Use of Metabolomics in Improving Assessment of Dietary Intake. Clin. Chem. 2018, 64, 82–98. [Google Scholar] [CrossRef] [Green Version]
- Lim, A.; Hippchen, T.; Unger, I.; Heinze, O.; Welker, A.; Kräusslich, H.-G.; Weigand, M.A.; Merle, U. An Outpatient Management Strategy Using a Coronataxi Digital Early Warning System Reduces Coronavirus Disease 2019 Mortality. Open Forum Infect. Dis. 2022, 9, ofac063. [Google Scholar] [CrossRef]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Stringer, D.; Braude, P.; Myint, P.K.; Evans, L.; Collins, J.T.; Verduri, A.; Quinn, T.J.; Vilches-Moraga, A.; Stechman, M.J.; Pearce, L.; et al. The role of C-reactive protein as a prognostic marker in COVID-19. Int. J. Epidemiol. 2021, 50, 420–429. [Google Scholar] [CrossRef]
- Cheng, L.; Li, H.; Li, L.; Liu, C.; Yan, S.; Chen, H.; Li, Y. Ferritin in the coronavirus disease 2019 (COVID-19): A systematic review and meta-analysis. J. Clin. Lab. Anal. 2020, 34, e23618. [Google Scholar] [CrossRef]
- Teahan, O.; Gamble, S.; Holmes, E.; Waxman, J.; Nicholson, J.K.; Bevan, C.; Keun, H.C. Impact of Analytical Bias in Metabonomic Studies of Human Blood Serum and Plasma. Anal. Chem. 2006, 78, 4307–4318. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Robert Koch Institut. Bericht zu Virusvarianten von SARS-CoV-2 in Deutschland. 26 May 2021. Available online: https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/DESH/Bericht_VOC_2021-05-26.pdf?__blob=publicationFile (accessed on 13 October 2022).
- B.I. Methods—Enabling Standardization of IVDr by NMR: Using Bruker IVDr Methods for Urine/ Plasma/ Serum Quantification. Available online: https://www.bruker.com/de/products-and-solutions/mr/nmr-clinical-research-solutions/b-i-methods.html (accessed on 12 December 2022).
- Wu, D.H.; Chen, A.D.; Johnson, C.S. An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. J. Magn. Reson. Ser. A 1995, 115, 260–264. [Google Scholar] [CrossRef]
- Nitschke, P.; Lodge, S.; Hall, D.; Schaefer, H.; Spraul, M.; Embade, N.; Millet, O.; Holmes, E.; Wist, J.; Nicholson, J.K. Direct Low Field J-Edited DIffusional Proton NMR Spectroscopic Measurement of COVID-19 Inflammatory Biomarkers in Human Serum. Analyst 2022, 147, 4213–4221. [Google Scholar] [CrossRef]
- Jiménez, B.; Holmes, E.; Heude, C.; Tolson, R.F.; Harvey, N.; Lodge, S.L.; Chetwynd, A.J.; Cannet, C.; Fang, F.; Pearce, J.T.M.; et al. Quantitative Lipoprotein Subclass and Low Molecular Weight Metabolite Analysis in Human Serum and Plasma by 1H NMR Spectroscopy in a Multilaboratory Trial. Anal. Chem. 2018, 90, 11962–11971. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37 (Suppl. 2), W652–W660. [Google Scholar] [CrossRef] [Green Version]
- Armitage, E.G.; Godzien, J.; Alonso-Herranz, V.; López-Gonzálvez, Á.; Barbas, C. Missing value imputation strategies for metabolomics data. Electrophoresis 2015, 36, 3050–3060. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- la Pena, J.E.D.; Rasc, R.A.; Ascencio-Montiel, I.D.; Gonz, E.; Fern, J.E.; Medina-G, O.S.; Borja-Bustamante, P.; Santill, J.A.; Borja-Aburto, V.H. Hypertension, Diabetes and Obesity, Major Risk Factors for Death in Patients with COVID-19 in Mexico. Arch. Med. Res. 2021, 52, 443–449. [Google Scholar] [CrossRef]
- Landstra, C.P.; de Koning, E.J.P. COVID-19 and Diabetes: Understanding the Interrelationship and Risks for a Severe Course. Front. Endocrinol. 2021, 12, 649525. [Google Scholar] [CrossRef]
- Critchley, J.A.; Carey, I.M.; Harris, T.; DeWilde, S.; Hosking, F.J.; Cook, D.G. Glycemic Control and Risk of Infections Among People With Type 1 or Type 2 Diabetes in a Large Primary Care Cohort Study. Diabetes Care 2018, 41, 2127–2135. [Google Scholar] [CrossRef]
- Liu, S.P.; Zhang, Q.; Wang, W.; Zhang, M.; Liu, C.; Xiao, X.F.; Liu, Z.D.; Hu, W.M.; Jin, P. Hyperglycemia is a strong predictor of poor prognosis in COVID-19. Diabetes Res. Clin. Pract. 2020, 167, 108338. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tian, S.; Chen, T.; Cui, Z.; Shi, N.; Zhong, X.; Qiu, K.; Zhang, J.; Zeng, T.; Chen, L.; et al. Newly diagnosed diabetes is associated with a higher risk of mortality than known diabetes in hospitalized patients with COVID-19. Diabetes Obes. Metab. 2020, 22, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Bae, J.H.; Kwon, H.-S.; Nauck, M.A. COVID-19 and diabetes mellitus: From pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021, 17, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-T.; Lidsky, P.V.; Xiao, Y.; Lee, I.T.; Cheng, R.; Nakayama, T.; Jiang, S.; Demeter, J.; Bevacqua, R.J.; Chang, C.A.; et al. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 2021, 33, 1565–1576.e5. [Google Scholar] [CrossRef] [PubMed]
- Rotter, V.; Nagaev, I.; Smith, U. Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-α, overexpressed in human fat cells from insulin-resistant subjects. J. Biol. Chem. 2003, 278, 45777–45784. [Google Scholar] [CrossRef] [Green Version]
- Schultz, O.; Oberhauser, F.; Saech, J.; Rubbert-Roth, A.; Hahn, M.; Krone, W.; Laudes, M. Effects of Inhibition of Interleukin-6 Signalling on Insulin Sensitivity and Lipoprotein (A) Levels in Human Subjects with Rheumatoid Diseases. PLoS ONE 2010, 5, e14328. [Google Scholar] [CrossRef] [Green Version]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L.D.B.; De Souza, G.F.; Muraro, S.P.; Virgilio-Da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; De Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 437–446.e5. [Google Scholar] [CrossRef]
- Icard, P.; Lincet, H.; Wu, Z.R.; Coquerel, A.; Forgez, P.; Alifano, M.; Fournel, L. The key role of Warburg effect in SARS-CoV-2 replication and associated inflammatory response. Biochimie 2021, 180, 169–177. [Google Scholar] [CrossRef]
- Icard, P.; Lincet, H. The reduced concentration of citrate in cancer cells: An indicator of cancer aggressiveness and a possible therapeutic target. Drug Resist. Updat. 2016, 29, 47–53. [Google Scholar]
- Ceperuelo-Mallafré, V.; Reverté, L.; Peraire, J.; Madeira, A.; Maymó-Masip, E.; López-Dupla, M.; Gutierrez-Valencia, A.; Ruiz-Mateos, E.; Buzón, M.J.; Jorba, R.; et al. Circulating pyruvate is a potent prognostic marker for critical COVID-19 outcomes. Front. Immunol. 2022, 13, 912579. [Google Scholar] [CrossRef]
- Rodas, P.C.; Rooyackers, O.; Hebert, C.; Norberg, Å.; Wernerman, J. Glutamine and glutathione at ICU admission in relation to outcome. Clin. Sci. 2012, 122, 591–597. [Google Scholar] [CrossRef]
- Matsuyama, T.; Yoshinaga, S.K.; Shibue, K.; Mak, T.W. Comorbidity-associated glutamine deficiency is a predisposition to severe COVID-19. Cell Death Differ. 2021, 28, 3199–3213. [Google Scholar] [CrossRef]
- Newsholme, P. Why Is L-Glutamine Metabolism Important to Cells of the Immune System in Health, Postinjury, Surgery or Infection? J. Nutr. 2001, 131, 2515S–2522S. [Google Scholar] [CrossRef] [Green Version]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Hippchen, T.; Altamura, S.; Muckenthaler, M.U.; Merle, U. Hypoferremia is Associated With Increased Hospitalization and Oxygen Demand in COVID-19 Patients. HemaSphere 2020, 4, e492. [Google Scholar] [CrossRef]
- Liu, W.; Li, H. COVID-19: Captures iron and generates reactive oxygen species to damage the human immune system. Autoimmunity 2021, 54, 1–12. [Google Scholar]
- Neurauter, G.; Grahmann, A.V.; Klieber, M.; Zeimet, A.; Ledochowski, M.; Sperner-Unterweger, B.; Fuchs, D. Serum phenylalanine concentrations in patients with ovarian carcinoma correlate with concentrations of immune activation markers and of isoprostane-8. Cancer Lett. 2008, 272, 141–147. [Google Scholar] [CrossRef]
- Geisler, S.; Gostner, J.M.; Becker, K.; Ueberall, F.; Fuchs, D. Immune activation and inflammation increase the plasma phenylalanine-to-tyrosine ratio. Pteridines 2013, 24, 27–31. [Google Scholar] [CrossRef]
- Wilhelm, C.; Surendar, J.; Karagiannis, F. Enemy or ally? Fasting as an essential regulator of immune responses. Trends Immunol. 2021, 42, 389–400. [Google Scholar] [CrossRef]
- Stubbs, B.J.; Koutnik, A.P.; Goldberg, E.L.; Upadhyay, V.; Turnbaugh, P.J.; Verdin, E.; Newman, J.C. Investigating Ketone Bodies as Immunometabolic Countermeasures against Respiratory Viral Infections. Med 2020, 1, 43–65. [Google Scholar] [CrossRef]
- Karagiannis, F.; Peukert, K.; Surace, L.; Michla, M.; Nikolka, F.; Fox, M.; Weiss, P.; Feuerborn, C.; Maier, P.; Schulz, S.; et al. Impaired ketogenesis ties metabolism to T cell dysfunction in COVID-19. Nature 2022, 609, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, P.C.; Seeds, W.A.; Miller, A.C.; Mahajan, V.R.; Curtis, W.M. COVID-19: Proposing a Ketone-Based Metabolic Therapy as a Treatment to Blunt the Cytokine Storm. Oxidative Med. Cell. Longev. 2020, 2020, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Hirschberger, S.; Gellert, L.; Effinger, D.; Muenchhoff, M.; Herrmann, M.; Briegel, J.M.; Zwißler, B.; Kreth, S. Ketone Bodies Improve Human CD8(+) Cytotoxic T-Cell Immune Response During COVID-19 Infection. Front. Med. 2022, 9, 923502. [Google Scholar] [CrossRef] [PubMed]
- Khovidhunkit, W.; Kim, M.-S.; Memon, R.A.; Shigenaga, J.K.; Moser, A.H.; Feingold, K.R.; Grunfeld, C. Effects of infection and inflammation on lipid and lipoprotein metabolism: Mechanisms and consequences to the host. J. Lipid Res. 2004, 45, 1169–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Q.; Huang, D.; Yu, H.; Zhu, Z.; Xia, Z.; Su, Y.; Li, Z.; Zhou, G.; Gou, J.; Qu, J.; et al. COVID-19: Abnormal liver function tests. J. Hepatol. 2020, 73, 566–574. [Google Scholar] [CrossRef]
- Cichoż-Lach, H.; Michalak, A. Liver injury in the era of COVID-19. World J. Gastroenterol. 2021, 27, 377–390. [Google Scholar] [CrossRef]
- Taneva, G.; Dimitrov, D.; Velikova, T. Liver dysfunction as a cytokine storm manifestation and prognostic factor for severe COVID-19. World J. Hepatol. 2021, 13, 2005–2012. [Google Scholar] [CrossRef]
- Gray, N.; Lawler, N.; Zeng, A.; Ryan, M.; Bong, S.; Boughton, B.; Bizkarguenaga, M.; Bruzzone, C.; Embade, N.; Wist, J.; et al. Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection. Metabolites 2021, 11, 467. [Google Scholar] [CrossRef]
- Namiri-Kalantari, R.; Gao, F.; Chattopadhyay, A.; Wheeler, A.A.; Navab, K.D.; Farias-Eisner, R.; Reddy, S.T. The dual nature of HDL: Anti-Inflammatory and pro-Inflammatory. BioFactors 2015, 41, 153–159. [Google Scholar] [CrossRef]
- Rao, V.S.; Bindu, H.G.; Kakkar, V.V. Friend turns foe: Transformation of anti-inflammatory HDL to proinflammatory HDL during acute-phase response. Cholesterol 2011, 2011, 274629. [Google Scholar]
- Begue, F.; Tanaka, S.; Mouktadi, Z.; Rondeau, P.; Veeren, B.; Diotel, N.; Tran-Dinh, A.; Robert, T.; Vélia, E.; Mavingui, P.; et al. Altered high-density lipoprotein composition and functions during severe COVID-19. Sci. Rep. 2021, 11, 2291. [Google Scholar] [CrossRef]
- Freedman, D.S.; Otvos, J.D.; Jeyarajah, E.J.; Shalaurova, I.; Cupples, L.A.; Parise, H.; D’Agostino, R.B.; Wilson, P.W.F.; Schaefer, E.J. Sex and Age Differences in Lipoprotein Subclasses Measured by Nuclear Magnetic Resonance Spectroscopy: The Framingham Study. Clin. Chem. 2004, 50, 1189–1200. [Google Scholar] [CrossRef]
- Lucero, D.; Islam, P.; Freeman, L.A.; Jin, X.; Pryor, M.; Tang, J.; Kruth, H.S.; Remaley, A.T. Interleukin 10 promotes macrophage uptake of HDL and LDL by stimulating fluid-phase endocytosis. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2020, 1865, 158537. [Google Scholar] [CrossRef]
- Li, C.; Ye, J.; Chen, Q.; Hu, W.; Wang, L.; Fan, Y.; Lu, Z.; Chen, J.; Chen, Z.; Chen, S.; et al. Elevated Lactate Dehydrogenase (LDH) level as an independent risk factor for the severity and mortality of COVID-19. Aging 2020, 12, 15670–15681. [Google Scholar] [CrossRef]
- Schienkiewitz, A.; Mensink, G.B.M.; Kuhnert, R.; Lange, C. Übergewicht und Adipositas bei Erwachsenen in Deutschland. J. Health Monit. 2017. [Google Scholar] [CrossRef]
- Subramanian, A.; Nirantharakumar, K.; Hughes, S.; Myles, P.; Williams, T.; Gokhale, K.M.; Taverner, T.; Chandan, J.S.; Brown, K.; Simms-Williams, N.; et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat. Med. 2022, 28, 1706–1714. [Google Scholar] [CrossRef]
- Xu, J.; Pan, T.; Qi, X.; Tan, R.; Wang, X.; Liu, Z.; Tao, Z.; Qu, H.; Zhang, Y.; Chen, H.; et al. Increased mortality of acute respiratory distress syndrome was associated with high levels of plasma phenylalanine. Respir. Res. 2020, 21, 99. [Google Scholar] [CrossRef]
- Luporini, R.L.; Pott-Junior, H.; Di Medeiros Leal, M.C.B.; Castro, A.; Ferreira, A.G.; Cominetti, M.R.; De Freitas Anibal, F. Phenylalanine and COVID-19: Tracking disease severity markers. Int. Immunopharmacol. 2021, 101, 108313. [Google Scholar] [CrossRef]
- Malo, A.-I.; Rull, A.; Girona, J.; Domingo, P.; Fuertes-Martín, R.; Amigó, N.; Rodríguez-Borjabad, C.; Martínez-Micaelo, N.; Leal, M.; Peraire, J.; et al. Glycoprotein Profile Assessed by 1H-NMR as a Global Inflammation Marker in Patients with HIV Infection. A Prospective Study. J. Clin. Med. 2020, 9, 1344. [Google Scholar] [CrossRef]
- Rodríguez-Carrio, J.; Alperi-López, M.; López, P.; Pérez-Álvarez, Á.I.; Gil-Serret, M.; Amigó, N.; Ulloa, C.; Benavente, L.; Ballina-García, F.J.; Suárez, A. GlycA Levels during the Earliest Stages of Rheumatoid Arthritis: Potential Use as a Biomarker of Subclinical Cardiovascular Disease. J. Clin. Med. 2020, 9, 2472. [Google Scholar] [CrossRef]
- Chung, C.P.; Ormseth, M.J.; Connelly, M.A.; Oeser, A.; Solus, J.F.; Otvos, J.D.; Raggi, P.; Stein, C.M. GlycA, a novel marker of inflammation, is elevated in systemic lupus erythematosus. Lupus 2016, 25, 296–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dierckx, T.; Verstockt, B.; Vermeire, S.; Van Weyenbergh, J. GlycA, a Nuclear Magnetic Resonance Spectroscopy Measure for Protein Glycosylation, is a Viable Biomarker for Disease Activity in IBD. J. Crohn’s Colitis 2019, 13, 389–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dungan, K.; Binkley, P.; Osei, K. GlycA is a Novel Marker of Inflammation Among Non-Critically Ill Hospitalized Patients with Type 2 Diabetes. Inflammation 2015, 38, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Gruppen, E.G.; Riphagen, I.J.; Connelly, M.A.; Otvos, J.D.; Bakker, S.J.L.; Dullaart, R.P.F. GlycA, a Pro-Inflammatory Glycoprotein Biomarker, and Incident Cardiovascular Disease: Relationship with C-Reactive Protein and Renal Function. PLoS ONE 2015, 10, e0139057. [Google Scholar] [CrossRef] [PubMed]
- Gruppen, E.G.; Connelly, M.A.; Otvos, J.D.; Bakker, S.J.L.; Dullaart, R.P.F. A novel protein glycan biomarker and LCAT activity in metabolic syndrome. Eur. J. Clin. Investig. 2015, 45, 850–859. [Google Scholar] [CrossRef]
- Fuertes-Martín, R.; Moncayo, S.; Insenser, M.; Martínez-García, M.Á.; Luque-Ramírez, M.; Grau, N.A.; Blanchar, X.C.; Escobar-Morreale, H.F. Glycoprotein A and B Height-to-Width Ratios as Obesity-Independent Novel Biomarkers of Low-Grade Chronic Inflammation in Women with Polycystic Ovary Syndrome (PCOS). J. Proteome Res. 2019, 18, 4038–4045. [Google Scholar] [CrossRef]
- Akira, S.; Kishimoto, T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol. Rev. 1992, 127, 25–50. [Google Scholar] [CrossRef]
- Ritchie, S.C.; Würtz, P.; Nath, A.P.; Abraham, G.; Havulinna, A.S.; Fearnley, L.G.; Sarin, A.P.; Kangas, A.J.; Soininen, P.; Aalto, K.; et al. The Biomarker GlycA Is Associated with Chronic Inflammation and Predicts Long-Term Risk of Severe Infection. Cell Syst. 2015, 1, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Fischer, K.; Kettunen, J.; Würtz, P.; Haller, T.; Havulinna, A.S.; Kangas, A.J.; Soininen, P.; Esko, T.; Tammesoo, M.-L.; Mägi, R.; et al. Biomarker Profiling by Nuclear Magnetic Resonance Spectroscopy for the Prediction of All-Cause Mortality: An Observational Study of 17,345 Persons. PLoS Med. 2014, 11, e1001606. [Google Scholar] [CrossRef]
- Julkunen, H.; Cichońska, A.; Slagboom, P.E.; Würtz, P. Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. eLife 2021, 10, e63033. [Google Scholar] [CrossRef]
- Nitschke, P.; Lodge, S.; Kimhofer, T.; Masuda, R.; Bong, S.-H.; Hall, D.; Schäfer, H.; Spraul, M.; Pompe, N.; Diercks, T.; et al. J-Edited DIffusional Proton Nuclear Magnetic Resonance Spectroscopic Measurement of Glycoprotein and Supramolecular Phospholipid Biomarkers of Inflammation in Human Serum. Anal. Chem. 2022, 94, 1333–1341. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pahan, K. Is COVID-19 Gender-sensitive? J. Neuroimmune Pharmacol. 2021, 16, 38–47. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rössler, T.; Berezhnoy, G.; Singh, Y.; Cannet, C.; Reinsperger, T.; Schäfer, H.; Spraul, M.; Kneilling, M.; Merle, U.; Trautwein, C. Quantitative Serum NMR Spectroscopy Stratifies COVID-19 Patients and Sheds Light on Interfaces of Host Metabolism and the Immune Response with Cytokines and Clinical Parameters. Metabolites 2022, 12, 1277. https://doi.org/10.3390/metabo12121277
Rössler T, Berezhnoy G, Singh Y, Cannet C, Reinsperger T, Schäfer H, Spraul M, Kneilling M, Merle U, Trautwein C. Quantitative Serum NMR Spectroscopy Stratifies COVID-19 Patients and Sheds Light on Interfaces of Host Metabolism and the Immune Response with Cytokines and Clinical Parameters. Metabolites. 2022; 12(12):1277. https://doi.org/10.3390/metabo12121277
Chicago/Turabian StyleRössler, Titus, Georgy Berezhnoy, Yogesh Singh, Claire Cannet, Tony Reinsperger, Hartmut Schäfer, Manfred Spraul, Manfred Kneilling, Uta Merle, and Christoph Trautwein. 2022. "Quantitative Serum NMR Spectroscopy Stratifies COVID-19 Patients and Sheds Light on Interfaces of Host Metabolism and the Immune Response with Cytokines and Clinical Parameters" Metabolites 12, no. 12: 1277. https://doi.org/10.3390/metabo12121277
APA StyleRössler, T., Berezhnoy, G., Singh, Y., Cannet, C., Reinsperger, T., Schäfer, H., Spraul, M., Kneilling, M., Merle, U., & Trautwein, C. (2022). Quantitative Serum NMR Spectroscopy Stratifies COVID-19 Patients and Sheds Light on Interfaces of Host Metabolism and the Immune Response with Cytokines and Clinical Parameters. Metabolites, 12(12), 1277. https://doi.org/10.3390/metabo12121277