Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Reagents
2.3. Sample Preparation
2.4. Biomolecules Quantification and Agronomic Characters
2.5. UPLC-MS (Ultraperformance Liquid Chromatography-Tandem Mass Spectrometry)
- ESI+: Heater Temp 300 °C; Sheath Gas Flow rate, 45 arb; Aux Gas Flow Rate, 15 arb; Sweep Gas
- Flow Rate, 1 arb; spray voltage, 3.0 kV; Capillary Temp, 350 °C; S-Lens RF Level, 30%.
- ESI−: Heater Temp 300 °C, Sheath Gas Flow rate, 45 arb; Aux Gas Flow Rate, 15arb; Sweep Gas
- Flow Rate, 1 arb; spray voltage, 3.2 kV; Capillary Temp, 350 °C; S-Lens RF Level, 60%.
2.6. Statistical Analysis
3. Results
3.1. Morphological Traits and Analysis of Starch and Fat Content
3.2. Metabolic Profiling and Quality Control
3.3. Metabolic Changes in the Genotypes
3.4. Variable Importance Projection (VIP)
3.5. Volcano Plot
3.6. Cluster Analysis
3.7. Identification of Potential Metabolites
3.8. Dot Plot
3.9. Correlation Network of Metabolites
4. Discussion
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mecha, E.; Erny, G.L.; Guerreiro, A.C.L.; Feliciano, R.P.; Barbosa, I.; Bento da Silva, A.; Leitao, S.T.; Veloso, M.M.; Rubiales, D.; Rodriguez-Mateos, A.; et al. Metabolomics Profile Responses to Changing Environments in a Common Bean (Phaseolus vulgaris L.) Germplasm Collection. Food Chem. 2022, 370, 131003. [Google Scholar] [CrossRef] [PubMed]
- Bueno, P.C.P.; Lopes, N.P. Metabolomics to Characterize Adaptive and Signaling Responses in Legume Crops under Abiotic Stresses. ACS Omega 2020, 5, 1752–1763. [Google Scholar] [CrossRef]
- Keurentjes, J.J.B.; Fu, J.; de Vos, C.H.R.; Lommen, A.; Hall, R.D.; Bino, R.J.; van der Plas, L.H.W.; Jansen, R.C.; Vreugdenhil, D.; Koornneef, M. The Genetics of Plant Metabolism. Nat. Genet. 2006, 38, 842–849. [Google Scholar] [CrossRef]
- Teklić, T.; Paradiković, N.; Spoljarević, M.; Zeljkovic, S.; Loncaric, Z.; Lisjak, M. Linking Abiotic Stress, Plant Metabolites, Biostimulants and Functional Food. Ann. Appl. Biol. 2021, 178, 169–191. [Google Scholar] [CrossRef]
- Hassan, F.A.S.; Ali, E.; Gaber, A.; Fetouh, M.I.; Mazrou, R. Chitosan Nanoparticles Effectively Combat Salinity Stress by Enhancing Antioxidant Activity and Alkaloid Biosynthesis in Catharanthus roseus (L.) G. Don. Plant Physiol. Biochem. 2021, 162, 291–300. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Balfagon, D.; Gómez-Cadenas, A.; Mittler, R. Plant Responses to Climate Change: Metabolic Changes under Combined Abiotic Stresses. J. Exp. Bot. 2022, 73, 3339–3354. [Google Scholar] [CrossRef] [PubMed]
- Banothu, V.; Uma, A. Effect of Biotic and Abiotic Stresses on Plant Metabolic Pathways; Intechopen: London, UK, 2021. [Google Scholar] [CrossRef]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Weckwerth, W.; Varshney, R.K. Proteomics and Metabolomics: Two Emerging Areas for Legume Improvement. Front. Plant Sci. 2015, 6, 1116. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Gundaraniya, S.A.; Ambalam, P.S.; Tomar, R.S. Metabolomic Profiling of Drought-Tolerant and Susceptible Peanut (Arachis hypogaea L.) Genotypes in Response to Drought Stress. ACS Omega 2020, 5, 31209–31219. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A.R. The Use of Metabolomics to Dissect Plant Responses to Abiotic Stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Aninbon, C.; Jogloy, S.; Vorasoot, N.; Nuchadomrong, S.; Senawong, T.; Holbrook, C.C.; Patanothai, A. Effect of Mid Season Drought on Phenolic Compounds in Peanut Genotypes with Different Levels of Resistance to Drought. Field Crops Res. 2016, 187, 127–134. [Google Scholar] [CrossRef]
- Lima, L.L.; Balbi, B.P.; Mesquita, R.O.; da Silva, J.C.F.; Coutinho, F.S.; Carmo, F.M.S.; Vital, C.E.; Mehta, A.; Loureiro, M.E.; Fontes, E.B.P. Proteomic and Metabolomic Analysis of a Drought Tolerant Soybean Cultivar from Brazilian Savanna. Crop Breed. Genet. Genom. 2019, 1, e190022. [Google Scholar] [CrossRef][Green Version]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic Responses to Drought Stress in the Tissues of Drought-Tolerant and Drought-Sensitive Wheat Genotype Seedlings. AoB PLANTS 2018, 10, ply016. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Martínez, S.E.; Ferriz-Martínez, R.A.; Campos-Vega, R.; Elton-Puente, J.E.; de La Torre Carbot, K.; García-Gasca, T. Bean Seeds: Leading Nutraceutical Source for Human Health. CYTA J. Food 2016, 14, 131–137. [Google Scholar] [CrossRef]
- Zhang, H.; Yasmin, F.; Song, B.H. Neglected Treasures in the Wild—Legume Wild Relatives in Food Security and Human Health. Curr. Opin. Plant Biol. 2019, 49, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Urrea, C.A.; Yonts, C.D.; Lyon, D.J.; Koehler, A.E. Selection for Drought Tolerance in Dry Bean Derived from the Mesoamerican Gene Pool in Western Nebraska. Crop Sci. 2009, 49, 2005–2010. [Google Scholar] [CrossRef]
- Rao, I.M.; Beebe, S.E.; Polania, J.; Grajales, M.; Cajiao, C.; Ricaurte, J.; García, R.; Rivera, M. Evidence for Genotypic Differences among Elite Lines of Common Bean in the Ability to Remobilize Photosynthate to Increase Yield under Drought. J. Agric. Sci. 2017, 155, 857–875. [Google Scholar] [CrossRef]
- Berny Mier Y Teran, J.C.; Konzen, E.R.; Palkovic, A.; Tsai, S.M.; Rao, I.M.; Beebe, S.; Gepts, P. Effect of Drought Stress on the Genetic Architecture of Photosynthate Allocation and Remobilization in Pods of Common Bean (Phaseolus vulgaris L.), a Key Species for Food Security. BMC Plant Biol. 2019, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Bayati, P.; Karimmojeni, H.; Razmjoo, J. Changes in Essential Oil Yield and Fatty Acid Contents in Black Cumin (Nigella sativa L.) Genotypes in Response to Drought Stress. Ind. Crops Prod. 2020, 155, 112764. [Google Scholar] [CrossRef]
- Goufo, P.; Moutinho-Pereira, J.M.; Jorge, T.F.; Correia, C.M.; Oliveira, M.R.; Rosa, E.A.S.; António, C.; Trindade, H. Cowpea (Vigna unguiculata L. Walp.) Metabolomics: Osmoprotection as a Physiological Strategy for Drought Stress Resistance and Improved Yield. Front. Plant Sci. 2017, 8, 586. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gonzalez, J.J.; Guttikonda, S.K.; Tran, L.S.P.; Aldrich, D.L.; Zhong, R.; Yu, O.; Nguyen, H.T.; Sleper, D.A. Differential Expression of Isoflavone Biosynthetic Genes in Soybean during Water Deficits. Plant Cell Physiol. 2010, 51, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Yeloojeh, K.A.; Saeidi, G.; Sabzalian, M.R. Drought Stress Improves the Composition of Secondary Metabolites in Safflower Flower at the Expense of Reduction in Seed Yield and Oil Content. Ind. Crops Prod. 2020, 154, 112496. [Google Scholar] [CrossRef]
- Šamec, D.; Karalija, E.; Šola, I.; Vujčić Bok, V.; Salopek-Sondi, B. The Role of Polyphenols in Abiotic Stress Response: The Influence of Molecular Structure. Plants 2021, 10, 118. [Google Scholar] [CrossRef]
- Salem, N.; Msaada, K.; Dhifi, W.; Sriti, J.; Mejri, H.; Limam, F.; Marzouk, B. Effect of drought on safflower natural dyes and their biological activities. EXCLI J. 2014, 13, 1–18. [Google Scholar]
- Gharibi, S.; Tabatabaei, B.E.S.; Saeidi, G.; Goli, S.A.H. Effect of Drought Stress on Total Phenolic, Lipid Peroxidation, and Antioxidant Activity of Achillea Species. Appl. Biochem. Biotechnol. 2016, 178, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.D.; Hosfield, G.L.; Varner, G.V.; Uebersax, M.A.; Taylor, J. Registration of ‘Matterhorn’ Great Northern Bean. Crop Sci. 1999, 39, 589–590. [Google Scholar] [CrossRef]
- Urrea, C.A.; Smith, J.R.; Porch, T.G. Release of Drought-Tolerant Pinto SB-DT2 and Small Red SB-DT3 Common Bean Germplasm from a Shuttle Breeding Program between Nebraska and Puerto Rico. J. Plant Regist. 2021, 16, 400–409. [Google Scholar] [CrossRef]
- Singh, S.P.; Terán, H.; Lema, M.; Dennis, M.F.; Hayes, R.; Robinson, C. Development of Large-Seeded High-Quality, High-Yielding Great Northern Dry Bean ‘Hungerford’ and ‘Sawtooth’. J. Plant Regist. 2008, 2, 174–179. [Google Scholar] [CrossRef]
- Hosfield, G.L.; Varner, G.V.; Uebersax, M.A.; Kelly, J.D. Registration of ‘Merlot’ Small Red Bean. Crop Sci. 2004, 44, 351. [Google Scholar] [CrossRef]
- Osorno, J.M.; Grafton, K.F.; Rojas-Cifuentes, G.A.; Gelin, R.; Wal, A.J. vander Registration of ‘Lariat’ and ‘Stampede’ Pinto Beans. J. Plant Regist. 2010, 4, 5–11. [Google Scholar] [CrossRef]
- Becker, W.; Eriksson, A.; Haglund, M.; Wretling, S. Contents of Total Fat, Fatty Acids, Starch, Sugars and Dietary Fibre in Swedish Market Basket Diets. Br. J. Nutr. 2015, 113, 1453. [Google Scholar] [CrossRef]
- Madala, N.E.; Piater, L.A.; Steenkamp, P.A.; Dubery, I.A. Multivariate Statistical Models of Metabolomic Data Reveals Different Metabolite Distribution Patterns in Isonitrosoacetophenone-Elicited Nicotiana Tabacum and Sorghum Bicolor Cells. SpringerPlus 2014, 3, 254. [Google Scholar] [CrossRef]
- Bhargava, S.; Sawant, K. Drought Stress Adaptation: Metabolic Adjustment and Regulation of Gene Expression. Plant Breed. 2013, 132, 21–32. [Google Scholar] [CrossRef]
- Wagner, G.; Laperche, A.; Lariagon, C.; Marnet, N.; Renault, D.; Guitton, Y.; Bouchereau, A.; Delourme, R.; Manzanares-Dauleux, M.J.; Gravot, A. Resolution of Quantitative Resistance to Clubroot into QTL-Specific Metabolic Modules. J. Exp. Bot. 2019, 70, 5375. [Google Scholar] [CrossRef]
- Guo, X.; Xin, Z.; Yang, T.; Ma, X.; Zhang, Y.; Wang, Z.; Ren, Y.; Lin, T. Metabolomics Response for Drought Stress Tolerance in Chinese Wheat Genotypes (Triticum aestivum). Plants 2020, 9, 520. [Google Scholar] [CrossRef]
- Marček, T.; Hamow, K.Á.; Végh, B.; Janda, T.; Darko, E. Metabolic Response to Drought in Six Winter Wheat Genotypes. PLoS ONE 2019, 14, e0212411. [Google Scholar] [CrossRef]
- Jiménez-Arias, D.; García-Machado, F.J.; Morales-Sierra, S.; Luis, J.C.; Suarez, E.; Hernández, M.; Valdés, F.; Borges, A.A.; Rodríguez, B. Lettuce Plants Treated with L-Pyroglutamic Acid Increase Yield under Water Deficit Stress. Environ. Exp. Bot. 2019, 158, 215–222. [Google Scholar] [CrossRef]
- Pinheiro, S.S.; de Morais Cardoso, L.; Anunciação, P.C.; de Menezes, C.B.; Queiroz, V.A.V.; Martino, H.S.D.; della Lucia, C.M.; Pinheiro Sant’Ana, H.M. Water Stress Increased the Flavonoid Content in Tannin-Free Sorghum Grains. J. Food Compos. Anal. 2021, 100, 103892. [Google Scholar] [CrossRef]
- Watanabe, S.; Nakagawa, A.; Izumi, S.; Shimada, H.; Sakamoto, A. RNA Interference-Mediated Suppression of Xanthine Dehydrogenase Reveals the Role of Purine Metabolism in Drought Tolerance in Arabidopsis. FEBS Lett. 2010, 584, 1181–1186. [Google Scholar] [CrossRef]
- Foti, C.; Kalampokis, I.F.; Aliferis, K.A.; Pavli, O.I. Metabolic Responses of Two Contrasting Lentil Genotypes to PEG-Induced Drought Stress. Agronomy 2021, 11, 1190. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Drought Stress Enhances Nutritional and Bioactive Compounds, Phenolic Acids and Antioxidant Capacity of Amaranthus Leafy Vegetable. BMC Plant Biol. 2018, 18, 258. [Google Scholar] [CrossRef]
- Galieni, A.; di Mattia, C.; de Gregorio, M.; Speca, S.; Mastrocola, D.; Pisante, M.; Stagnari, F. Effects of Nutrient Deficiency and Abiotic Environmental Stresses on Yield, Phenolic Compounds and Antiradical Activity in Lettuce (Lactuca sativa L.). Sci. Hortic. 2015, 187, 93–101. [Google Scholar] [CrossRef]
- Shi, J.; Arunasalam, K.; Yeung, D.; Kakuda, Y.; Mittal, G.; Jiang, Y. Saponins from Edible Legumes: Chemistry, Processing, and Health Benefits. J. Med. Food 2004, 7, 67–78. [Google Scholar] [CrossRef]
- Tzeng, Y.M.; Rao, Y.K.; Lee, M.J.; Chen, K.; Lee, Y.C.; Wu, W.S. Insulin-Mimetic Action of Rhoifolin and Cosmosiin Isolated from Citrus grandis (L.) Osbeck Leaves: Enhanced Adiponectin Secretion and Insulin Receptor Phosphorylation in 3T3-L1 Cells. Evid. Based Complement. Altern. Med. 2011, 2011, 624375. [Google Scholar] [CrossRef]
- Srinivasulu, C.; Ramgopal, M.; Ramanjaneyulu, G.; Anuradha, C.M.; Suresh Kumar, C. Syringic Acid (SA)—A Review of Its Occurrence, Biosynthesis, Pharmacological and Industrial Importance. Biomed. Pharmacother. 2018, 108, 547–557. [Google Scholar] [CrossRef]
- Hessini, K.; Wasli, H.; Al-Yasi, H.M.; Ali, E.F.; Issa, A.A.; Hassan, F.A.S.; Siddique, K.H.M.; Toscano, S.; Franzoni, G.; Álvarez, S.; et al. Graded Moisture Deficit Effect on Secondary Metabolites, Antioxidant, and Inhibitory Enzyme Activities in Leaf Extracts of Rosa Damascena Mill. Var. Trigentipetala. Horticulturae 2022, 8, 177. [Google Scholar] [CrossRef]
- Zhang, M.; Hong, L.Z.; Gu, M.F.; Wu, C.D.; Zhang, G. Transcriptome Analyses Revealed Molecular Responses of Cynanchum Auriculatum Leaves to Saline Stress. Sci. Rep. 2020, 10, 449. [Google Scholar] [CrossRef]
- Zhang, R.; Zhi, H.; Li, Y.; Guo, E.; Feng, G.; Tang, S.; Guo, W.; Zhang, L.; Jia, G.; Diao, X. Response of Multiple Tissues to Drought Revealed by a Weighted Gene Co-Expression Network Analysis in Foxtail Millet [Setaria italica (L.) P. Beauv.]. Front. Plant Sci. 2022, 12, 3164. [Google Scholar] [CrossRef]
- Ma, H.L.; Xua, X.H.; Zhao, X.Y.; Liu, H.J.; Chen, H. Impacts of Drought Stress on Soluble Carbohydrates and Respiratory Enzymes in Fruit Body of Auricularia Auricula. Biotechnol. Biotechnol. Equip. 2015, 29, 10. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-Based Untargeted Metabolic Profiling Reveals Changes in Chickpea (Cicer arietinum) Metabolome Following Long-Term Drought Stress. Plant Cell Environ. 2019, 42, 115–132. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of Flavonoids and Effects of Stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]


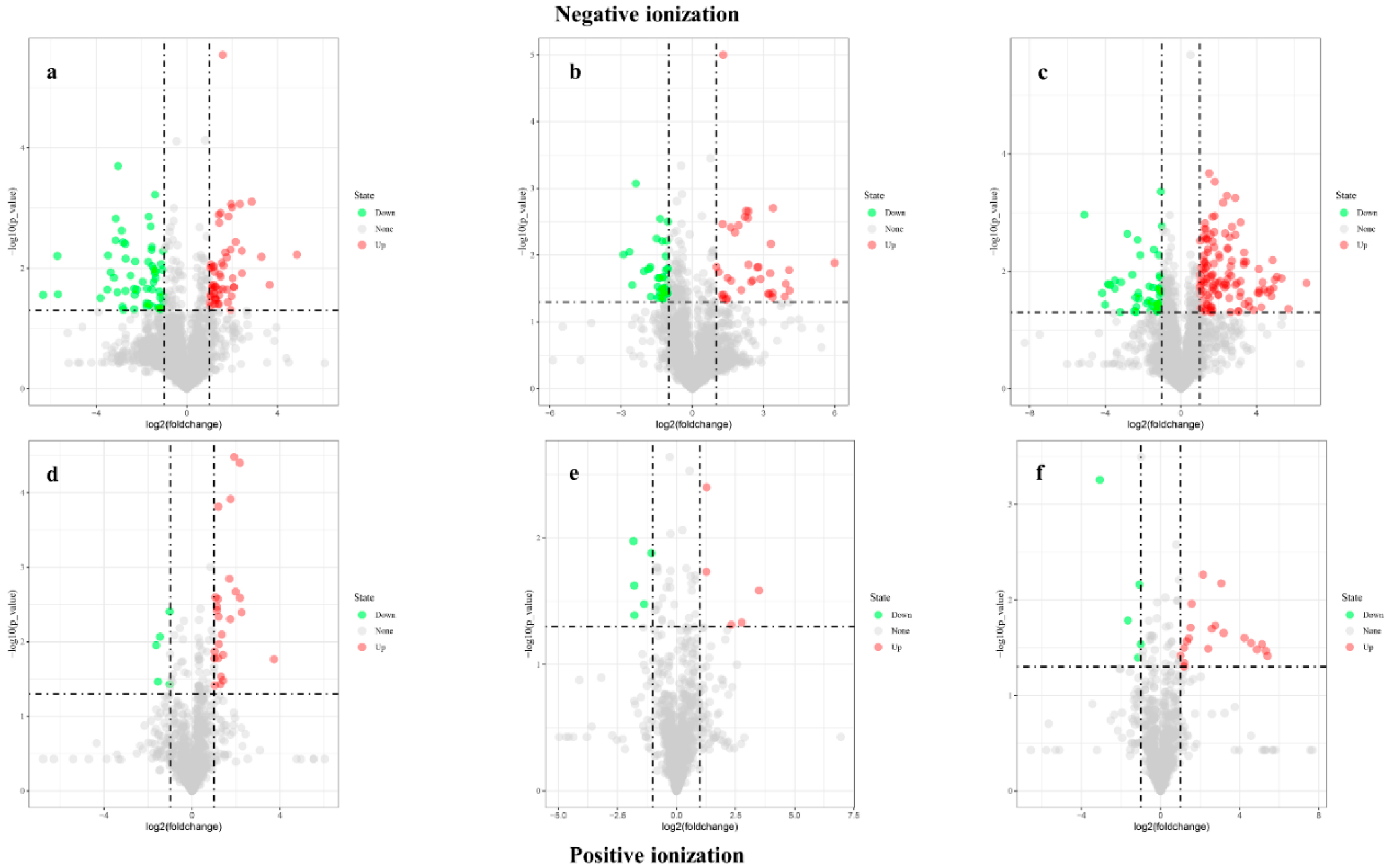
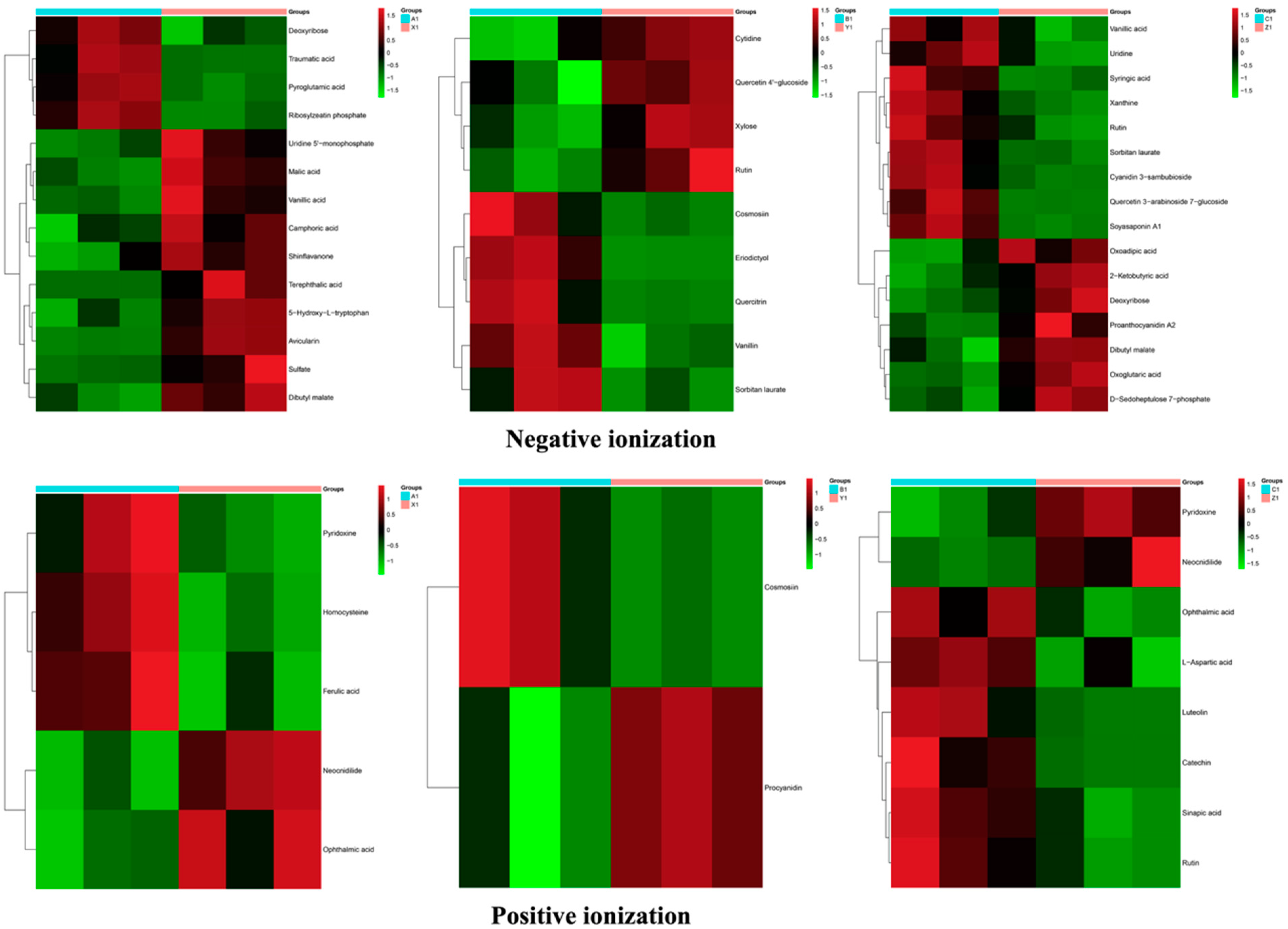


| Negative Ionization | Positive Ionization | ||||
|---|---|---|---|---|---|
| A1 vs. X1 (Matterhorn) | B1 vs. Y1 (SB-DT3) | C1 vs. Z1 (SB-DT2) | A1 vs. X1 (Matterhorn) | B1 vs. Y1 (SB-DT3) | C1 vs. Z1 (SB-DT2) |
| Ribosylzeatin phosphate | Quercitrin | Cyanidin 3-sambubioside | Homocysteine | Cosmosiin | Catechin |
| Cosmosiin | Quercetin 3-arabinoside 7-glucoside | Pyridoxine | Luteolin | ||
| Syringic acid | Eriodictyol | Sorbitan laurate | Ferulic acid | Rutin | |
| Pyroglutamic acid | Vanillin | Xanthine | Sinapic acid | ||
| Deoxyribose | Sorbitan laurate | Soyasaponin A1 | Ophthalmic acid | ||
| Uridine | L-Aspartic acid | ||||
| Rutin | |||||
| Syringic acid Vanillic acid | |||||
| Negative Ionization | Positive Ionization | |||
|---|---|---|---|---|
| A1 vs. X1 | B1 vs. Y1 | C1 vs. Z1 | A1 vs. X1 | C1 vs. Z1 |
| Pyroglutamic acid | Cytidine | Oxoglutaric acid | Homocysteine | L-Aspartic acid |
| L-malic acid | Vanillin | 2-Ketobutyric acid | Pyridoxine | Pyridoxine |
| Uridine 5′-monophosphate | Eriodictyol | Xanthine | Trans-Ferulic acid | Luteolin |
| Vanillic acid | D-Xylose | Uridine | ||
| Oxoadipic acid | ||||
| Vanillic acid | ||||
| Syringic acid | ||||
| Pathways | p < 0.05 | Genotypes Comparisons |
|---|---|---|
| Monobactam biosynthesis | 0.044826 | Matterhorn (A1 vs. X1) |
| Flavone and flavonol biosynthesis | 9.88 × 10−4 | SB-DT3 (B1 vs. Y1), SB-DT2 (C1 vs. Z1) |
| Pentose phosphate pathway | 0.0073327 | SB-DT2 (C1 vs. Z1) |
| C5-Branched dibasic acid metabolism | 0.042084 | SB-DT2 (C1 vs. Z1) |
| Cysteine and methionine metabolism | 0.0098777 | Matterhorn (A1 vs. X1), SB-DT2 (C1 vs. Z1) |
| Vitamin B6 metabolism | 0.038646 | Matterhorn (A1 vs. X1) |
| Flavonoid biosynthesis | 0.027047 | SB-DT2 (C1 vs. Z1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subramani, M.; Urrea, C.A.; Kalavacharla, V. Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress. Metabolites 2022, 12, 944. https://doi.org/10.3390/metabo12100944
Subramani M, Urrea CA, Kalavacharla V. Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress. Metabolites. 2022; 12(10):944. https://doi.org/10.3390/metabo12100944
Chicago/Turabian StyleSubramani, Mayavan, Carlos A. Urrea, and Venu Kalavacharla. 2022. "Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress" Metabolites 12, no. 10: 944. https://doi.org/10.3390/metabo12100944
APA StyleSubramani, M., Urrea, C. A., & Kalavacharla, V. (2022). Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress. Metabolites, 12(10), 944. https://doi.org/10.3390/metabo12100944





