Time-Restricted Feeding Could Not Reduce Rainbow Trout Lipid Deposition Induced by Artificial Night Light
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish
2.2. Experiment Design
2.3. Sample Collection
2.4. Biochemical Analysis
2.5. Gene Expression
2.6. Statistical Analysis
3. Results
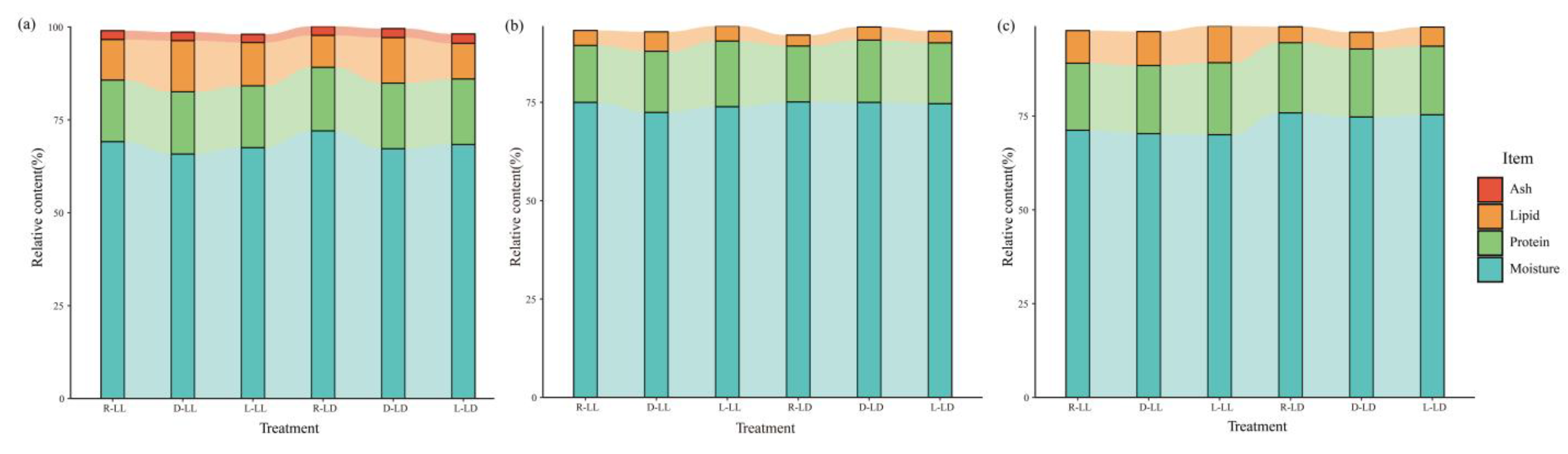
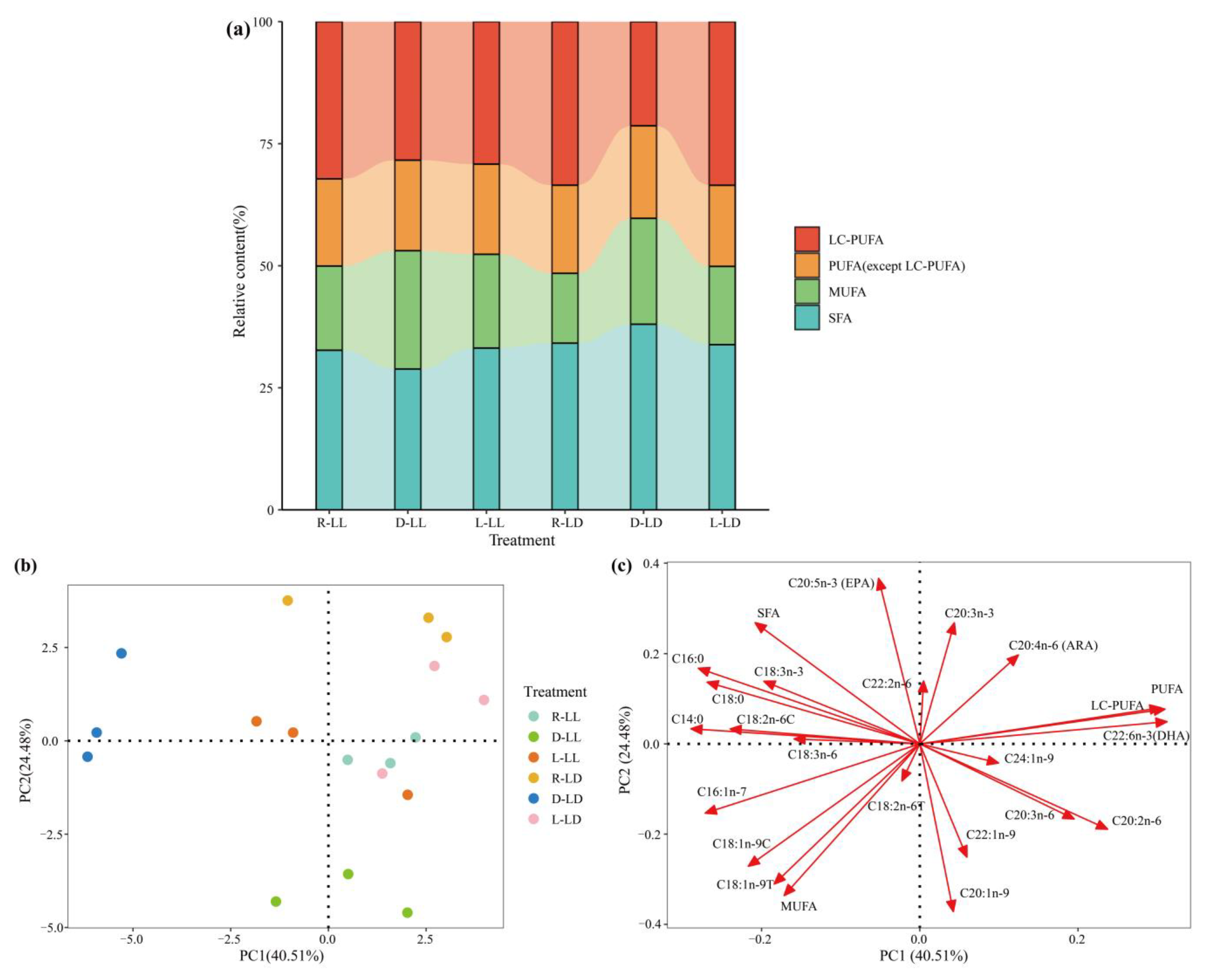
3.1. Body Composition
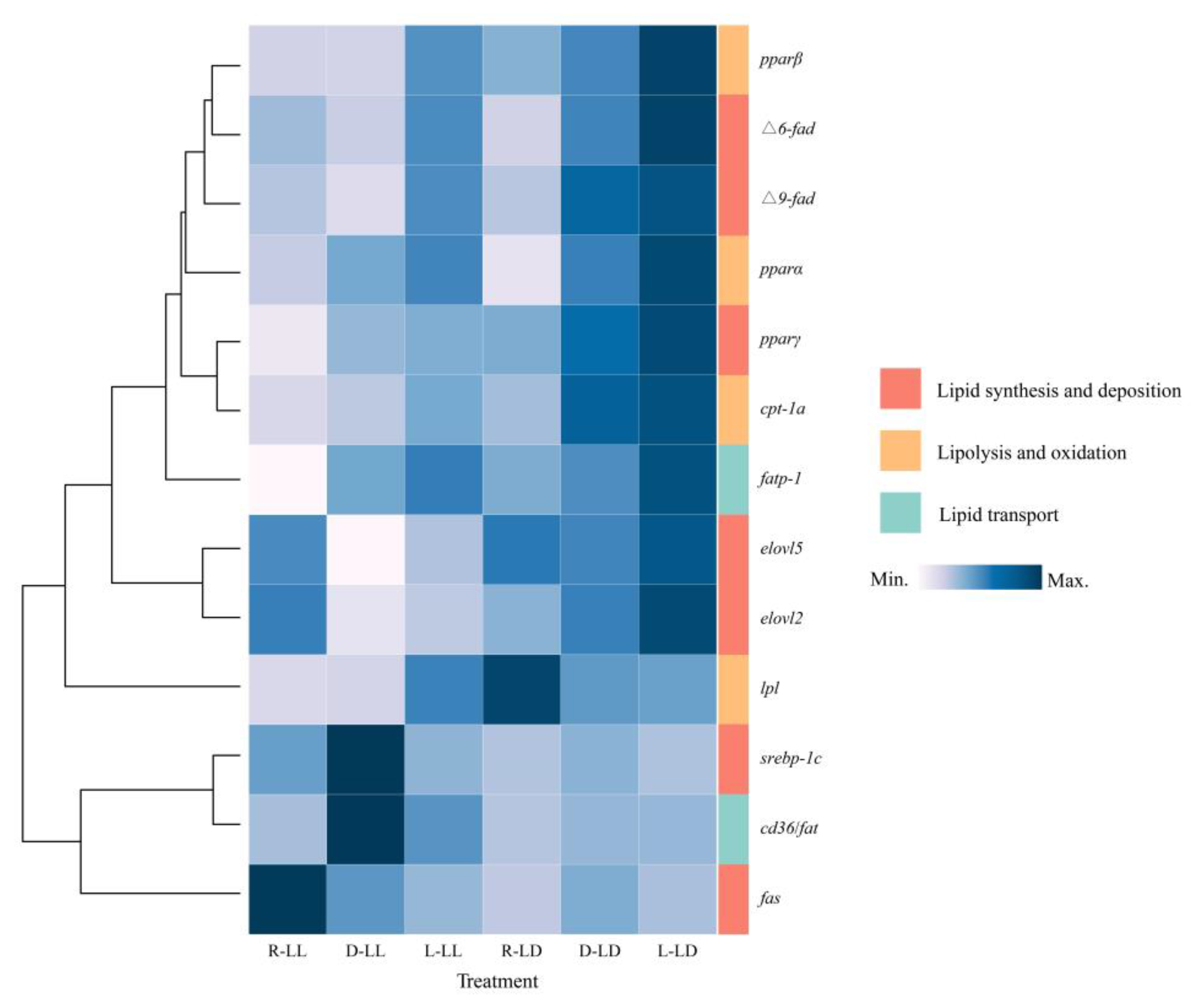
3.2. Lipid Metabolism Genes
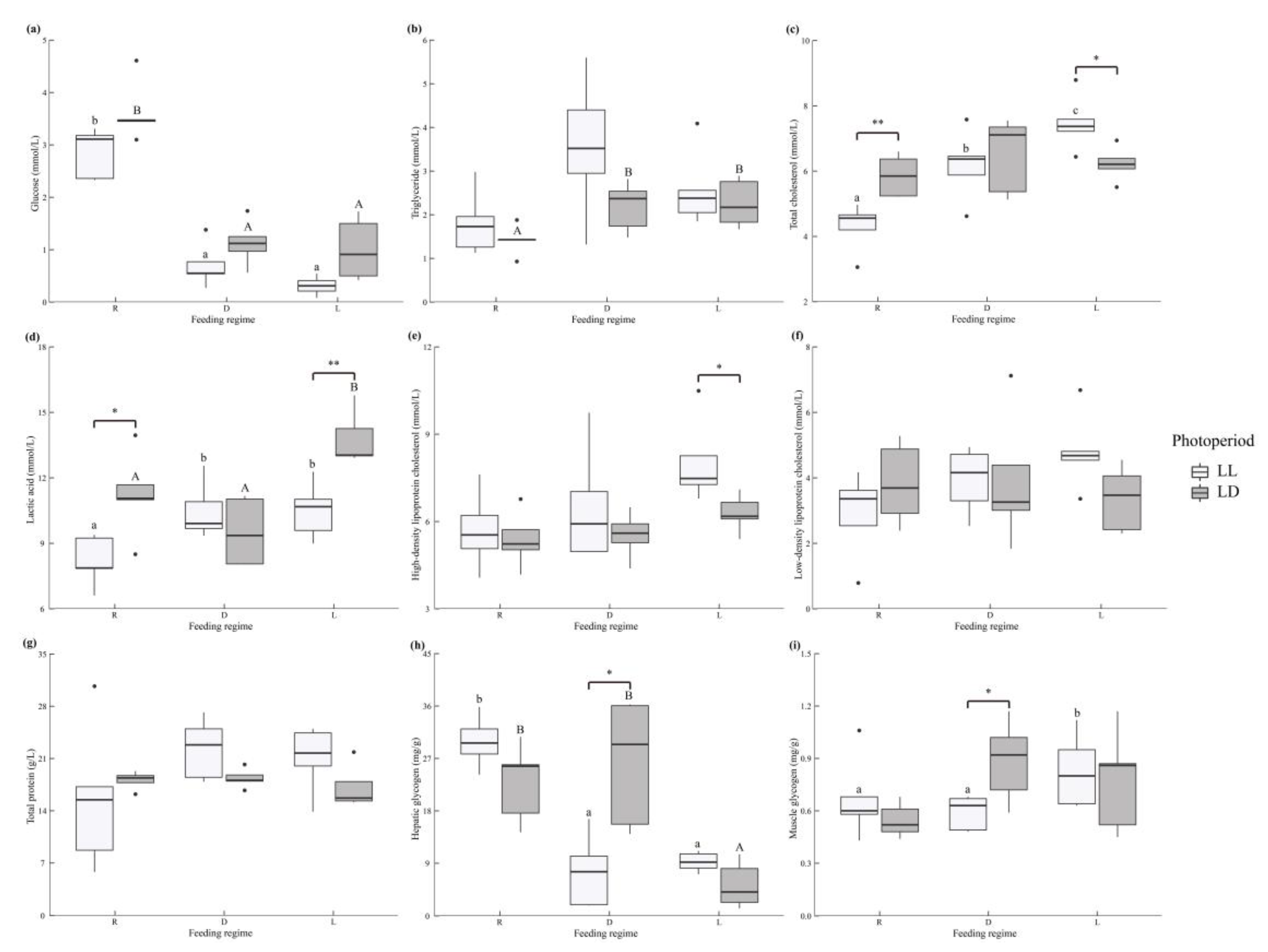
3.3. Serum Metabolites
3.4. Structural Equation Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cinzano, P.; Falchi, F.; Elvidge, C.D. The first World Atlas of the artificial night sky brightness. Mon. Not. R. Astron. Soc. 2001, 328, 689–707. [Google Scholar] [CrossRef]
- Longcore, T.; Rich, C. Ecological light pollution. Front. Ecol. Environ. 2004, 2, 191–198. [Google Scholar] [CrossRef]
- Tancredi, S.; Urbano, T.; Vinceti, M.; Filippini, T. Artificial light at night and risk of mental disorders: A systematic review. Sci. Total Environ. 2022, 833, 155185. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.G.; Helfrich-Förster, C. The regulation of circadian clocks by light in fruitflies and mice. Philos. Trans. R. Soc. B-Biol. Sci. 2001, 356, 1779–1789. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, S.M.; Molusky, M.M.; Lin, J.D. Circadian autophagy rhythm: A link between clock and metabolism? Trends Endocrinol. Metab. 2012, 23, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.Y.; Sarkar, C.; Ni, M.Y.; Gallacher, J.; Webster, C. Exposure to light at night (LAN) and risk of obesity: A systematic review and meta-analysis of observational studies. Environ. Res. 2020, 187, 109637. [Google Scholar] [CrossRef]
- Obayashi, K.; Yamagami, Y.; Kurumatani, N.; Saeki, K. Bedroom lighting environment and incident diabetes mellitus: A longitudinal study of the HEIJO-KYO cohort. Sleep Med. 2020, 65, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Ramsey, K.M.; Marcheva, B.; Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Investig. 2011, 121, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Fleury, G.; Masis-Vargas, A.; Kalsbeek, A. Metabolic Implications of Exposure to Light at Night: Lessons from Animal and Human Studies. Obesity 2020, 28, S18–S28. [Google Scholar] [CrossRef]
- Watanabe, T. Lipid nutrition in fish. Comp. Biochem. Physiol. Part B Comp. Biochem. 1982, 73, 3–15. [Google Scholar] [CrossRef]
- Xu, H.Y.; Han, T.; Li, X.Y.; Wang, J.T.; Zheng, P.Q.; Yin, F.; Wang, C.L. Effects of dietary lipid levels on survival, growth performance, and antioxidant ability of the early juvenile Scylla paramamosain. Aquaculture 2020, 528, 735559. [Google Scholar] [CrossRef]
- Wilson, R.P. Utilization of dietary carbohydrate by fish. Aquaculture 1994, 124, 67–80. [Google Scholar] [CrossRef]
- Stone, D.A.J. Dietary carbohydrate utilization by fish. Rev. Fish. Sci. 2003, 11, 337–369. [Google Scholar] [CrossRef]
- Greene, D.H.; Selivonchick, D.P. Lipid metabolism in fish. Prog. Lipid Res. 1987, 26, 53–85. [Google Scholar] [CrossRef]
- Turchini, G.M.; Francis, D.S.; Du, Z.Y.; Olsen, R.E.; Ringø, E.; Tocher, D.R. The lipids. In Fish Nutrition, 4th ed.; Hardy, R.W., Kaushik, S.J., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 303–467. [Google Scholar] [CrossRef]
- Shimano, H. Sterol Regulatory Element-binding Protein-1 as a Dominant Transcription Factor for Gene Regulation of Lipogenic Enzymes in the Liver. Trends Cardiovasc. Med. 2000, 10, 275–278. [Google Scholar] [CrossRef]
- Pai, W.Y.; Hsu, C.C.; Lai, C.Y.; Chang, T.Z.; Tsai, Y.L.; Her, G.M. Cannabinoid receptor 1 promotes hepatic lipid accumulation and lipotoxicity through the induction of SREBP-1c expression in zebrafish. Transgenic Res. 2013, 22, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Nilsson-Ehle, P.; Garfinkel, A.S.; Schotz, M.C. Lipolytic enzymes and plasma lipoprotein metabolism. Annu. Rev. Biochem. 1980, 49, 667–693. [Google Scholar] [CrossRef]
- Wang, L.; Kaneko, G.; Takahashi, S.I.; Watabe, S.; Ushio, H. Identification and gene expression profile analysis of a major type of lipoprotein lipase in adult medaka Oryzias latipes. Fish. Sci. 2015, 81, 163–173. [Google Scholar] [CrossRef]
- Wang, C.C.; Si, L.F.; Li, W.Y.; Zheng, J.L. A functional gene encoding carnitine palmitoyltransferase 1 and its transcriptional and kinetic regulation during fasting in large yellow croaker. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2019, 231, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Nassir, F.; Wilson, B.; Han, X.L.; Gross, R.W.; Abumrad, N.A. CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J. Biol. Chem. 2007, 282, 19493–19501. [Google Scholar] [CrossRef]
- Stahl, A. A current review of fatty acid transport proteins (SLC27). Pflug. Arch.-Eur. J. Physiol. 2004, 447, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Schoonjans, K.; Staels, B.; Auwerx, J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res. 1996, 37, 907–925. [Google Scholar] [CrossRef]
- Tsai, M.L.; Chen, H.Y.; Tseng, M.C.; Chang, R.C. Cloning of peroxisorne proliferators activated receptors in the cobia (Rachycentron canadum) and their expression at different life-cycle stages under cage aquaculture. Gene 2008, 425, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Zeb, F.; Wu, X.Y.; Fatima, S.; Zaman, M.H.; Khan, S.A.; Safdar, M.; Alam, I.; Feng, Q. Time-restricted feeding regulates molecular mechanisms with involvement of circadian rhythm to prevent metabolic diseases. Nutrition 2021, 89, 111244. [Google Scholar] [CrossRef] [PubMed]
- Chaix, A.; Manoogian, E.N.C.; Melkani, G.C.; Panda, S. Time-restricted eating to prevent and manage chronic metabolic diseases. Annu. Rev. Nutr. 2019, 39, 291–315. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.J.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Chaix, A.; Zarrinpar, A.; Miu, P.; Panda, S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014, 20, 991–1005. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.Q.; Xu, H.P.; Xie, Z.B.; Wang, L.; Sun, Y.N.; Yang, H.Y.; Hu, D.D.; Mao, Y.L. Time-restricted feeding reduces the detrimental effects of a high-fat diet, possibly by modulating the circadian rhythm of hepatic lipid metabolism and gut microbiota. Front. Nutr. 2020, 7, 596285. [Google Scholar] [CrossRef] [PubMed]
- Chaix, A.; Lin, T.; Le, H.D.; Chang, M.W.; Panda, S. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 2019, 29, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, J.E.; Livelo, C.; Trujillo, A.S.; Chandran, S.; Woodworth, B.; Andrade, L.; Le, H.D.; Manor, U.; Panda, S.; Melkani, G.C. Time-restricted feeding restores muscle function in Drosophila models of obesity and circadian-rhythm disruption. Nat. Commun. 2019, 10, 2700. [Google Scholar] [CrossRef] [PubMed]
- Che, T.T.; Yan, C.; Tian, D.Y.; Zhang, X.; Liu, X.J.; Wu, Z.M. Time-restricted feeding improves blood glucose and insulin sensitivity in overweight patients with type 2 diabetes: A randomised controlled trial. Nutr. Metab. 2021, 18, 88. [Google Scholar] [CrossRef]
- Yamamuro, D.; Takahashi, M.; Nagashima, S.; Wakabayashi, T.; Yamazaki, H.; Takei, A.; Takei, S.; Sakai, K.; Ebihara, K.; Iwasaki, Y.; et al. Peripheral circadian rhythms in the liver and white adipose tissue of mice are attenuated by constant light and restored by time-restricted feeding. PLoS ONE 2020, 15, e0234439. [Google Scholar] [CrossRef]
- Flanagan, A.; Bechtold, D.A.; Pot, G.K.; Johnston, J.D. Chrono-nutrition: From molecular and neuronal mechanisms to human epidemiology and timed feeding patterns. J. Neurochem. 2021, 157, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Ratamess, N.A.; Faigenbaum, A.D.; Bush, J.A.; Beller, N.; Vargas, A.; Fardman, B.; Andriopoulos, T. Effect of Time-Restricted Feeding on Anthropometric, Metabolic, and Fitness Parameters: A Systematic Review. J. Am. Coll. Nutr. 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Paredes, J.F.; López-Olmeda, J.F.; Martínez, F.J.; Sánchez-Vázquez, F.J. Daily rhythms of lipid metabolic gene expression in zebra fish liver: Response to light/dark and feeding cycles. Chronobiol. Int. 2015, 32, 1438–1448. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2020; Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Davies, T.W.; Duffy, J.P.; Bennie, J.; Gaston, K.J. The nature, extent, and ecological implications of marine light pollution. Front. Ecol. Environ. 2014, 12, 347–355. [Google Scholar] [CrossRef]
- Bolton, D.; Mayer-Pinto, M.; Clark, G.F.; Dafforn, K.A.; Brassil, W.A.; Becker, A.; Johnston, E.L. Coastal urban lighting has ecological consequences for multiple trophic levels under the sea. Sci. Total Environ. 2017, 576, 1–9. [Google Scholar] [CrossRef]
- Li, D.; Huang, J.; Zhou, Q.M.; Gu, L.; Sun, Y.F.; Zhang, L.; Yang, Z. Artificial light pollution with different wavelengths at night interferes with development, reproduction, and antipredator defenses of Daphnia magna. Environ. Sci. Technol. 2022, 56, 1702–1712. [Google Scholar] [CrossRef]
- Bassi, A.; Love, O.P.; Cooke, S.J.; Warriner, T.R.; Harris, C.M.; Madliger, C.L. Effects of artificial light at night on fishes: A synthesis with future research priorities. Fish Fish. 2022, 23, 631–647. [Google Scholar] [CrossRef]
- Becker, A.; Whitfield, A.K.; Cowley, P.D.; Järnegren, J.; Naesje, T.F. Potential effects of artificial light associated with anthropogenic infrastructure on the abundance and foraging behaviour of estuary-associated fishes. J. Appl. Ecol. 2013, 50, 43–50. [Google Scholar] [CrossRef]
- Pulgar, J.; Zeballos, D.; Vargas, J.; Aldana, M.; Manriquez, P.H.; Manriquez, K.; Quijón, P.A.; Widdicombe, S.; Anguita, C.; Quintanilla, D.; et al. Endogenous cycles, activity patterns and energy expenditure of an intertidal fish is modified by artificial light pollution at night (ALAN). Environ. Pollut. 2019, 244, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.R.; Michel, C.J.; Gary, M.P.; Lehman, B.M.; Demetras, N.J.; Hammen, J.J.; Horn, M.J. Effects of artificial lighting at night on predator density and salmonid predation. Trans. Am. Fish. Soc. 2021, 150, 147–159. [Google Scholar] [CrossRef]
- Zapata, M.J.; Sullivan SM, P.; Gray, S.M. Artificial lighting at night in estuaries-Implications from individuals to ecosystems. Estuaries Coasts 2019, 42, 309–330. [Google Scholar] [CrossRef]
- Brüning, A.; Hölker, F.; Franke, S.; Kleiner, W.; Kloas, W. Influence of light intensity and spectral composition of artificial light at night on melatonin rhythm and mRNA expression of gonadotropins in roach Rutilus rutilus. Fish Physiol. Biochem. 2018, 44, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Labala, R.K.; Yumnamcha, T.; Devi, S.D.; Mondal, G.; Devi, H.S.; Rajiv, C.; Bharali, R.; Chattoraj, A. Artificial Light at Night (ALAN), an alarm to ovarian physiology: A study of possible chronodisruption on zebrafish (Danio rerio). Sci. Total Environ. 2018, 628, 1407–1421. [Google Scholar] [CrossRef]
- O’Connor, J.J.; Fobert, E.K.; Besson, M.; Jacob, H.; Lecchini, D. Live fast, die young: Behavioural and physiological impacts of light pollution on a marine fish during larval recruitment. Mar. Pollut. Bull. 2019, 146, 908–914. [Google Scholar] [CrossRef]
- Wei, H.; Cai, W.J.; Liu, H.K.; Han, D.; Zhu, X.M.; Yang, Y.X.; Jin, J.Y.; Xie, S.Q. Effects of photoperiod on growth, lipid metabolism and oxidative stress of juvenile gibel carp (Carassius auratus). J. Photochem. Photobiol. B Biol. 2019, 198, 111552. [Google Scholar] [CrossRef]
- Basili, D.; Lutfi, E.; Falcinelli, S.; Balbuena-Pecino, S.; Navarro, I.; Bertolucci, C.; Capilla, E.; Carnevali, O. Photoperiod Manipulation Affects Transcriptional Profile of Genes Related to Lipid Metabolism and Apoptosis in Zebrafish (Danio rerio) Larvae: Potential Roles of Gut Microbiota. Microb. Ecol. 2020, 79, 933–946. [Google Scholar] [CrossRef]
- Kupprat, F.; Hölker, F.; Kloas, W. Can skyglow reduce nocturnal melatonin concentrations in Eurasian perch? Environ. Pollut. 2020, 262, 114324. [Google Scholar] [CrossRef]
- Mondal, G.; Devi, S.D.; Khan, Z.A.; Yumnamcha, T.; Rajiv, C.; Devi, H.S.; Chattoraj, A. The influence of feeding on the daily rhythm of mRNA expression on melatonin bio-synthesizing enzyme genes and clock associated genes in the zebrafish (Danio rerio) gut. Biol. Rhythm. Res. 2021, 53, 1073–1090. [Google Scholar] [CrossRef]
- Nemova, N. Different photoperiod regimes affect the fatty acid profile of juvenile Atlantic salmon Salmo salar L.: Aquaculture and conservation approach to restoration of wild populations. J. Am. Oil Chem. Soc. 2021, 98, 236–237. [Google Scholar]
- Ytrestøyl, T.; Hjelle, E.; Kolarevic, J.; Takle, H.; Rebl, A.; Afanasyev, S.; Krasnov, A.; Brunsvik, P.; Terjesen, B.F. Photoperiod in recirculation aquaculture systems and timing of seawater transfer affect seawater growth performance of Atlantic salmon (Salmo salar). J. World Aquac. Soc. 2022, 1–23. [Google Scholar] [CrossRef]
- FAO. FAO Yearbook. Fishery and Aquaculture Statistics 2018; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Polakof, S.; Míguez, J.M.; Soengas, J.L. Daily changes in parameters of energy metabolism in liver, white muscle, and gills of rainbow trout: Dependence on feeding. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2007, 147, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, J.; Míguez, J.M.; Librán-Pérez, M.; Otero-Rodiño, C.; Naderi, F.; Soengas, J.L.; López-Patiño, M.A. Daily rhythms in activity and mRNA abundance of enzymes involved in glucose and lipid metabolism in liver of rainbow trout, Oncorhynchus mykiss. Influ. Light Food Availab. Chronobiol. Int. 2015, 32, 1391–1408. [Google Scholar] [CrossRef] [PubMed]
- Nisembaum, L.G.; Velarde, E.; Tinoco, A.B.; Azpeleta, C.; de Pedro, N.; Alonso-Gómez, A.L.; Delgado, M.J.; Isorna, E. Light-dark cycle and feeding time differentially entrains the gut molecular clock of the goldfish (Carassius auratus). Chronobiol. Int. 2012, 29, 665–673. [Google Scholar] [CrossRef]
- AOAC. Official methods of analysis of AOAC international. In Official Analytical Chemists, 16th ed.; Cunniff, P., Ed.; AOAC International: Arlington, VA, USA, 1995; pp. 1141–1154. [Google Scholar]
- Liu, T.; Xu, H.Y.; Han, T.; Wang, J.T.; Yin, F.; Wang, C.L. Effect of dietary egg yolk lecithin levels on survival, growth, lipid metabolism, and antioxidant capacity of early juvenile green mud crab Scylla paramamosain. Aquaculture 2021, 540, 736706. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sanchez, G. PLS Path Modeling with R. Trowchez Editions. Berkeley. 2013. Available online: http://www.gastonsanchez.com/PLSPathModelingwithR.pdf (accessed on 25 April 2022).
- Fjelldal, P.G.; Nordgarden, U.; Berg, A.; Grotmol, S.; Totland, G.K.; Wargelius, A.; Hansen, T. Vertebrae of the trunk and tail display different growth rates in response to photoperiod in Atlantic salmon, Salmo salar L., post-smolts. Aquaculture 2005, 250, 516–524. [Google Scholar] [CrossRef]
- Wargelius, A.; Fjelldal, P.G.; Nordgarden, U.; Hansen, T. Continuous light affects mineralization and delays osteoid incorporation in vertebral bone of Atlantic salmon (Salmo salar L.). J. Exp. Biol. 2009, 212, 656–661. [Google Scholar] [CrossRef][Green Version]
- Fjelldal, P.G.; Lock, E.J.; Hansen, T.; Waagbo, R.; Wargelius, A.; Martens, L.G.; El-Mowafi, A.; Onsrud, R. Continuous light induces bone resorption and affects vertebral morphology in Atlantic salmon (Salmo salar L.) fed a phosphorous deficient diet. Aquac. Nutr. 2012, 18, 610–619. [Google Scholar] [CrossRef]
- Villamizar, N.; Garciá-Alcazar, A.; Sánchez-Vázquez, F.J. Effect of light spectrum and photoperiod on the growth, development and survival of European sea bass (Dicentrarchus labrax) larvae. Aquaculture 2009, 292, 80–86. [Google Scholar] [CrossRef]
- Mhalhel, K.; Germanà, A.; Abbate, F.; Guerrera, M.C.; Levanti, M.; Laurà, R.; Montalbano, G. The Effect of Orally Supplemented Melatonin on Larval Performance and Skeletal Deformities in Farmed Gilthead Seabream (Sparus aurata). Int. J. Mol. Sci. 2020, 21, 9597. [Google Scholar] [CrossRef]
- Mistlberger, R.E. Circadian food-anticipatory activity: Formal models and physiological mechanisms. Neurosci. Biobehav. Rev. 1994, 18, 171–195. [Google Scholar] [CrossRef]
- Reebs, S.G.; Gallant, B.Y. Food-anticipatory activity as a cue for local enhancement in golden shiners (Pisces: Cyprinidae, Notemigonus crysoleucas). Ethology 1997, 103, 1060–1069. [Google Scholar] [CrossRef]
- Chen, W.M.; Purser, G.J. The effect of feeding regime on growth, locomotor activity pattern and the development of food anticipatory activity in greenback flounder. J. Fish Biol. 2001, 58, 177–187. [Google Scholar] [CrossRef]
- Stephan, F.K. The “other” circadian system: Food as zeitgeber. J. Biol. Rhythm. 2002, 17, 284–292. [Google Scholar] [CrossRef]
- Vera, L.M.; De Pedro, N.; Gomez-Milán, E.; Delgado, M.J.; Sánchez-Muros, M.J.; Madrid, J.A.; Sánchez-Vázquez, F.J. Feeding entrainment of locomotor activity rhythms, digestive enzymes and neuroendocrine factors in goldfish. Physiol. Behav. 2007, 90, 518–524. [Google Scholar] [CrossRef]
- Montoya, A.; López-Olmeda, J.F.; Yúfera, M.; Sánchez-Muros, M.J.; Sánchez-Vázquez, F.J. Feeding time synchronises daily rhythms of behaviour and digestive physiology in gilthead seabream (Sparus aurata). Aquaculture 2010, 306, 315–321. [Google Scholar] [CrossRef]
- Sánchez, J.A.; López-Olmeda, J.F.; Blanco-Vives, B.; Sánchez-Vázquez, F.J. Effects of feeding schedule on locomotor activity rhythms and stress response in sea bream. Physiol. Behav. 2009, 98, 125–129. [Google Scholar] [CrossRef]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T.W. Glucose metabolism in fish: A review. J. Comp. Physiol. B-Biochem. Syst. Environ. Physiol. 2012, 182, 1015–1045. [Google Scholar] [CrossRef] [PubMed]
- Capilla, E.; Medale, F.; Navarro, I.; Panserat, S.; Vachot, C.; Kaushik, S.; Gutierrez, J. Muscle insulin binding and plasma levels in relation to liver glucokinase activity, glucose metabolism and dietary carbohydrates in rainbow trout. Regul. Pept. 2003, 110, 123–132. [Google Scholar] [CrossRef]
- Gong, Y.; Chen, W.; Dong, H.; Zhu, X.; Yang, Y.; Jin, J.; Liu, H.; Xie, S. Effects of food restriction on growth, body composition and gene expression related in regulation of lipid metabolism and food intake in grass carp. Aquaculture 2017, 469, 28–35. [Google Scholar] [CrossRef]
- Koonen, D.P.Y.; Jacobs, R.L.; Febbraio, M.; Young, M.E.; Soltys, C.L.M.; Ong, H.; Vance, D.E.; Dyck, J.R.B. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 2007, 56, 2863–2871. [Google Scholar] [CrossRef] [PubMed]
- Frohnert, B.I.; Hui, T.Y.; Bernlohr, D.A. Identification of a functional peroxisome proliferator-responsive element in the murine fatty acid transport protein gene. J. Biol. Chem. 1999, 274, 3970–3977. [Google Scholar] [CrossRef] [PubMed]
- Vieira, R.F.L.; Munoz, V.R.; Junqueira, R.L.; de Oliveira, F.; Gaspar, R.C.; Nakandakari, S.C.B.R.; Costa, S.D.; Torsoni, M.A.; da Silva, A.S.R.; Cintra, D.E.; et al. Time-restricted feeding combined with aerobic exercise training can prevent weight gain and improve metabolic disorders in mice fed a high-fat diet. J. Physiol. 2021, 600, 797–813. [Google Scholar] [CrossRef]
- Wagner, T.; Congleton, J.L. Blood chemistry correlates of nutritional condition, tissue damage, and stress in migrating juvenile chinook salmon (Oncorhynchus tshawytscha). Can. J. Fish. Aquat. Sci. 2004, 61, 1066–1074. [Google Scholar] [CrossRef]
- Sánchez-Bretaño, A.; Gueguen, M.M.; Cano-Nicolau, J.; Kah, O.; Alonso-Gomez, A.L.; Delgado, M.J.; Isorna, E. Anatomical distribution and daily profile of gper1b gene expression in brain and peripheral structures of goldfish (Carassius auratus). Chronobiol. Int. 2015, 32, 889–902. [Google Scholar] [CrossRef]
- López-Olmeda, J.F.; Sánchez-Vázquez, F.J. Feeding Rhythms in Fish: From Behavioural to Molecular Approach. In Biological Clock in Fish; Kulczykowska, E., Popek, W., Kapoor, B.G., Eds.; Science Publishers: Enfield, CT, USA, 2010; pp. 155–184. [Google Scholar]
- Vera, L.M.; Negrini, P.; Zagatti, C.; Frigato, E.; Sanchez-Vazquez, F.J.; Bertolucci, C. Light and feeding entrainment of the molecular circadian clock in a marine teleost (Sparus aurata). Chronobiol. Int. 2013, 30, 649–661. [Google Scholar] [CrossRef]
- Iigo, M.; Tabata, M. Circadian Rhythms of Locomotor Activity in the Rainbow Trout Oncorhynchus mykiss. Fish. Sci. 1997, 63, 77–80. [Google Scholar] [CrossRef]
- Sánchez-Vázquez, F.J.; Tabata, M. Circadian rhythms of demand-feeding and locomotor activity in rainbow trout. J. Fish Biol. 1998, 52, 255–267. [Google Scholar] [CrossRef]
- Bolliet, V.; Aranda, A.; Boujard, T. Demand-feeding rhythm in rainbow trout and European catfish: Synchronisation by photoperiod and food availability. Physiol. Behav. 2001, 73, 625–633. [Google Scholar] [CrossRef]
- Salgado-Delgado, R.C.; Saderi, N.; Basualdo, M.D.; Guerrero-Vargas, N.N.; Escobar, C.; Buijs, R.M. Shift Work or Food Intake during the Rest Phase Promotes Metabolic Disruption and Desynchrony of Liver Genes in Male Rats. PLoS ONE 2013, 8, e60052. [Google Scholar] [CrossRef]
- López-Olmeda, J.F. Nonphotic entrainment in fish. Comp. Biochem. Physiol. 2017, 203 Pt A, 133–143. [Google Scholar] [CrossRef]
- Fortes-Silva, R.; Do Valle, S.V.; Lopez-Olmeda, J.F. Daily rhythms of swimming activity, synchronization to different feeding times and effects on anesthesia practice in an Amazon fish species (Colossoma macropomum). Chronobiol. Int. 2018, 35, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Frolow, J.; Milligan, C.L. Hormonal regulation of glycogen metabolism in white muscle slices from rainbow trout (Oncorhynchus mykiss Walbaum). Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2004, 287, R1344–R1353. [Google Scholar] [CrossRef] [PubMed]
- Sargent, J.R.; Tocher, D.R.; Bell, J.G. The Lipid. In Fish Nutrition; Halver, J.E., Hardy, R.W., Eds.; Academic Press: Cambridge, MA, USA, 2002; pp. 181–257. [Google Scholar]
- Tocher, D.R. Metabolism and functions of lipids and fatty acids in teleost fish. Rev. Fish. Sci. 2003, 11, 107–184. [Google Scholar] [CrossRef]
- Hillyer, K.E.; Beale, D.J.; Shima, J.S. Artificial light at night interacts with predatory threat to alter reef fish metabolite profiles. Sci. Total Environ. 2021, 769, 144482. [Google Scholar] [CrossRef]
- Henderson, R.J.; Sargent, J.R.; Hopkins, C.C.E. Changes in the content and fatty acid composition of lipid in an isolated population of the capelin Mallotus villosus during sexual maturation and spawning. Mar. Biol. 1984, 78, 255–263. [Google Scholar] [CrossRef]
- Valenzuela, A.; Rodríguez, I.; Schulz, B.; Cortés, R.; Acosta, J.; Campos, V.; Escobar-Aguirre, S. Effects of continuous light (LD24:0) modulate the expression of lysozyme, mucin and peripheral blood cells in rainbow trout. Fishes 2022, 7, 28. [Google Scholar] [CrossRef]
- Qin, C.J.; Gong, Q.; Wen, Z.Y.; Zou, Y.C.; Yuan, D.Y.; Shao, T.; Li, H.T. Comparative analysis of the liver transcriptome of Pelteobagrus vachellii with an alternative feeding time. Comp. Biochem. Physiol. D-Genom. Proteom. 2017, 22, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.Z.; Wang, S.Q.; You, C.H.; Chen, F.; Zhang, Q.H.; Li, Y.Y. Influencing factors and mechanisms on HUFA biosynthesis in teleosts. J. Fish. Sci. China 2013, 20, 456–466. [Google Scholar] [CrossRef]
- Calabretti, A.; Cateni, F.; Procida, G.; Favretto, L.G. Influence of environmental temperature on composition of lipids in edible flesh of rainbow trout (Oncorhynchus mykiss). J. Sci. Food Agric. 2003, 83, 1493–1498. [Google Scholar] [CrossRef]
- Cengiz, E.I.; Unlu, E.; Bashan, M.; Satar, A.; Uysal, E. Effect of seasonal variations on the fatty acid composition of total lipid phospholipid and triacylglicerol in the dorsal muscle of Mesopotamian catfish (Silurus triostegus Heckel, 1843) in Tigris River (Turkey). Turk. J. Fish. Aquat. Sci. 2012, 2012, 31–37. [Google Scholar] [CrossRef]
- Skiba-Cassy, S.; Geurden, I.; Panserat, S.; Seiliez, I. Dietary methionine imbalance alters the transcriptional regulation of genes involved in glucose, lipid and amino acid metabolism in the liver of rainbow trout (Oncorhynchus mykiss). Aquaculture 2016, 454, 56–65. [Google Scholar] [CrossRef]
- Song, X.R.; Marandel, L.; Skiba-Cassy, S.; Corraze, G.; Dupont-Nivet, M.; Quillet, E.; Geurden, I.; Panserat, S. Regulation by dietary carbohydrates of intermediary metabolism in liver and muscle of two isogenic lines of rainbow trout. Front. Physiol. 2018, 9, 1579. [Google Scholar] [CrossRef]
- Kamalam, B.S.; Médale, F.; Larroquet, L.; Corraze, G.; Panserat, S. Metabolism and fatty acid profile in fat and lean rainbow trout lines fed with vegetable oil: Effect of carbohydrates. PLoS ONE 2013, 8, e76570. [Google Scholar] [CrossRef] [PubMed]
- Velasco, C.; Comesaña, S.; Conde-Sieira, M.; Míguez, J.M.; Soengas, J.L. Effects of CCK-8 and GLP-1 on fatty acid sensing and food intake regulation in trout. J. Mol. Endocrinol. 2019, 62, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Conde-Sieira, M.; Capelli, V.; Álvarez-Otero, R.; Díaz-Rúa, A.; Velasco, C.; Comesaña, S.; López, M.; Soengas, J.L. Hypothalamic AMPKα2 regulates liver energy metabolism in rainbow trout through vagal innervation. Am. J. Physiol-Reg. I. 2020, 318, R122–R134. [Google Scholar] [CrossRef]
- Sánchez-Gurmaches, J.; Cruz-Garcia, L.; Gutiérrez, J.; Navarro, I. Adiponectin effects and gene expression in rainbow trout: An in vivo and in vitro approach. J. Exp. Biol. 2012, 215, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Naderi, F.; Míguez, J.M.; Soengas, J.L.; López-Patiño, M.A. SIRT1 mediates the effect of stress on hypothalamic clock genes and food intake regulators in rainbow trout, Oncorhynchus mykiss. Comp. Biochem. Phys. A 2019, 235, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Panserat, S.; Plagnes-Juan, E.; Gazzola, E.; Palma, M.; Magnoni, L.J.; Marandel, L.; Viegas, I. Hepatic glycerol metabolism-related genes in carnivorous rainbow trout (Oncorhynchus mykiss): Insights into molecular characteristics, ontogenesis, and nutritional regulation. Front. Physiol. 2020, 11, 882. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Shi, C.; Ye, Y.; Song, C.; Mu, C.; Wang, C. Time-Restricted Feeding Could Not Reduce Rainbow Trout Lipid Deposition Induced by Artificial Night Light. Metabolites 2022, 12, 904. https://doi.org/10.3390/metabo12100904
Xu H, Shi C, Ye Y, Song C, Mu C, Wang C. Time-Restricted Feeding Could Not Reduce Rainbow Trout Lipid Deposition Induced by Artificial Night Light. Metabolites. 2022; 12(10):904. https://doi.org/10.3390/metabo12100904
Chicago/Turabian StyleXu, Hanying, Ce Shi, Yangfang Ye, Changbin Song, Changkao Mu, and Chunlin Wang. 2022. "Time-Restricted Feeding Could Not Reduce Rainbow Trout Lipid Deposition Induced by Artificial Night Light" Metabolites 12, no. 10: 904. https://doi.org/10.3390/metabo12100904
APA StyleXu, H., Shi, C., Ye, Y., Song, C., Mu, C., & Wang, C. (2022). Time-Restricted Feeding Could Not Reduce Rainbow Trout Lipid Deposition Induced by Artificial Night Light. Metabolites, 12(10), 904. https://doi.org/10.3390/metabo12100904






