Metabolomic Analysis of Exosomes Derived from Lung Cancer Cell Line H460 Treated with SH003 and Docetaxel
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cell Viability Assay
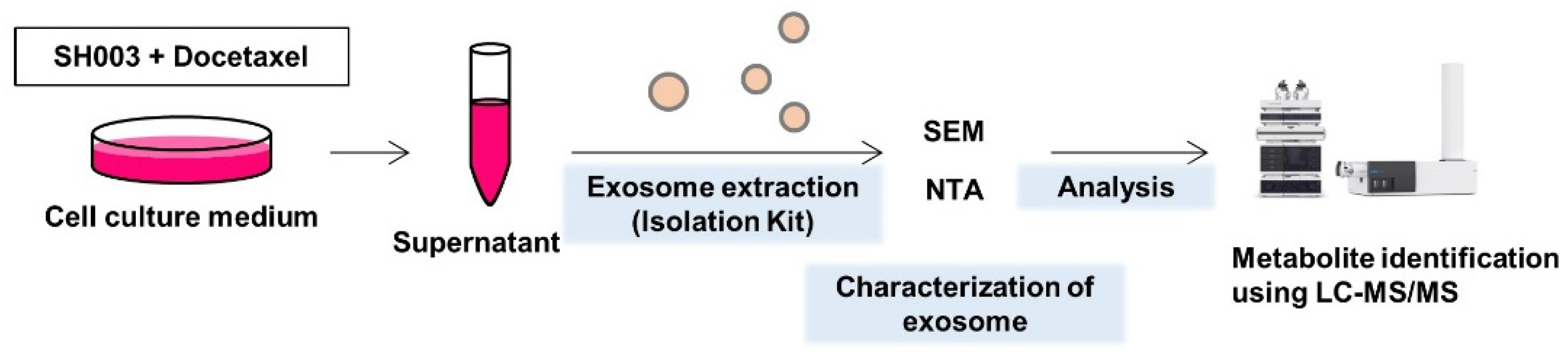
2.2. Isolation of Exosomes
2.3. Nanoparticle Tracking Analysis (NTA)
2.4. Scanning Electron Microscopy (SEM)
2.5. Western Blotting
2.6. LC-MS/MS Analysis
2.7. Quantification of Exosomal Metabolites and Statistical Analysis
3. Results
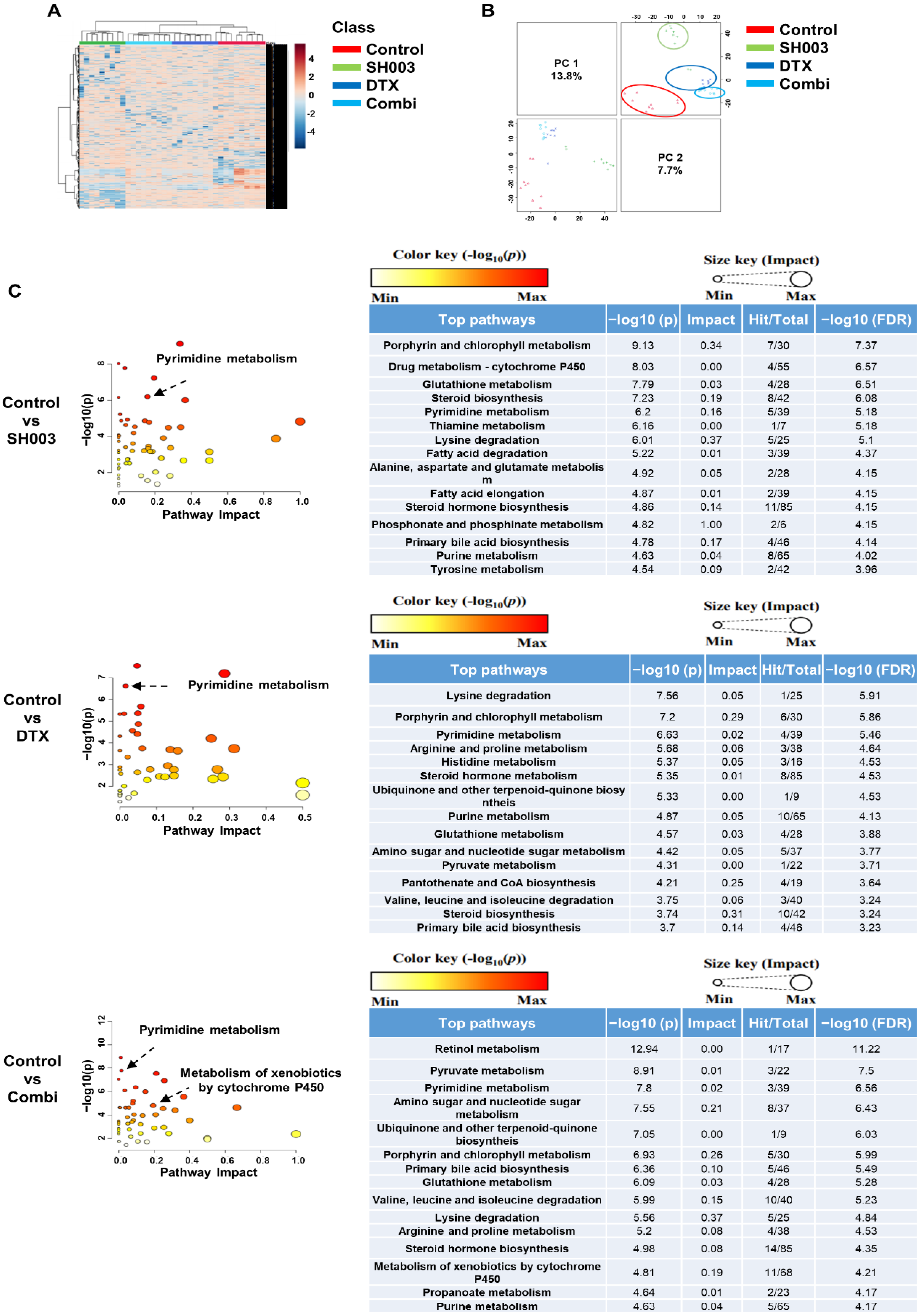
3.1. Characterization of Exosomes Released from H460 Cells Treated with Both SH003 and DTX
3.2. Metabolome Analysis of Exosomes after Combinatorial Treatment
3.3. Identification of Metabolites Altered by Combinatorial Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. 2020, 5, 145. [Google Scholar] [CrossRef]
- Huang, T.; Deng, C.X. Current Progresses of Exosomes as Cancer Diagnostic and Prognostic Biomarkers. Int. J. Biol. Sci. 2019, 15, 1–11. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Ab Razak, N.S.; Ab Mutalib, N.S.; Mohtar, M.A.; Abu, N. Impact of Chemotherapy on Extracellular Vesicles: Understanding the Chemo-EVs. Front. Oncol. 2019, 9, 1113. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. Opinion: Understanding ‘global’ systems biology: Metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2003, 2, 668–676. [Google Scholar] [CrossRef]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef]
- Gao, D.; Wang, Y.; Xie, W.; Yang, T.; Jiang, Y.; Guo, Y.; Guan, J.; Liu, H. Metabolomics study on the antitumor effect of marine natural compound flexibilide in HCT-116 colon cancer cell line. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1014, 17–23. [Google Scholar] [CrossRef]
- Cai, A.; Zheng, H.; Chen, Z.; Lin, X.; Li, C.; Yao, Q.; Bhutia, Y.D.; Ganapathy, V.; Chen, R.; Kou, L. Synergism between SLC6A14 blockade and gemcitabine in pancreactic cancer: A 1H-NMR-based metabolomic study in pancreatic cancer cells. Biochem. J. 2020, 477, 1923–1937. [Google Scholar] [CrossRef]
- Del Coco, L.; Majellaro, M.; Boccarelli, A.; Cellamare, S.; Altomare, C.D.; Fanizzi, F.P. Novel Antiproliferative Biphenyl Nicotinamide: NMR Metabolomic Study of its Effect on the MCF-7 Cell in Comparison with Cisplatin and Vinblastine. Molecules 2020, 25, 3502. [Google Scholar] [CrossRef]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.C.; He, X.; Kamp, M.A.; Sabel, M.; Barker, R.A.; Steiger, H.J.; et al. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef]
- Wei, B.; Wang, C.; Teng, T.; Guo, P.; Chen, M.; Xia, F.; Liu, H.; Xie, J.; Feng, J.; Huang, H. Chemotherapeutic efficacy of cucurmosin for pancreatic cancer as an alternative of gemcitabine: A comparative metabolomic study. Gland Surg. 2020, 9, 1428–1442. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Lu, R.; Song, H.; Wu, J.; Liu, X.; Zhou, X.; Yang, J.; Zhang, H.; Tang, C.; Guo, H.; et al. Metabolomic study of natrin-induced apoptosis in SMMC-7721 hepatocellular carcinoma cells by ultra-performance liquid chromatography-quadrupole/time-of-flight mass spectrometry. Int. J. Biol. Macromol. 2019, 124, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lee, K.; Yoon, J.H.; Cho, S.G.; Kim, Y.G.; Jeong, M.; Hwang, H.H.; Lee, S.Y.; Jung, S.E.; Ko, S.G. SH003 and Docetaxel Show Synergistic Anticancer Effects by Inhibiting EGFR Activation in Triple-Negative Breast Cancer. BioMed Res. Int. 2022, 2022, 3647900. [Google Scholar] [CrossRef]
- Jeong, M.S.; Lee, K.W.; Choi, Y.J.; Kim, Y.G.; Hwang, H.H.; Lee, S.Y.; Jung, S.E.; Park, S.A.; Lee, J.H.; Joo, Y.J.; et al. Synergistic Antitumor Activity of SH003 and Docetaxel via EGFR Signaling Inhibition in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 8405. [Google Scholar] [CrossRef]
- Kim, T.W.; Cheon, C.; Ko, S.G. SH003 activates autophagic cell death by activating ATF4 and inhibiting G9a under hypoxia in gastric cancer cells. Cell Death Dis. 2020, 11, 717. [Google Scholar] [CrossRef]
- Lee, K.M.; Lee, K.; Choi, Y.K.; Choi, Y.J.; Seo, H.S.; Ko, S.G. SH003induced G1 phase cell cycle arrest induces apoptosis in HeLa cervical cancer cells. Mol. Med. Rep. 2017, 16, 8237–8244. [Google Scholar] [CrossRef]
- Seo, H.S.; Ku, J.M.; Lee, H.J.; Woo, J.K.; Cheon, C.; Kim, M.; Jang, B.H.; Shin, Y.C.; Ko, S.G. SH003 reverses drug resistance by blocking signal transducer and activator of transcription 3 (STAT3) signaling in breast cancer cells. Biosci. Rep. 2017, 37, BSR20170125. [Google Scholar] [CrossRef]
- Choi, Y.J.; Choi, Y.K.; Lee, K.M.; Cho, S.G.; Kang, S.Y.; Ko, S.G. SH003 induces apoptosis of DU145 prostate cancer cells by inhibiting ERK-involved pathway. BMC Complement. Altern. Med. 2016, 16, 507. [Google Scholar] [CrossRef]
- Choi, H.S.; Cho, S.G.; Kim, M.K.; Lee, H.J.; Moon, S.H.; Jang, H.J.; Ko, S.G. SH003 enhances paclitaxel chemosensitivity in MCF-7/PAX breast cancer cells through inhibition of MDR1 activity. Mol. Cell Biochem. 2017, 426, 1–8. [Google Scholar] [CrossRef]
- Woo, S.M.; Kim, A.J.; Choi, Y.K.; Shin, Y.C.; Cho, S.G.; Ko, S.G. Synergistic Effect of SH003 and Doxorubicin in Triple-negative Breast Cancer. Phytother. Res. 2016, 30, 1817–1823. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.K.; Kim, S.M.; Hong, S.W.; Moon, J.H.; Shin, J.S.; Kim, J.H.; Hwang, I.Y.; Jung, S.A.; Lee, D.H.; Lee, E.Y.; et al. SH003 selectively induces p73dependent apoptosis in triplenegative breast cancer cells. Mol. Med. Rep. 2016, 14, 3955–3960. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, Y.K.; Cho, S.G.; Choi, Y.J.; Yun, Y.J.; Lee, K.M.; Lee, K.; Yoo, H.H.; Shin, Y.C.; Ko, S.G. SH003 suppresses breast cancer growth by accumulating p62 in autolysosomes. Oncotarget 2017, 8, 88386–88400. [Google Scholar] [CrossRef]
- Choi, H.S.; Kim, M.K.; Lee, K.; Lee, K.M.; Choi, Y.K.; Shin, Y.C.; Cho, S.G.; Ko, S.G. SH003 represses tumor angiogenesis by blocking VEGF binding to VEGFR2. Oncotarget 2016, 7, 32969–32979. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Cho, S.G.; Woo, S.M.; Yun, Y.J.; Park, S.; Shin, Y.C.; Ko, S.G. Herbal extract SH003 suppresses tumor growth and metastasis of MDA-MB-231 breast cancer cells by inhibiting STAT3-IL-6 signaling. Mediat. Inflamm. 2014, 2014, 492173. [Google Scholar] [CrossRef]
- Lee, K.; Youn, B.Y.; Choi, Y.J.; Moon, S.; Im, J.; Cho, K.; Ko, S.G.; Cheon, C. State of the Art and Future Implications of SH003: Acting as a Therapeutic Anticancer Agent. Cancers 2022, 14, 1089. [Google Scholar] [CrossRef]
- Cheon, C.; Ko, S.G. Phase I study to evaluate the maximum tolerated dose of the combination of SH003 and docetaxel in patients with solid cancer: A study protocol. Medicine 2020, 99, e22228. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, S.; Xie, C.; Fang, J. Uridine Metabolism and Its Role in Glucose, Lipid, and Amino Acid Homeostasis. BioMed Res. Int. 2020, 2020, 7091718. [Google Scholar] [CrossRef]
- Armitage, E.G.; Barbas, C. Metabolomics in cancer biomarker discovery: Current trends and future perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. [Google Scholar] [CrossRef]
- Miyamoto, S.; Taylor, S.L.; Barupal, D.K.; Taguchi, A.; Wohlgemuth, G.; Wikoff, W.R.; Yoneda, K.Y.; Gandara, D.R.; Hanash, S.M.; Kim, K.; et al. Systemic Metabolomic Changes in Blood Samples of Lung Cancer Patients Identified by Gas Chromatography Time-of-Flight Mass Spectrometry. Metabolites 2015, 5, 192–210. [Google Scholar] [CrossRef] [PubMed]
- Mollick, T.; Lain, S. Modulating pyrimidine ribonucleotide levels for the treatment of cancer. Cancer Metab. 2020, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Hartinger, J.; Vesely, P.; Netikova, I.; Matouskova, E.; Petruzelka, L. The protective effect of pyrimidine nucleosides on human HaCaT keratinocytes treated with 5-FU. Anticancer. Res. 2015, 35, 1303–1310. [Google Scholar]
- Connolly, G.P.; Duley, J.A. Uridine and its nucleotides: Biological actions, therapeutic potentials. Trends Pharm. Sci. 1999, 20, 218–225. [Google Scholar] [CrossRef]
- van Schaik, R.H. Cancer treatment and pharmacogenetics of cytochrome P450 enzymes. Investig. New Drugs 2005, 23, 513–522. [Google Scholar] [CrossRef] [PubMed]
- St Helen, G.; Benowitz, N.L.; Ahluwalia, J.S.; Tyndale, R.F.; Addo, N.; Gregorich, S.E.; Perez-Stable, E.J.; Cox, L.S. Black Light Smokers: How Nicotine Intake and Carcinogen Exposure Differ Across Various Biobehavioral Factors. J. Natl. Med. Assoc. 2019, 111, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Esteves, F.; Rueff, J.; Kranendonk, M. The Central Role of Cytochrome P450 in Xenobiotic Metabolism-A Brief Review on a Fascinating Enzyme Family. J. Xenobiot. 2021, 11, 94–114. [Google Scholar] [CrossRef]



| Peak Diameter (nm) | Particles/mL | % | Mean (nm) | |
|---|---|---|---|---|
| Control | 163.4 | 1.5 × 106 | 51.2 | 158.5 |
| 144.3 | 1.5 × 106 | 46.1 | ||
| 303.8 | 1.5 × 105 | 2.7 | ||
| SH003 | 153.8 | 1.2 × 106 | 100 | 164.3 |
| 145.5 | 4.2 × 106 | 72.2 | 159.9 | |
| Docetaxel | 114.2 | 3.4 × 106 | 21.5 | |
| 53.9 | 4.2 × 105 | 6.3 | ||
| Combination * | 147.9 | 1.3 × 106 | 96.9 | 163 |
| 339 | 1.9 × 105 | 3.1 |
| Control | SH003 | DTX | Combi | T test (p Value) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mz | time | Mean | SD | SEM | Mean | SD | SEM | Mean | SD | SEM | Mean | SD | SEM | Control vs. SH003 | Control vs. DTX | Control vs. Combi | ||||
| Retinol metabolism | ||||||||||||||||||||
| beta-Carotene | 575.3958 | 786.2536 | 2305 | 6914 | 2305 | 206,037 | 279,956 | 93,319 | 285,444 | 120,658 | 40,219 | 272,445 | 30,756 | 10,252 | 0.0294 | * | 0.0004 | *** | <0.0001 | *** |
| Pyrimidine metabolism | ||||||||||||||||||||
| 2′-Deoxy-5-hydroxymethylcytidine-5′-diphosphate | 418.0422 | 769.836 | 122,100 | 55,140 | 18,380 | 96,258 | 151,995 | 50,665 | 24,520 | 50,510 | 16,837 | 0 | 0 | 0 | 0.0336 | * | 0.0063 | ** | <0.0001 | *** |
| 2′-Deoxy-5-hydroxymethylcytidine-5′-triphosphate | 535.9679 | 554.8825 | 206,112 | 85,006 | 28,335 | 164,458 | 316,409 | 105,470 | 94,471 | 204,050 | 68,017 | 220,157 | 165,718 | 55,239 | 0.0336 | * | 0.0063 | ** | 0.1259 | |
| Uridine | 245.0761 | 316.7708 | 1,019,626 | 227,421 | 75,807 | 4,013,268 | 1,098,555 | 366,185 | 5,464,452 | 1,485,010 | 495,003 | 7,548,178 | 967,850 | 322,617 | <0.0001 | *** | <0.0001 | *** | <0.0001 | *** |
| dUDP | 371.0011 | 604.2127 | 67,650 | 51,578 | 17,193 | 58,477 | 118,058 | 39,353 | 131,200 | 55,963 | 18,654 | 107,978 | 16,564 | 5521 | 0.0063 | ** | 0.0336 | * | 0.0063 | ** |
| dUMP | 347.0017 | 350.6969 | 79,779 | 61,204 | 20,401 | 86,368 | 133,099 | 44,366 | 77,786 | 78,168 | 26,056 | 87,092 | 40,605 | 13,535 | 0.2882 | 0.9737 | 0.3517 | |||
| dUTP | 506.9332 | 705.5207 | 13,140 | 30,905 | 10,302 | 33,207 | 99,622 | 33,207 | 124,109 | 81,905 | 27,302 | 40,246 | 60,053 | 20,018 | 0.7176 | 0.0019 | ** | 0.848 | ||
| Lysine degradation | ||||||||||||||||||||
| L-Lysine | 129.1022 | 873.5476 | 5860 | 8855 | 2952 | 32,586 | 10,740 | 3580 | 32,740 | 43,308 | 14,436 | 24,262 | 6844 | 2281 | 0.0007 | *** | 0.093 | 0.0007 | *** | |
| N6-(L-1,3-Dicarboxypropyl)-L-lysine | 277.1371 | 44.96933 | 105,655 | 20,446 | 6815 | 69,513 | 27,810 | 9270 | 34,041 | 19,798 | 6599 | 39,090 | 15,954 | 5318 | 0.0063 | ** | <0.0001 | *** | <0.0001 | *** |
| 5-Phosphonooxy-L-lysine | 281.0275 | 461.769 | 75,761 | 39,205 | 13,068 | 121,458 | 8788 | 2929 | 141,050 | 37,636 | 12,545 | 113,290 | 21,591 | 7197 | 0.0063 | ** | 0.0336 | * | 0.0063 | ** |
| 2-Oxoadipate | 199.0014 | 888.6948 | 41,562 | 18,121 | 6040 | 157,442 | 66,139 | 22,046 | 62,238 | 26,272 | 8757 | 39,224 | 10,768 | 3589 | 0.0007 | *** | 0.1259 | 0.9895 | ||
| N-Acetylputrescine | 131.1173 | 361.3031 | 126,478 | 58,625 | 19,542 | 211,008 | 158,323 | 52,774 | 57,457 | 68,535 | 22,845 | 30,973 | 2945 | 981.6 | 0.1259 | 0.0336 | * | 0.0007 | *** | |
| Glutaryl-CoA | 882.1579 | 600.7987 | 145,656 | 107,450 | 35,817 | 140,440 | 226,181 | 75,394 | 260,080 | 116,159 | 38,720 | 57,769 | 85,988 | 28,663 | 0.1146 | 0.1259 | 0.1259 | |||
| Arginine and proline metabolism | ||||||||||||||||||||
| 4-Aminobutyraldehyde | 88.07495 | 175.7108 | 143,178 | 56,241 | 18,747 | 53,268 | 110,275 | 36,758 | 173,923 | 81,752 | 27,251 | 127,881 | 78,918 | 26,306 | 0.0063 | ** | 0.1259 | 0.9895 | ||
| 4-Guanidinobutanoate | 146.0915 | 55.58422 | 59,007 | 60,056 | 20,019 | 7716 | 23,148 | 7716 | 0 | 0 | 0 | 27,310 | 33,490 | 11,163 | 0.0007 | *** | <0.0001 | *** | 0.1259 | |
| Spermidine | 146.1648 | 46.3017 | 170,411 | 52,614 | 17,538 | 200,397 | 57,593 | 19,198 | 119,336 | 62,308 | 20,769 | 73,007 | 72,973 | 24,324 | 0.0007 | *** | <0.0001 | *** | 0.1259 | |
| N-Acetylputrescine | 113.1069 | 874.4732 | 52,133 | 45,246 | 15,082 | 7599 | 22,797 | 7599 | 9669 | 11,657 | 3886 | 65,659 | 63,978 | 21,326 | 0.0007 | *** | 0.1259 | 0.3517 | ||
| L-Glutamate 5-semialdehyde | 154.0467 | 17.7201 | 92,822 | 37,286 | 12,429 | 95,409 | 131,679 | 43,893 | 154,858 | 22,740 | 7580 | 100,759 | 44,569 | 14,856 | 0.1259 | <0.0001 | *** | 0.7301 | ||
| trans-3-Hydroxy-L-proline | 154.0467 | 17.7201 | 92,822 | 37,286 | 12,429 | 95,409 | 131,679 | 43,893 | 154,858 | 22,740 | 7580 | 100,759 | 44,569 | 14,856 | 0.1259 | <0.0001 | *** | 0.7301 | ||
| cis-4-Hydroxy-D-proline | 154.0467 | 17.7201 | 92,822 | 37,286 | 12,429 | 95,409 | 131,679 | 43,893 | 154,858 | 22,740 | 7580 | 100,759 | 44,569 | 14,856 | 0.1259 | <0.0001 | *** | 0.7301 | ||
| (R)-3-Amino-2-Methylpropanoate | 86.06042 | 821.6563 | 132,513 | 42,946 | 14,315 | 31,0233 | 44,331 | 14,777 | 151,340 | 48,679 | 16,226 | 116,901 | 34,186 | 11,395 | <0.0001 | *** | 0.1259 | 0.7301 | ||
| Metabolism of xenobiotics by cytochrome P450 | ||||||||||||||||||||
| Benzo[a]pyrene-7,8-diol | 269.0944 | 791.3047 | 201,542 | 121,100 | 40,367 | 77,150 | 144,628 | 48,209 | 97,513 | 116,385 | 38,795 | 140,782 | 114,059 | 38,020 | 0.029 | * | 0.1146 | 0.3517 | ||
| Benzo[a]pyrene-4,5-epoxide | 269.0944 | 791.3047 | 201,542 | 121,100 | 40,367 | 77,150 | 144,628 | 48,209 | 97,513 | 116,385 | 38,795 | 140,782 | 114,059 | 38,020 | 0.029 | * | 0.1146 | 0.3517 | ||
| Benzo[a]pyrene-7,8-epoxide | 269.0944 | 791.3047 | 201,542 | 121,100 | 40,367 | 77,150 | 144,628 | 48,209 | 97,513 | 116,385 | 38,795 | 140,782 | 114,059 | 38,020 | 0.029 | * | 0.1146 | 0.3517 | ||
| Benzo[a]pyrene-9,10-epoxide | 269.0944 | 791.3047 | 201,542 | 121,100 | 40,367 | 77,150 | 144,628 | 48,209 | 97,513 | 116,385 | 38,795 | 140,782 | 114,059 | 38,020 | 0.029 | * | 0.1146 | 0.3517 | ||
| 9-Hydroxylbenzo[a]pyrene | 269.0944 | 791.3047 | 201,542 | 121,100 | 40,367 | 77,150 | 144,628 | 48,209 | 97,513 | 116,385 | 38,795 | 140,782 | 114,059 | 38,020 | 0.029 | * | 0.1146 | 0.3517 | ||
| 1,1-Dichloroethylene | 96.96057 | 203.2261 | 533,199 | 119,133 | 39,711 | 516,083 | 325,207 | 108,402 | 590,390 | 25,927 | 8642 | 472,406 | 45,756 | 15,252 | 0.7301 | 0.0336 | * | 0.1259 | ||
| 1,1-Dichloroethylene epoxide | 112.9561 | 887.2592 | 158,296 | 66,239 | 22,080 | 165,189 | 157,468 | 52,489 | 143,482 | 99,269 | 33,090 | 175,132 | 59,608 | 19,869 | 0.1259 | 0.7301 | 0.7301 | |||
| Chloroacetyl chloride | 112.9561 | 887.2592 | 158,296 | 66,239 | 22,080 | 165,189 | 157,468 | 52,489 | 143,482 | 99,269 | 33,090 | 175,132 | 59,608 | 19,869 | 0.1259 | 0.7301 | 0.7301 | |||
| 4,5-Dihydro-4-hydroxy-5-S-glutathionyl-benzo[a]pyrene | 558.1676 | 787.7941 | 72,075 | 51,507 | 17,169 | 60,519 | 100,124 | 33,375 | 86,611 | 89,559 | 29,853 | 67,481 | 51,654 | 17,218 | 0.0336 | * | 0.3517 | 0.7301 | ||
| 7,8-Dihydro-4-hydroxy-5-S-glutathionyl-benzo[a]pyrene | 558.1676 | 787.7941 | 72,075 | 51,507 | 17,169 | 60,519 | 100,124 | 33,375 | 86,611 | 89,559 | 29,853 | 67,481 | 51,654 | 17,218 | 0.0336 | * | 0.3517 | 0.7301 | ||
| 2,2-Dichloroacetaldehyde | 112.9561 | 887.2592 | 158,296 | 66,239 | 22,080 | 165,189 | 157,468 | 52,489 | 143,482 | 99,269 | 33,090 | 175,132 | 59,608 | 19,869 | 0.1259 | 0.7301 | 0.7301 | |||
| (1S,2R)-Naphthalene 1,2-oxide | 145.0652 | 50.46406 | 170,470 | 54,915 | 18,305 | 252,364 | 76,667 | 25,556 | 124,575 | 102,539 | 34,180 | 97,295 | 68,028 | 22,676 | 0.0336 | * | 0.0063 | ** | 0.0063 | ** |
| (1R,2S)-Naphthalene 1,2-oxide | 145.0652 | 50.46406 | 170,470 | 54,915 | 18,305 | 252,364 | 76,667 | 25,556 | 124,575 | 102,539 | 34,180 | 97,295 | 68,028 | 22,676 | 0.0336 | * | 0.0063 | ** | 0.0063 | ** |
| 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol(NNAL) | 248.0794 | 317.7357 | 2321 | 4919 | 1640 | 0 | 0 | 0 | 0 | 0 | 0 | 97,397 | 35,068 | 11,689 | 0.4706 | 0.4706 | <0.0001 | *** | ||
| 1-(Methylnitrosoamino)-4-(3-pyridinyl)-1,4-butanediol | 226.1207 | 367.8325 | 52,277 | 21,209 | 7070 | 77,463 | 3554 | 1185 | 50,549 | 6684 | 2526 | 48,744 | 5941 | 1980 | 0.0063 | ** | 0.6476 | 0.7301 | ||
| alpha-[3-[(Hydroxymethyl)nitrosoamino]propyl]-3-pyridinemethanol | 226.1207 | 367.8325 | 52,277 | 21,209 | 7070 | 77,463 | 3554 | 1185 | 50,549 | 6684 | 2526 | 48,744 | 5941 | 1980 | 0.0063 | ** | 0.6476 | 0.7301 | ||
| Propanoate metabolism | ||||||||||||||||||||
| (S)-methylmalonate semialdehyde | 140.9936 | 875.0135 | 129,827 | 64,967 | 21,656 | 91,637 | 105,217 | 35,072 | 128,205 | 62,406 | 20,802 | 114,891 | 59,161 | 19,720 | 0.1259 | 0.9895 | 0.9895 | |||
| 2-Oxobutanoate | 140.9936 | 875.0135 | 129,827 | 64,967 | 21,656 | 91,637 | 105,217 | 35,072 | 128,205 | 62,406 | 20,802 | 114,891 | 59,161 | 19,720 | 0.1259 | 0.9895 | 0.9895 | |||
| Thiamin diphosphate | 408.0386 | 125.1114 | 3220 | 9660 | 3220 | 47,367 | 38,566 | 12,855 | 122,786 | 71,811 | 23,937 | 119,663 | 67,467 | 22,489 | 0.009 | ** | 0.004 | ** | 0.0007 | *** |
| Acetoacetyl-CoA | 890.0926 | 600.5392 | 80,604 | 62,873 | 20,958 | 389,992 | 161,620 | 53,873 | 234,119 | 101,068 | 33,689 | 218,945 | 59,623 | 19,874 | 0.0007 | *** | 0.0007 | *** | <0.0001 | *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.-J.; Lee, K.; Jeong, M.; Shin, Y.C.; Ko, S.-G. Metabolomic Analysis of Exosomes Derived from Lung Cancer Cell Line H460 Treated with SH003 and Docetaxel. Metabolites 2022, 12, 1037. https://doi.org/10.3390/metabo12111037
Choi Y-J, Lee K, Jeong M, Shin YC, Ko S-G. Metabolomic Analysis of Exosomes Derived from Lung Cancer Cell Line H460 Treated with SH003 and Docetaxel. Metabolites. 2022; 12(11):1037. https://doi.org/10.3390/metabo12111037
Chicago/Turabian StyleChoi, Yu-Jeong, Kangwook Lee, Miso Jeong, Yong Cheol Shin, and Seong-Gyu Ko. 2022. "Metabolomic Analysis of Exosomes Derived from Lung Cancer Cell Line H460 Treated with SH003 and Docetaxel" Metabolites 12, no. 11: 1037. https://doi.org/10.3390/metabo12111037
APA StyleChoi, Y.-J., Lee, K., Jeong, M., Shin, Y. C., & Ko, S.-G. (2022). Metabolomic Analysis of Exosomes Derived from Lung Cancer Cell Line H460 Treated with SH003 and Docetaxel. Metabolites, 12(11), 1037. https://doi.org/10.3390/metabo12111037





