Transcriptome and Metabolome Studies on Pre-Harvest Nitrogen Impact on Fruit Yield and Quality of Peach (Prunus persica L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Experimental Design
2.2. Plant Sampling and Measurements
2.3. Sample Preparation and Metabolite Extraction
2.4. RNA-Seq and Annotation
2.5. qRT-PCR Analysis
2.6. Statistical Analysis
3. Results
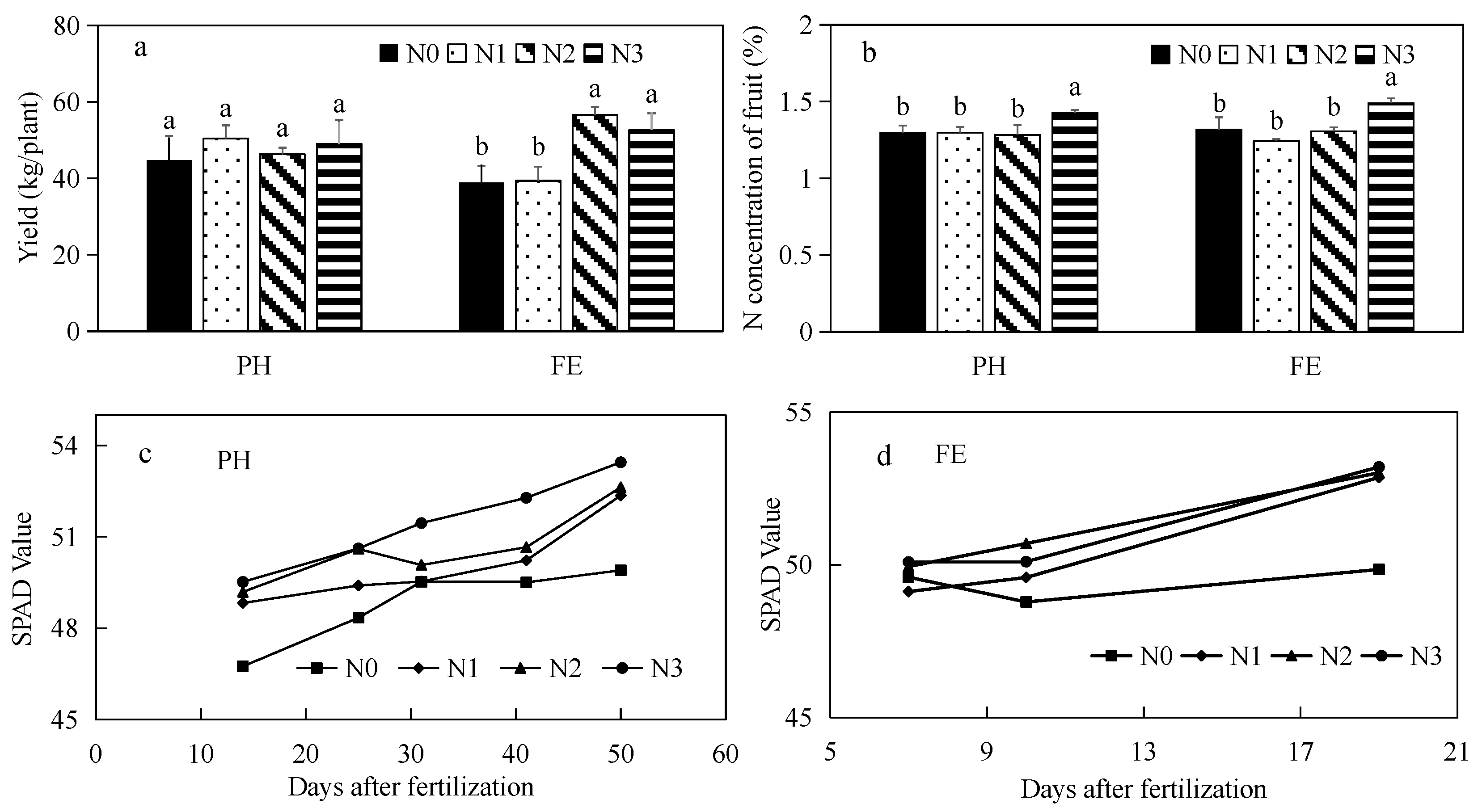
3.1. Effect of Nitrogen Supply on Yield Attributes and Chlorophyll Contents in Peach Leaves at Different Growth Stages
3.2. Effect of Nitrogen Supply on Fruit Quality at Different Growth Stages
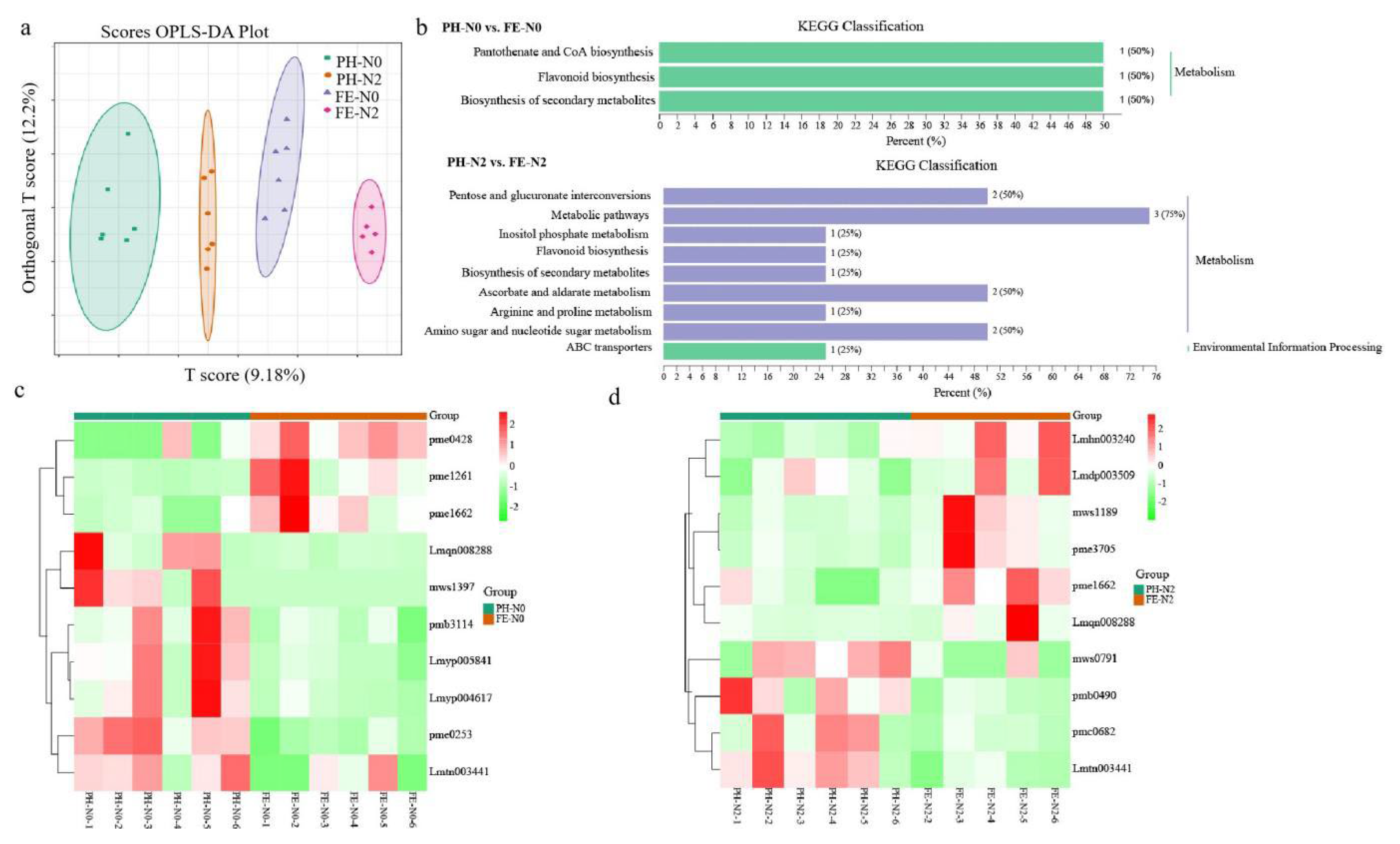
3.3. Metabolite Profiles of Fruit in Response to Nitrogen Availability
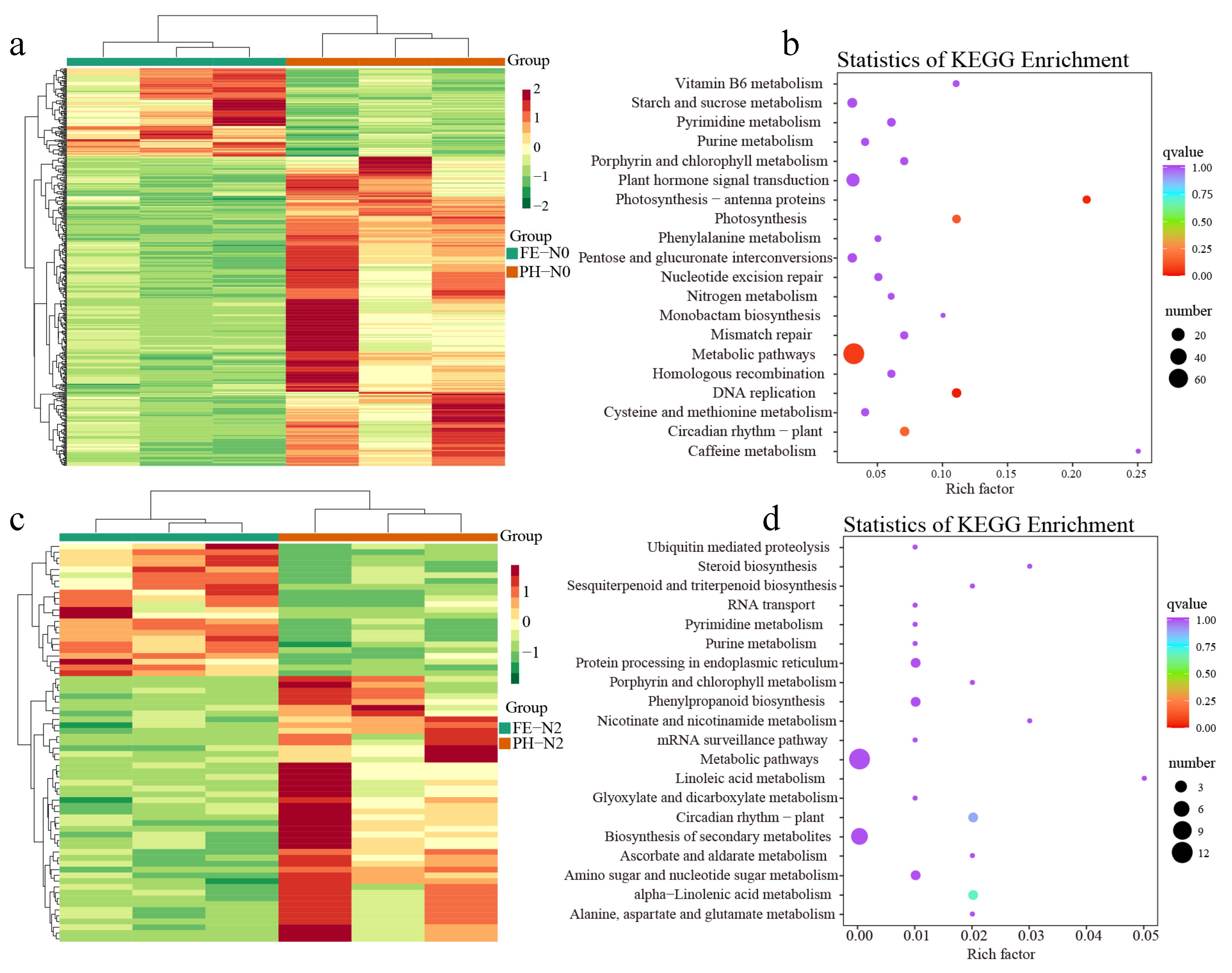
3.4. Impact of Nitrogen on Differentially Expressed Genes in Fruit
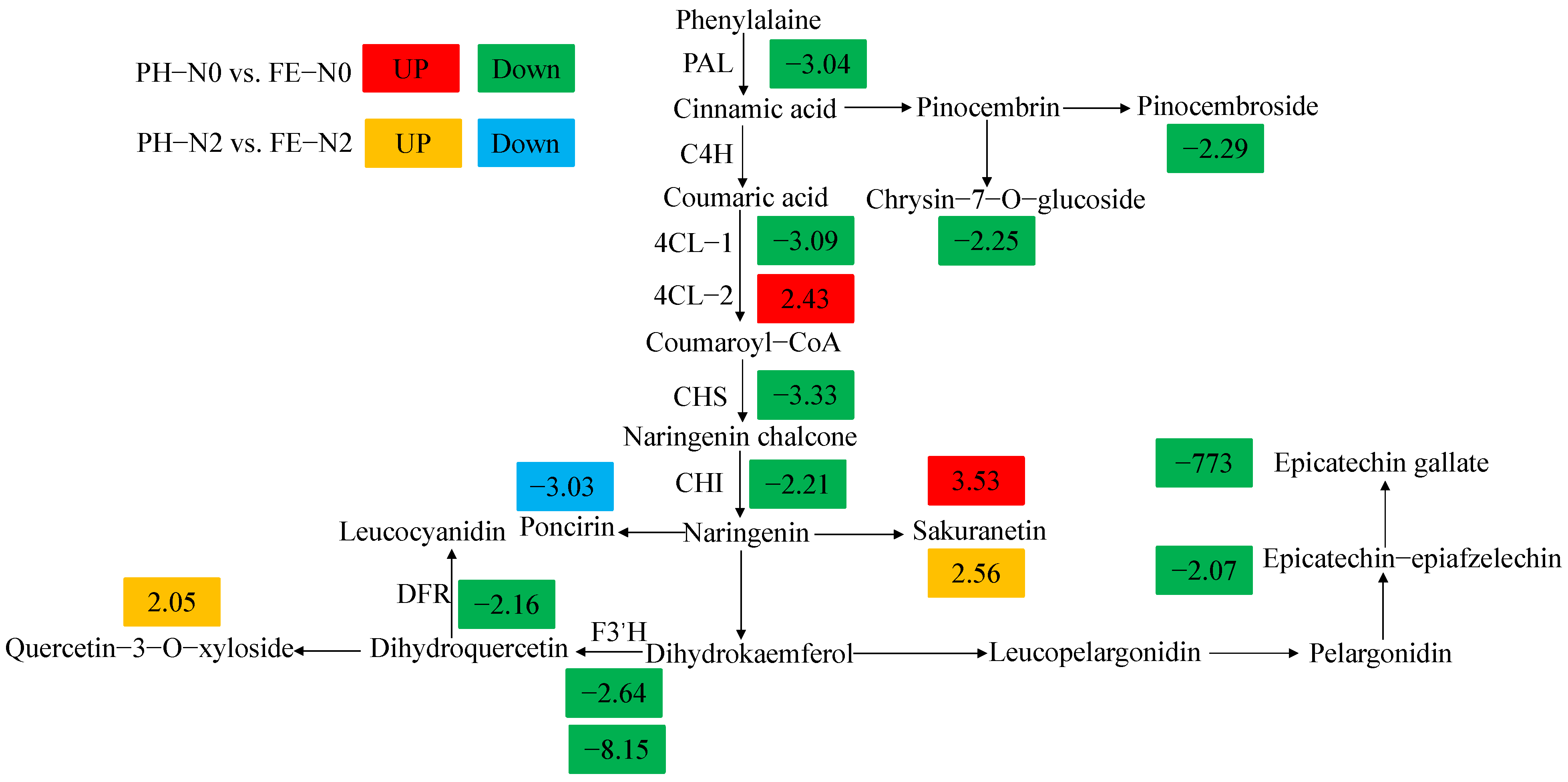
3.5. Profiles of DEGs and DAMs in Flavonoid Biosynthetic Pathways in Developing Fruits
3.6. Validation of DEGs by qRT-PCR
4. Discussion
4.1. Effects of Application of Nitrogen Fertilizer at Different Growth Stages on Yield and Fruit Quality of Peach
4.2. Effects of Application of Nitrogen Fertilizer at Different Growth Stages on Transcriptome and Metabolome of Peach
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, D.P.; Li, M.X.; Liu, Y.; Shi, L.X. Integration of the metabolome and transcriptome reveals the resistance mechanism to low nitrogen in wild soybean seedling roots. Environ. Exp. Bot. 2020, 175, 104043. [Google Scholar] [CrossRef]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The evolution and future of earth’s nitrogen cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef]
- Law, B. Nitrogen deposition and forest carbon. Nature 2013, 496, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, P.; Ding, W.; Ullah, S.; Abbas, T.; Li, M.; Ai, C.; Zhou, W. Identifying the critical nitrogen fertilizer rate for optimum yield and minimum nitrate leaching in a typical field radish cropping system in China. Environ. Pollut. 2021, 268, 115004. [Google Scholar] [CrossRef] [PubMed]
- Dimkpa, C.O.; Fugice, J.; Singh, U.; Lewis, T.D. Development of fertilizers for enhanced nitrogen use efficiency-trends and perspectives. Sci. Total Environ. 2020, 731, 139113. [Google Scholar] [CrossRef]
- Sun, T.T.; Zhang, J.K.; Zhang, Q.; Li, X.L.; Li, M.J.; Yang, Y.Z.; Zhou, J.; Wei, Q.P.; Zhou, B.B. Integrative physiological, transcriptome, and metabolome analysis reveals the effects of nitrogen sufficiency and deficiency conditions in apple leaves and roots. Environ. Exp. Bot. 2021, 192, 104633. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Ren, F.; Jiang, Q.; Zhou, X.; Zhao, J.; Liu, X. Integrated physiological, transcriptomic, and metabolomic analyses of the response of peach to nitrogen levels during different growth stages. Int. J. Mol. Sci. 2022, 23, 10876. [Google Scholar] [CrossRef]
- Ristova, D.; Carre, C.; Pervent, M.; Medici, A.; Kim, G.J.; Scalia, D.; Ruffel, S.; Birnbaum, K.D.; Lacombe, B.; Busch, W. Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci. Signal. 2016, 9, rs13. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.; Zhang, W.; Gao, J.; Yi, J.; Zhen, X.; Li, Z.; Zhao, Y.; Peng, C.; Zhao, C. An integrated analysis of the rice transcriptome and metabolome reveals differential regulation of carbon and nitrogen metabolism in response to nitrogen availability. Int. J. Mol. Sci. 2019, 20, 2349. [Google Scholar] [CrossRef]
- Xia, G.; Cheng, L.; Lakso, A.; Goffinet, M. Effects of nitrogen supply on source-sink balance and fruit size of ‘Gala’ apple trees. J. Am. Soc. Hortic. Sci. 2009, 134, 126–133. [Google Scholar] [CrossRef]
- Jakopic, J.; Schmitzer, V.; Veberic, R.; Smrke, T.; Stampar, F. Metabolic response of ‘Topaz’ apple fruit to minimal application of nitrogen during cell enlargement stage. Horticulturae 2021, 7, 266. [Google Scholar] [CrossRef]
- Ibell, P.; Maddox, C.; Wright, C.; Bally, I. How does pre-harvest applications of nitrogen fertiliser affect branch growth, leaf morphology and fruit, in Mangifera indica (Mango) orchards? Acta Hortic. 2020, 12, 123–132. [Google Scholar] [CrossRef]
- Monti, L.L.; Bustamante, C.A.; Osorio, S.; Gabilondo, J.; Borsani, J.; Lauxmann, M.A.; Maulión, E.; Valentini, G.; Budde, C.O.; Fernie, A.R.; et al. Metabolic profiling of a range of peach fruit varieties reveals high metabolic diversity and commonalities and differences during ripening. Food Chem. 2016, 190, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, D.J.; Ende, B.V.D. A reappraisal of the growth and development of peach fruit. Funct. Plant Biol. 1975, 2, 623–634. [Google Scholar] [CrossRef]
- Tonutti, P.; Casson, P.; Ramina, A. Ethylene biosynthesis during peach fruit development. J. Am. Soc. Hortic. Sci. 1991, 116, 274–279. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Kim, W.S.; El-Kereamy, A.; Jayasankar, S.; Svircev, A.M.; Brown, D.C.W. Isolation and characterization of four ethylene signal transduction elements in plums (Prunus salicina L.). J. Exp. Bot. 2007, 58, 3631–3643. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, V.A.; Osorio, S.; Borsani, J.; Lauxmann, M.A.; Bustamante, C.A.; Budde, C.O.; Andreo, C.S.; Lara, M.V.; Drincovich, F.M.F. Metabolie profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental Stage. Plant Physiol. 2011, 157, 1696–1710. [Google Scholar] [CrossRef]
- Dardick, C.D.; Callahan, A.M.; Chiozzotto, R.; Schaffer, R.J.; Piagnani, M.C.; Scorza, R. Stone formation in peach fruit exhibits spatial coordination of the lignin and flavonoid pathways and similarity to Arabidopsis dehiscence. BMC Biol. 2010, 8, 13. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, Y.; Yu, M.; Su, B.; Gong, W.; Baluska, F.; Komis, G.; Samaj, J.; Shan, X.; Lin, J. Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol. 2019, 181, 480–498. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Determination of total nitrogen in plant material. Agron. J. 1973, 65, 109–112. [Google Scholar] [CrossRef]
- Gilibowski, P.; Skrzypek, M.; Ćwiklińska, M.; Drozd, M.; Kowalska, A. Chemical stability of fructans in apple beverages and their influence on chronic constipation. Food Funct. 2020, 11, 3860–3866. [Google Scholar] [CrossRef] [PubMed]
- Loong, C.; Leo, L.; Loke, W.M. Sugar, vitamin C, and polyphenols in commercial apple beverages and their effects on antioxidant and anti-inflammatory activities in vitro. J. Food Bioactives 2021, 13, 13259. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT, a fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq, a matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Boussadia, O.; Steppe, K.; Zgallai, H.; Hadj, S.B.E.; Braham, M.; Lemeur, R.; Labeke, M.C.V. Effects of nitrogen deficiency on leaf photosynthesis, carbohydrate status and biomass production in two olive cultivars ‘Meski’ and ‘Koroneiki’. Sci. Hortic. 2010, 123, 336–342. [Google Scholar] [CrossRef]
- Heyneke, E.; Watanabe, M.; Erban, A.; Duan, G.; Buchner, P.; Walther, D.; Kopka, J.; Hawkesford, M.J.; Hoefgen, R. Characterization of the wheat leaf metabolome during grain filling and under varied N-supply. Front. Plant Sci. 2017, 8, 2048. [Google Scholar] [CrossRef]
- Falguera, V.; Lordan, J.; Gatius, F.; Pascual, M.; Villar, J.M.; Ibarza, A.; Rufat, J. Influence of nitrogen fertilization on polyphenol oxidase activity in peach fruits. Sci. Hortic. 2012, 142, 155–157. [Google Scholar] [CrossRef]
- Pascual, M.; Villar, J.M.; Lordan, J.; Fonseca, F.; Falguera, V.; Rufat, J. Relationship between polyphenol oxidase activity and nutrition, maturity and quality. J. Sci. Food Agric. 2013, 93, 3384–3389. [Google Scholar] [CrossRef]
- Kim, Y.X.; Kwon, M.C.; Lee, S.; Jung, E.S.; Lee, C.H.; Sung, J. Effects of nutrient and water supply during fruit development on metabolite composition in tomato fruits (Solanum lycopersicum L.) grown in magnesium excess soils. Front. Plant Sci. 2020, 11, 562399. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, V.; Sanchez, E.; Tognetti, J. Timing of nitrogen fertilization influences color and anthocyanin content of apple (Malus domestica Borkh. cv ‘Royal Gala’) fruits. Int. J. Fruit Sci. 2011, 11, 364–375. [Google Scholar] [CrossRef]
- Sharma, L.K. Effect of nutrients sprays on growth, yield and fruit quality of apple under cold desert condition of Himachal Pradesh. J. Appl. Nat. Sci. 2016, 8, 297–300. [Google Scholar] [CrossRef][Green Version]
- Dolinski, M.A.; Dangelo, J.W.D.O.; Cuquel, F.L.; Motta, A.C.V.; De Mio, L.L.M. Quality peach produced in fertilizer doses of nitrogen and green pruning. Bragantia 2018, 77, 134–140. [Google Scholar] [CrossRef]
- Titus, J.S.; Kang, S. Nitrogen metabolism, translocation, and recycling in apple trees. Hort. Rev. 1982, 4, 204–246. [Google Scholar] [CrossRef]
- Choi, S.H.; Ahn, J.B.; Kim, H.J.; Im, N.K.; Kozukue, N.; Levin, C.E.; Friedman, M. Changes in free amino acid, protein, and flavonoid content in jujube (Ziziphus jujube) fruit during eight stages of growth and antioxidative and cancer cell inhibitory effects by extracts. J. Agric. Food Chem. 2012, 60, 10245–10255. [Google Scholar] [CrossRef]
- Chhapekar, S.S.; Brahma, V.; Rawoof, A.; Kumar, N.; Gaur, R.; Jaiswal, V.; Kumar, A.; Yadava, S.K.; Kumar, R.; Sharma, V.; et al. Transcriptome profiling, simple sequence repeat markers development and genetic diversity analysis of potential industrial crops Capsicum chinense and C. frutescens of northeast India. Ind. Crop. Prod. 2020, 154, 112687. [Google Scholar] [CrossRef]
- Soltani, N.; Nazarian-Firouzabadi, F.; Shafeinia, A.; Sadr, A.S.; Shirali, M. The expression of terpenoid indole alkaloid (TIAs) pathway genes in Catharanthus roseus in response to salicylic acid treatment. Mol. Biol. Rep. 2020, 47, 7009–7016. [Google Scholar] [CrossRef]
- Hui, W.; Zhao, F.; Wang, J.; Chen, X.; Li, J.; Zhong, Y.; Li, H.; Zheng, J.; Zhang, L.; Que, Q.; et al. De novo transcriptome assembly for the five major organs of Zanthoxylum armatum and the identification of genes involved in terpenoid compound and fatty acid metabolism. BMC Genomics 2020, 21, 81. [Google Scholar] [CrossRef]
- Muroi, A.; Ishihara, A.; Tanaka, C.; Ishizuka, A.; Takabayashi, J.; Miyoshi, H.; Nishioka, T. Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta 2009, 230, 517–527. [Google Scholar] [CrossRef]
- Shinya, T.; Hojo, Y.; Desaki, Y.; Christeller, J.T.; Okada, K.; Shibuya, N.; Galis, I. Modulation of plant defense responses to herbivores by simultaneous recognition of different herbivore-associated elicitors in rice. Sci. Rep. 2016, 6, 32537. [Google Scholar] [CrossRef]
- Alamgir, K.M.; Hojo, Y.; Christeller, J.T.; Fukumoto, K.; Isshiki, R.; Shinya, T.; Baldwin, I.T.; Galis, I. Systematic analysis of rice (Oryza sativa) metabolic responses to herbivory. Plant Cell Environ. 2016, 39, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Yasuda, S.; Hyodo, K.; Tani, R.; Hojo, Y.; Fujiwara, Y.; Hiruma, K.; Ishizaki, T.; Fujita, Y.; Saijo, Y.; et al. Integration of danger peptide signals with herbivore-associated molecular pattern signaling amplifies anti-herbivore defense responses in rice. Plant J. 2018, 94, 626–637. [Google Scholar] [CrossRef]
- Wari, D.; Alamgir, K.M.; Mujiono, K.; Hojo, Y.; Shinya, T.; Tani, A.; Nakatani, H.; Galis, I. Honeydew-associated microbes elicit defense responses against brown planthopper in rice. J. Exp. Bot. 2019, 70, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Li, M.; Kong, W.J.; Bai, J.G.; Wang, X.F.; Wei, M.; Shi, Q.H. Nitric oxide, as a downstream signal, plays vital role in auxin induced cucumber tolerance to sodic alkaline stress. Plant Physiol. Biochem. 2014, 83, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.R.; Song, M.Y.; Wang, Z.; Chen, S.W.; Ma, H.Q. Metabolome and transcriptome analysis of flavor components and flavonoid biosynthesis in fig female flower tissues (Ficus carica L.) after bagging. BMC Plant Biol. 2021, 21, 396. [Google Scholar] [CrossRef]
- Sugiyama, A.; Yazaki, K. Flavonoids in plant rhizospheres, secretion, fate and their effects on biological communication. Plant Biotechnol. 2014, 31, 431–443. [Google Scholar] [CrossRef]
- Liu, J.C.; Zhang, C.L.; Chen, D.L.; Jiao, Z.G. Changes in phenolics, vitamin C and antioxidant capacity during development of different cultivars of Jujube Fruits. Food Sci. 2015, 36, 94–98. [Google Scholar] [CrossRef]
- Xia, L.H.; Chen, Y.L.; Feng, Y.B.; Jiao, Z.G.; Liu, H.; Wang, Q.H. Changes in flavonoids, total phenolics, triterpenoidic acids and antioxidant capacity during fruit development of different cultivars of apricot. J. Fruit Sci. 2016, 33, 425–435. [Google Scholar] [CrossRef]
- Pandey, A.; Alok, A.; Lakhwani, D.; Singh, J.; Asif, M.H.; Trivedi, P.K. Genome-wide expression analysis and metabolite profiling elucidate transcriptional regulation of flavonoid biosynthesis and modulation under abiotic stresses in banana. Sci. Rep. 2016, 6, 31361. [Google Scholar] [CrossRef] [PubMed]
- Park, H.L.; Yoo, Y.; Bhoo, S.H.; Lee, T.H.; Cho, M.H. Two chalcone synthase isozymes participate redundantly in UV-Induced sakuranetin synthesis in rice. Int. J. Mol. Sci. 2020, 21, 3777. [Google Scholar] [CrossRef] [PubMed]

| Treatment Name | N Supply (kg N/ha) | Urea (g) | KH2PO4 (g) | KCl (g) |
|---|---|---|---|---|
| N0 | 0 | 0 | 77 | 124 |
| N1 | 100 | 267 | 77 | 124 |
| N2 | 200 | 534 | 77 | 124 |
| N3 | 400 | 1070 | 77 | 124 |
| Quality Index | N Application at the Fruit Pit Hardening Stage (PH) | N Application at the Fruit Expansion Stage (FE) | ||||||
|---|---|---|---|---|---|---|---|---|
| N0 | N1 | N2 | N3 | N0 | N1 | N2 | N3 | |
| Soluble solid (%) | 15.51 ± 0.27 a | 15.20 ± 0.76 a | 15.16 ± 0.29 a | 15.62 ± 0.10 a | 13.99 ± 0.26 b | 15.36 ± 0.52 a | 14.25 ± 0.22 ab | 14.07 ± 0.54 ab |
| Skin hardness (kg/cm2) | 4.25 ± 0.33 ab | 3.67 ± 0.32 b | 4.44 ± 0.34 ab | 5.34 ± 0.47 a | 4.22 ± 0.34 a | 4.66 ± 0.32 a | 4.96 ± 0.32 a | 4.20 ± 0.46 a |
| Peel hardness (kg/cm2) | 2.71 ± 0.13 ab | 2.36 ± 0.16 b | 2.67 ± 0.16 ab | 2.89 ± 0.13 a | 2.70 ± 0.15 a | 2.81 ± 0.18 a | 2.75 ± 0.12 a | 2.68 ± 0.20 a |
| Reducing sugar (%) | 2.73 ± 0.11 b | 2.83 ± 0.05 ab | 2.79 ± 0.04 ab | 2.98 ± 0.13 a | 3.24 ± 0.22 a | 3.12 ± 0.18 a | 3.28 ± 0.07 a | 3.03 ± 0.16 a |
| VC (mg/100 g) | 8.00 ± 0.18 a | 7.47 ± 0.83 a | 7.77 ± 0.24 a | 8.13 ± 0.65 a | 9.44 ± 0.46 a | 9.09 ± 0.25 a | 8.04 ± 0.69 a | 4.90 ± 0.62 b |
| Component Name | Metabolite Name | Treatment | VIP | Log2FC | Type | |
|---|---|---|---|---|---|---|
| PH-N0 | FE-N0 | |||||
| Flavonoids | 5,4′-Dihydroxy-7-methoxyflavanone (Sakuranetin) | 1.26 × 103 | 4.71 × 103 | 13.9 | 18.2 | up |
| Chrysin-7-O-glucoside | 4.03 × 105 | 1.79 × 105 | 17.1 | 11.7 | down | |
| Pinocembrin-7-O-glucoside (Pinocembroside) | 1.25 × 105 | 4.27 × 104 | 16.8 | 15.5 | down | |
| Epicatechin gallate | 6.96 × 103 | 9.00 × 10 | 17.1 | 95.9 | down | |
| Epicatechin-epiafzelechin | 8.66 × 104 | 4.19 × 104 | 16.0 | 10.5 | down | |
| Phenolic acids | 3,4,5-Trimethoxycinnamic acid | 1.81 × 103 | 6.96 × 103 | 17.7 | 19.5 | up |
| 2-Acetyl-3-hydroxyphenyl-1-O-glucoside | 1.74 × 104 | 8.31 × 103 | 14.7 | 10.7 | down | |
| Amino acids and derivatives | N-Acetyl-L-leucine | 3.28 × 104 | 1.60 × 104 | 21.1 | 10.4 | down |
| Lipids | 3-Hydroxyoctadecanoic Acid | 4.28 × 105 | 1.54 × 105 | 13.9 | 14.8 | down |
| Saccharides and Alcohols | D-Panthenol | 6.67 × 103 | 3.54 × 104 | 18.8 | 24.1 | up |
| Component Name | Metabolite Name | Treatment | VIP | Log2FC | Type | |
|---|---|---|---|---|---|---|
| PH-N2 | FE-N2 | |||||
| Flavonoids | 5,4′-Dihydroxy-7-methoxyflavanone (Sakuranetin) | 1.38 × 103 | 3.53 × 103 | 13.8 | 13.6 | up |
| Quercetin-3-O-xyloside (Reynoutrin) | 8.66 × 103 | 1.77 × 104 | 11.7 | 10.3 | up | |
| Poncirin (Isosakuranetin-7-O-neohesperidoside) | 4.00 × 103 | 1.32 × 103 | 12.6 | 16.0 | down | |
| Phenolic acids | Benzoylmalic acid | 2.51 × 105 | 6.49 × 105 | 19.7 | 13.7 | up |
| 2-Acetyl-3-hydroxyphenyl-1-O-glucoside | 1.99 × 104 | 6.61 × 103 | 13.4 | 15.9 | down | |
| Alkaloids | 4-Aminoindole | 2.28 × 105 | 7.65 × 104 | 14.3 | 15.7 | down |
| p-Coumaroylputrescine | 6.02 × 104 | 2.58 × 104 | 16.1 | 12.3 | down | |
| Organic acids | D-Galacturonic acid | 5.56 × 105 | 1.50 × 106 | 18.9 | 14.3 | up |
| Lipids | 3-Hydroxyoctadecanoic acid | 1.78 × 105 | 3.79 × 105 | 11.8 | 10.9 | up |
| Saccharides and Alcohols | D-Glucoronic acid | 6.23 × 105 | 1.64 × 106 | 19.1 | 14.0 | up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Guo, J.; Zhou, X.; Zhao, J.; Liu, X.; Jiang, Q.; Ren, F. Transcriptome and Metabolome Studies on Pre-Harvest Nitrogen Impact on Fruit Yield and Quality of Peach (Prunus persica L.). Metabolites 2022, 12, 905. https://doi.org/10.3390/metabo12100905
Zhang Y, Guo J, Zhou X, Zhao J, Liu X, Jiang Q, Ren F. Transcriptome and Metabolome Studies on Pre-Harvest Nitrogen Impact on Fruit Yield and Quality of Peach (Prunus persica L.). Metabolites. 2022; 12(10):905. https://doi.org/10.3390/metabo12100905
Chicago/Turabian StyleZhang, Yu, Jiying Guo, Xin Zhou, Jianbo Zhao, Xin Liu, Quan Jiang, and Fei Ren. 2022. "Transcriptome and Metabolome Studies on Pre-Harvest Nitrogen Impact on Fruit Yield and Quality of Peach (Prunus persica L.)" Metabolites 12, no. 10: 905. https://doi.org/10.3390/metabo12100905
APA StyleZhang, Y., Guo, J., Zhou, X., Zhao, J., Liu, X., Jiang, Q., & Ren, F. (2022). Transcriptome and Metabolome Studies on Pre-Harvest Nitrogen Impact on Fruit Yield and Quality of Peach (Prunus persica L.). Metabolites, 12(10), 905. https://doi.org/10.3390/metabo12100905




