Metabolic Phenotypes in Asthmatic Adults: Relationship with Inflammatory and Clinical Phenotypes and Prognostic Implications
Abstract
1. Introduction
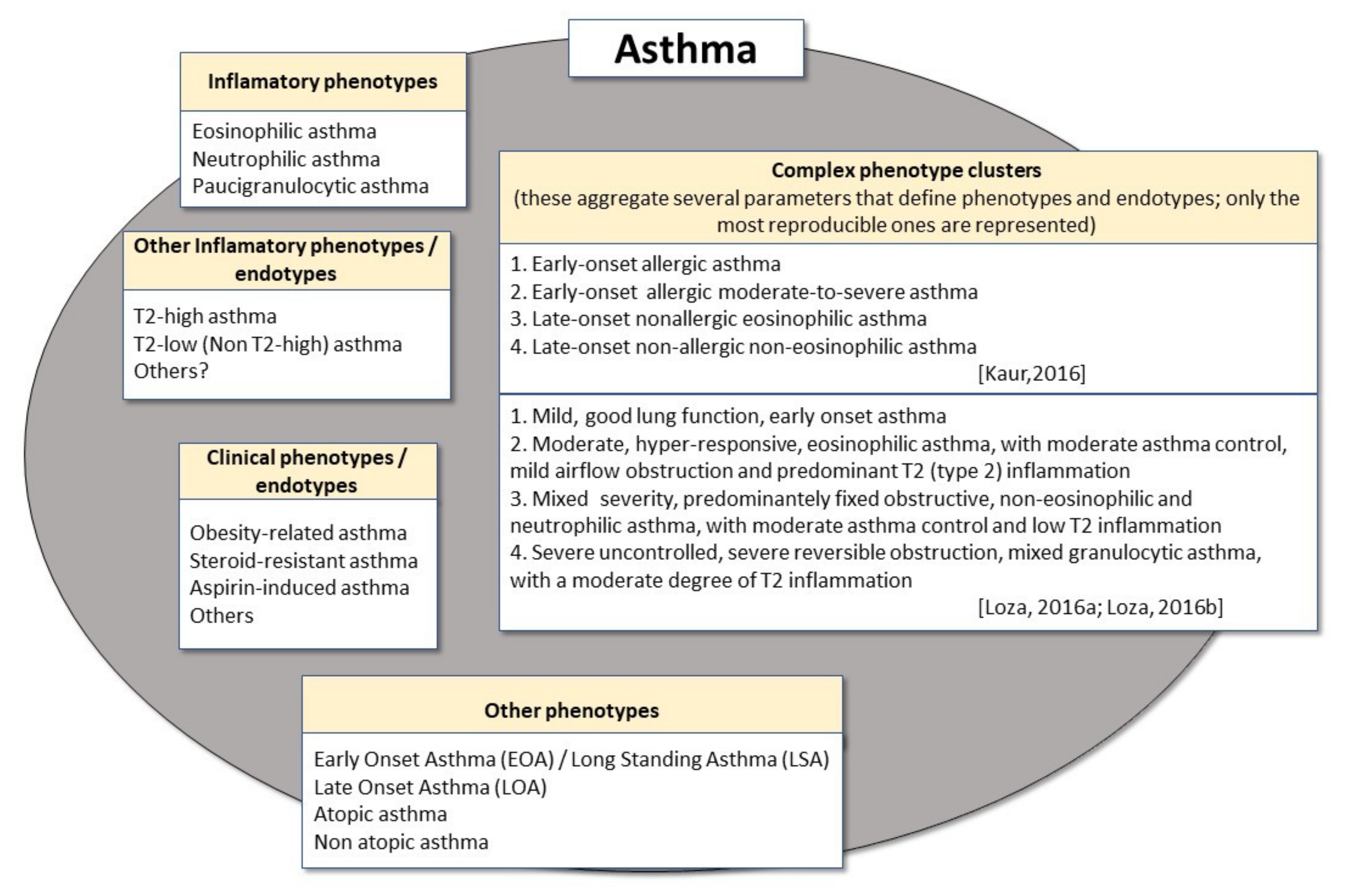
2. Phenotypes and Endotypes in Adult Asthma
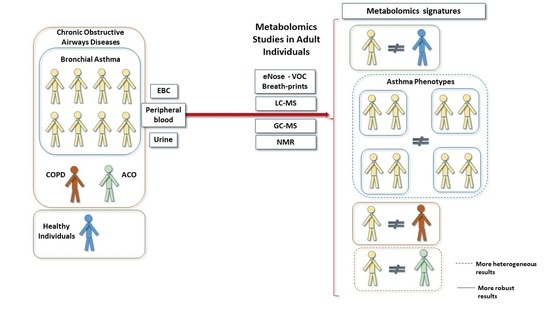
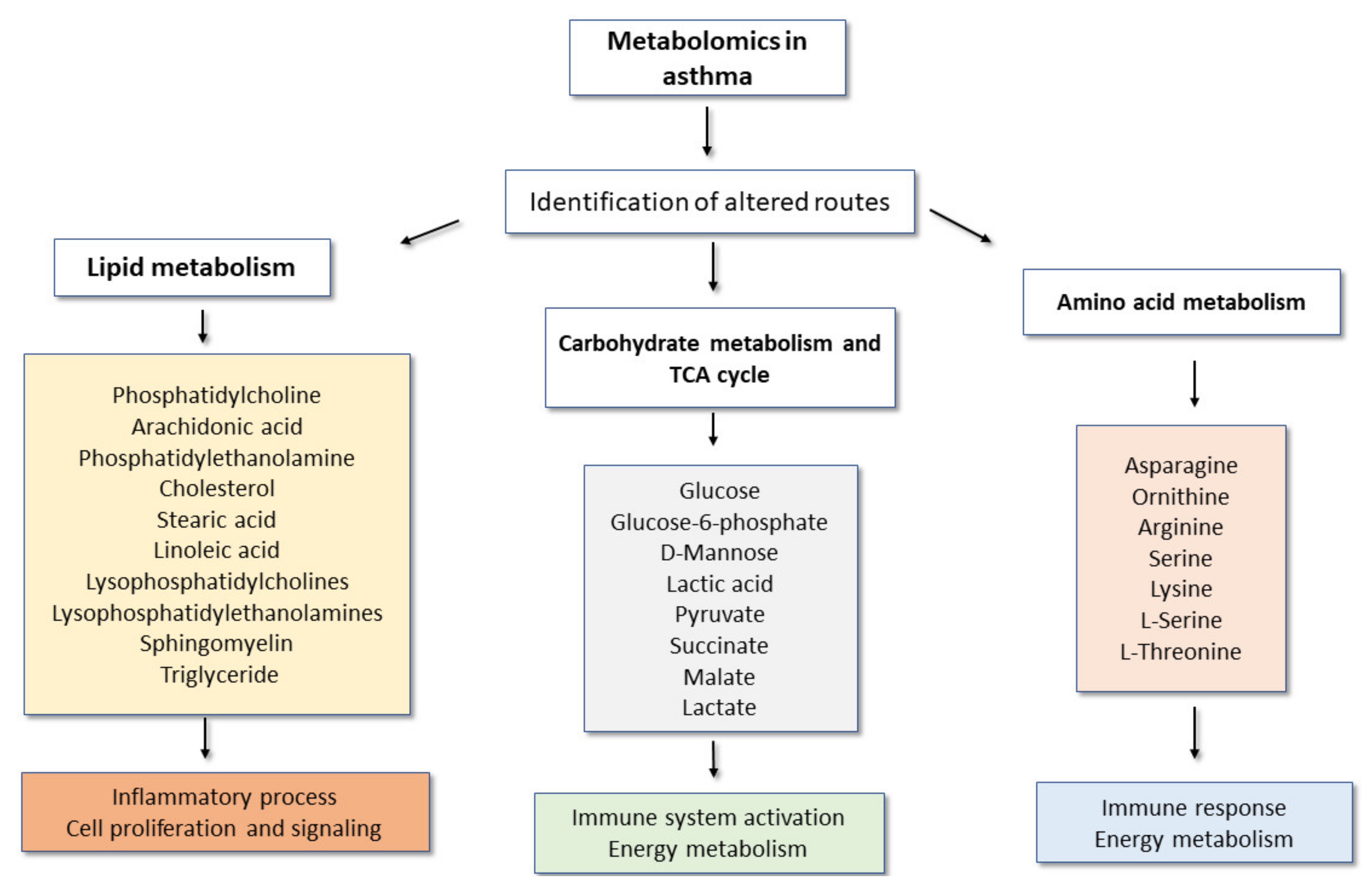
3. Main Metabolomic Signatures and Their Potential Implications in Adult Asthma
4. Assessment of Metabolic Changes in Inflammatory Asthma Phenotypes
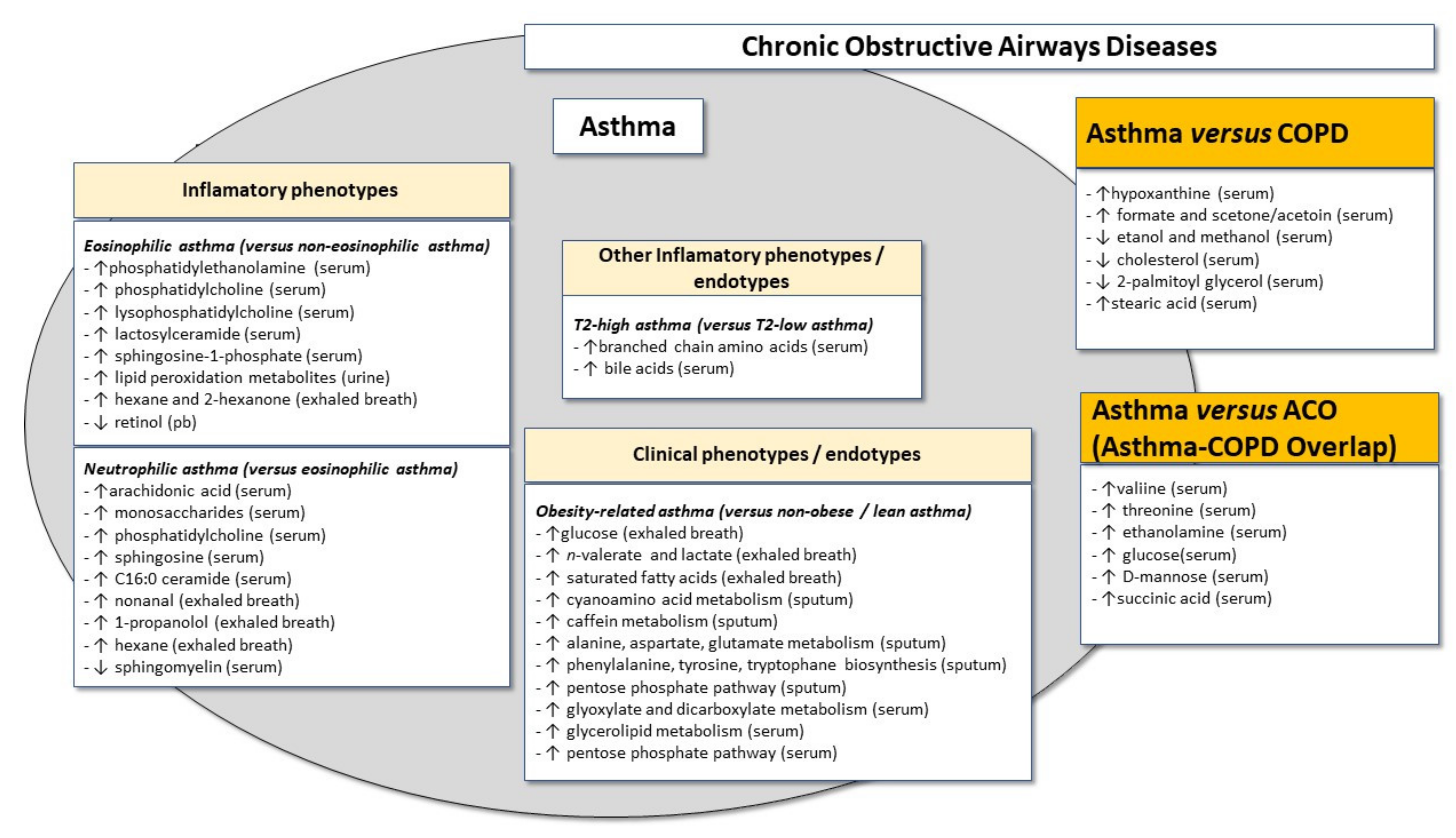
4.1. Global Metabolomic Signatures in Eosinophilic and Non-Eosinophilic Asthma Phenotypes
4.2. Lipidomics in Eosinophilic and Non-Eosinophilic Asthma Phenotypes
5. Assessment of Metabolic Changes in Clinical Asthma Phenotypes or Endotypes
5.1. Metabolomics Signature in the Atopic Asthma Phenotype
5.2. Metabolomics Signature in the Obese Asthma “Phenotype”
5.3. Assessment of Metabolomics in the Steroid-Resistant Asthma “Phenotype”
6. Revisiting the “Dutch Hypothesis”: Discriminating between the “Phenotypes” of Asthma and Other Chronic Obstructive Airways Diseases
7. Reproducibility and Stability of Asthma-Related Metabolic Signatures: Of Validation Cohorts, Time Stability, Age, Sex and Other Factors
8. Prognostic Value of Asthma-Related Metabolic Signatures
9. Concluding Remarks and Future Challenges
Author Contributions
Funding
Conflicts of Interest
References
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention. Updated 2021. Available online: Ginasthma.org/wp-content/uploads/2021/05/GINA-Main-Report-2021-V2-WMS.pdf (accessed on 2 March 2021).
- Global Asthma Network. The Global Asthma Report 2018. New Zealand 2018. Available online: http://www.globalasthmareport.org/ (accessed on 6 March 2021).
- Silkoff, P.E.; Strambu, I.; Laviolette, M.; Singh, D.; FitzGerald, J.M.; Lam, S.; Kelsen, S.; Eich, A.; Ludwig-Sengpiel, A.; Hupp, G.C.; et al. Asthma characteristics and biomarkers from the Airways Disease Endotyping for Personalized Therapeutics (ADEPT) longitudinal profiling study. Respir. Res. 2015, 16, 142. [Google Scholar] [CrossRef]
- Anderson, G.P. Endotyping asthma: New insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet 2008, 372, 1107–1119. [Google Scholar] [CrossRef]
- Lötvall, J.; Akdis, C.A.; Bacharier, L.B.; Bjermer, L.; Casale, T.B.; Custovic, A.; Lemanske, R.F., Jr.; Wardlaw, A.J.; Wenzel, S.E.; Greenberger, P.A. Asthma endotypes: A new approach to classification of disease entities within the asthma syndrome. J. Allergy Clin. Immunol. 2011, 127, 355–360. [Google Scholar] [CrossRef]
- Chung, K.F. Precision medicine in asthma: Linking phenotypes to targeted treatments. Curr. Opin. Pulm. Med. 2018, 24, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, M.E.; Lee, F.E.; Lee, G.B. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Rufo, J.; Taborda-Barata, L.; Lourenço, O. Serum biomarkers in elderly asthma. J. Asthma 2013, 50, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Peng, Y.-Q.; Agache, I.; Diamant, Z.; Eiwegger, T.; Fokkens, W.J.; Tradil-Hoffmann, C.; Nadeau, K.; O’Hehir, R.E.; O’Mahony, L.; et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy 2020, 75, 3039–3068. [Google Scholar] [CrossRef]
- Lee, Y.; Quoc, Q.L.; Park, H.S. Biomarkers for severe asthma: Lessons from longitudinal cohort studies. Allergy Asthma Immunol. Res. 2021, 13, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, C.; Kucuksezer, U.C.; Akdis, M.; Akdis, C.A. The concepts of asthma endotypes and phenotypes to guide current and novel treatment strategies. Expert Rev. Respir. Med. 2018, 12, 733–743. [Google Scholar] [CrossRef]
- Ribbenstedt, A.; Ziarrusta, H.; Benskin, J.P. Development, characterization and comparisons of targeted and non-targeted metabolomics methods. PLoS ONE 2018, 13, e0207082. [Google Scholar] [CrossRef]
- Zhu, Z.; Camargo, C.A., Jr.; Hasegawa, K. Metabolomics in the prevention and management of asthma. Expert Rev. Respir. Med. 2019, 13, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Peel, A.M.; Wilkinson, M.; Sinha, A.; Loke, Y.K.; Fowler, S.J.; Wilson, A.M. Volatile organic compounds associated with diagnosis and disease characteristics in asthma—A systematic review. Respir. Med. 2020, 169, 105984. [Google Scholar] [CrossRef]
- Gertsman, I.; Barshop, B.A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metab. Dis. 2018, 41, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Kurland, I.J.; Jones, M.R.; Dunn, W.B. How close are we to complete annotation of metabolomes? Curr. Opin. Chem. Biol. 2017, 36, 64–69. [Google Scholar] [CrossRef]
- Johnson, C.H.; Patterson, A.D.; Idle, J.R.; Gonzalez, F.J. Xenobiotic metabolomics: Major impact on the metabolome. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 37–56. [Google Scholar] [CrossRef]
- Eghbalnia, H.R.; Romero, P.R.; Westler, W.M.; Baskaran, K.; Ulrich, E.L.; Markley, J.L. Increasing rigor in NMR-based metabolomics through validated and open source tools. Curr. Opin. Biotechnol. 2017, 43, 56–61. [Google Scholar] [CrossRef]
- Dominick, T.M.; Gill, E.L.; Vedam-Mai, V.; Yost, R.A. Mass spectrometry-based cellular metabolomics: Current approaches, applications and future directions. Anal. Chem. 2021, 93, 546–566. [Google Scholar] [CrossRef]
- Crook, A.A.; Powers, R. Quantitative NMR-based biomedical metabolomics: Current status and applications. Molecules 2020, 25, 5128. [Google Scholar] [CrossRef]
- Alves, S.; Paris, A.; Rathahao-Paris, E. Chapter Four—Mass spectrometry-based metabolomics for an in-depth questioning of human health. Adv. Clin. Chem. 2020, 99, 147–191. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Advances in electronic-nose technologies for the detection of volatile biomarker metabolites in the human breath. Metabolites 2015, 5, 140–163. [Google Scholar] [CrossRef]
- Wilson, A.D. Biomarker metabolite signatures pave the way for electronic-nose applications in early clinical disease diagnoses. Curr. Metab. 2017, 5, 90–101. [Google Scholar] [CrossRef]
- Wilson, A.D.; Baietto, M. Advances in electronic-nose technologies developed for biomedical applications. Sensors 2011, 11, 1105–1176. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Noninvasive early disease diagnosis by electronic-nose and related VOC-detection devices. Biosensors 2020, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Lärstad, M.A.E.; Torén, K.; Bake, B.; Olin, A.-C. Determination of ethane, pentane and isoprene in exhaled air-effects of breath-holding, flow rate and purified air. Acta. Physiol. 2007, 189, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Cowan, J.O.; Filsell, S.; McLachlan, C.; Monti-Sheehan, G.; Jackson, P.; Robin Taylor, D. Diagnosing asthma: Comparisons between exhaled nitric oxide measurements and conventional tests. Am. J. Respir. Crit. Care Med. 2004, 169, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Cavaleiro Rufo, J.; Madureira, J.; Oliveira Fernandes, E.; Moreira, A. Volatile organic compounds in asthma diagnosis: A systematic review and meta-analysis. Allergy 2016, 71, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Corradi, M.; Majori, M.; Cacciani, G.C.; Consigli, G.F.; de’Munari, E.; Pesci, A. Increased exhaled nitric oxide in patients with stable chronic obstructive pulmonary disease. Thorax 1999, 54, 572–575. [Google Scholar] [CrossRef]
- Binson, V.A.; Subramoniam, M.; Mathew, L. Discrimination of COPD and lung cancer from controls through breath analysis using a self-developed e-nose. J. Breath Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ratiu, I.A.; Ligor, T.; Bocos-Bintintan, V.; Mayhew, C.A.; Buszewski, B. Volatile Organic Compounds in exhaled breath as fingerprints of lung cancer, asthma and COPD. J. Clin. Med. 2020, 10, 32. [Google Scholar] [CrossRef]
- Balint, B.; Kharitonov, S.A.; Hanazawa, T.; Donnelly, L.E.; Shah, P.L.; Hodson, M.E.; Barnes, P.J. Increased nitrotyrosine in exhaled breath condensate in cystic fibrosis. Eur. Respir. J. 2001, 17, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Kamboures, M.A.; Blake, D.R.; Cooper, D.M.; Newcomb, R.L.; Barker, M.; Larson, J.K.; Meinardi, S.; Nussbaum, E.; Rowland, F.S. Breath sulfides and pulmonary function in cystic fibrosis. Proc. Natl. Acad. Sci. USA 2005, 102, 15762–15767. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Hengst, M.; Schmid, J.; Buers, H.-J.; Mittermaier, B.; Klemp, D.; Koppmann, R. Volatile organic compounds in the exhaled breath of young patients with cystic fibrosis. Eur. Respir. J. 2006, 27, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Syhre, M.; Manning, L.; Phuanukoonnon, S.; Harino, P.; Chambers, S.T. The scent of Mycobacterium tuberculosis—Part II breath. Tuberculosis 2009, 89, 263–266. [Google Scholar] [CrossRef]
- Wilson, A.D. Applications of electronic-nose technologies for noninvasive early detection of plant, animal and human diseases. Chemosensors 2018, 6, 45. [Google Scholar] [CrossRef]
- Wilson, A.D. Developing electronic-nose technologies for clinical practice. J. Med. Surg. Pathol. 2018, 3, 4. [Google Scholar] [CrossRef]
- Paredi, P.; Kharitonov, S.A.; Barnes, P.J. Elevation of exhaled ethane concentration in asthma. Am. J. Respir. Crit. Care Med. 2000, 162, 140–1454. [Google Scholar] [CrossRef]
- Montuschi, P.; Santonico, M.; Mondino, C.; Pennazza, G.; Mantini, G.; Martinelli, E.; Capuano, R.; Ciabattoni, G.; Paolesse, R.; Di Natale, C.; et al. Diagnostic performance of an electronic nose, fractional exhaled nitric oxide, and lung function testing in asthma. Chest 2010, 137, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, B.; Basanta, M.; Cadden, P.; Singh, D.; Douce, D.; Woodcock, A.; Fowler, S.J. Non-invasive phenotyping using exhaled volatile organic compounds in asthma. Thorax 2011, 66, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Schivo, M.; Seichter, F.; Aksenov, A.A.; Pasamontes, A.; Peirano, D.J.; Mizaikoff, B.; Kenyon, N.J.; David, C.E. A mobile instrumentation platform to distinguish airway disorders. J. Breath Res. 2013, 7, 98–106. [Google Scholar] [CrossRef] [PubMed]
- van der Schee, M.P.; Palmay, R.; Cowan, J.O.; Taylor, D.R. Predictive steroid responsiveness in patients with asthma using exhaled breath profiling. Clin. Exp. Allergy 2013, 43, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Kim, S.-H.; Lee, H.S.; Choi, G.S.; Jung, Y.-S.; Ryu, D.H.; Park, H.-S.; Hwang, G.-S. Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clin. Exp. Allergy 2013, 43, 425–433. [Google Scholar] [CrossRef]
- Checkley, W.; Deza, M.P.; Klawitter, J.; Romero, K.M.; Klawitter, J.; Pollard, S.L.; Wise, R.A.; Christians, U.; Hansel, N.N. Identifying biomarkers for asthma diagnosis using targeted metabolomics approaches. Respir. Med. 2016, 121, 59–66. [Google Scholar] [CrossRef]
- Kelly, R.S.; Dahlin, A.; McGeachie, M.J.; Qiu, W.; Sordillo, J.; Wan, E.S.; Wu, A.C.; Lasky-Su, J. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest 2017, 151, 262–277. [Google Scholar] [CrossRef]
- Esteves, P.; Blanc, L.; Celle, A.; Dupin, I.; Maurat, E.; Amoedo, N.; Cardouat, G.; Ousova, O.; Gales, L.; Bellvert, F.; et al. Crucial role of fatty acid oxidation in asthmatic bronchial smooth muscle remodelling. Eur. Respir. J. 2021. [Google Scholar] [CrossRef]
- Jiang, T.; Dai, L.; Li, P.; Zhao, J.; Wang, X.; An, L.; Liu, M.; Wu, S.; Wang, Y.; Peng, Y.; et al. Lipid metabolism and identification of biomarkers in asthma by lipidomic analysis. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2021, 1866, 158853. [Google Scholar] [CrossRef]
- Maniscalco, M.; Paris, D.; Melck, D.J.; Molino, A.; Carone, M.; Ruggeri, P.; Caramori, G.; Motta, A. Differential diagnosis between newly diagnosed asthma and COPD using exhaled breath condensate metabolomics: A pilot study. Eur. Respir. J. 2018, 51, 1701825. [Google Scholar] [CrossRef]
- Saude, E.J.; Skappak, C.D.; Regush, S.; Cook, K.; Ben-Zevi, A.; Becker, A.; Moqbel, R.; Sykes, B.D.; Roew, B.H.; Adamkp, D.J. Metabolomic profiling of asthma: Diagnostic utility of urine nuvlear magnetic resonance spectroscopy. J. Allergy Clin. Immunol. 2011, 127, 757–764. [Google Scholar] [CrossRef]
- Comhair, S.A.A.; McDunn, J.; Bennett, C.; Fettig, J.; Erzurum, S.C.; Kalhan, S.C. Metabolic endotype of asthma. J. Immunol. 2015, 195, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, C.C.; Oliveira, A.S.; Santos, M.; Rudnitskaya, A.; Todo-Bom, A.; Bousquet, J.; Rocha, S.M. Urinary metabolomic profiling of asthmatics can be related to clinical characteristics. Allergy 2016, 71, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Carraro, S.; Giordano, G.; Reniero, F.; Carpi, D.; Stocchero, M.; Sterk, P.J.; Baraldi, E. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy 2013, 68, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Reinke, S.N.; Gallart-Ayala, H.; Gomez, C.; Checa, A.; Fauland, A.; Naz, S.; Kamleh, M.A.; Djukanovic, R.; Hinks, T.S.C.; Wheelock, C.E. Metabolomics analysis identifies different metabotypes of asthma severity. Eur. Respir. J. 2017, 49, 1601740. [Google Scholar] [CrossRef]
- Ntontsi, P.; Ntzoumanika, V.; Loukides, S.; Benaki, D.; Gkikas, E.; Mikros, E.; Bakakos, P. EBC metabolomics for asthma severity. J. Breath Res. 2020, 14, 036007. [Google Scholar] [CrossRef]
- Ibrahim, B.; Marsden, P.; Smith, J.A.; Custovic, A.; Nilsson, M.; Fowler, S.J. Breath metabolomic profiling by nuclear magnetic resonance spectroscopy in asthma. Allergy 2013, 68, 1050–1056. [Google Scholar] [CrossRef]
- Mattarucchi, E.; Baraldi, E.; Guillou, C. Metabolomics applied to urine samples in childhood asthma, differentiation between asthma phenotypes and identification of relevant metabolites. Biomed. Chromatogr. 2012, 26, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Maniscalco, M.; Paris, D.; Melck, D.J.; D’Amato, M.; Zedda, A.; Sofia, M.; Stellato, C.; Motta, A. Coexistence of obesity and asthma determines a distinct respiratory metabolic phenotype. J. Allergy Clin. Immunol. 2017, 139, 1536–1547. [Google Scholar] [CrossRef]
- Skloot, G.S.; Busse, P.J.; Braman, S.S.; Kovacs, E.J.; Dixon, A.E.; Vaz Fragoso, C.A.; Ragless, B.B. An official American Thoracic Society Workshop Report: Evaluation and management of asthma in the elderly. Ann. Am. Thorac. Soc. 2016, 13, 2064–2077. [Google Scholar] [CrossRef]
- Maricoto, T.; Santos, D.; Carvalho, C.; Teles, I.; Correia-de-Sousa, J.; Taborda-Barata, L. Assessment of poor inhaler technique in older patients with asthma or COPD: A predictive tool for clinical risk and inhaler performance. Drugs Aging 2020, 37, 605–616. [Google Scholar] [CrossRef]
- Papamichael, M.M.; Katsardis, C.; Sarandi, E.; Georgaki, S.; Frima, E.S.; Varvarigou, A.; Tsoukalas, D. Application of metabolomics in pediatric asthma: Prediction, diagnosis and personalized treatment. Metabolites 2021, 11, 251. [Google Scholar] [CrossRef]
- Schjodt, M.S.; Gürdeniz, G.; Chaws, B. The metabolomics of hildhood atopic diseases: A comprehensive pathway-specific review. Metabolites 2020, 10, 511. [Google Scholar] [CrossRef]
- Alves, R.S.A.A.; Vianna, F.A.F.; Pereira, C.A.C. Clinical phenotypes of asthma. J. Bras. Pneumol. 2008, 34, 646–653. [Google Scholar] [CrossRef][Green Version]
- Kim, S.-H. Blood molecular biomarkers of the inflammatory phenotypes of asthma. Korean J. Intern. Med. 2020, 35, 857–860. [Google Scholar] [CrossRef]
- Fitzpatrick, A.M.; Moore, W.C. Severe asthma phenotypes—How should they guide evaluation and treatment? J. Allergy Clin. Immunol. Pract. 2017, 5, 901–908. [Google Scholar] [CrossRef]
- Wenzel, S.E. Asthma: Defining of the persistent adult phenotypes. Lancet 2006, 368, 804–813. [Google Scholar] [CrossRef]
- Agache, I.; Akdis, C.; Jutel, M.; Virchow, J.C. Untangling asthma phenotypes and endotypes. Allergy 2012, 67, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Amaral, R.; Fonseca, J.A.; Jacinto, T.; Pereira, A.M.; Malinovschi, A.; Janson, C.; Alving, K. Having concomitant asthma phenotypes is commona and independently relates to poor lung function in NHANES 2007-2012. Clin. Transl. Allergy 2018, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Fahy, J.V. Type 2 inflammation in asthma—Present in most, absent in many. Nat. Rev. Immunol. 2015, 15, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Scott, R.; Boyle, M.J.; Gibson, P.G. Inflammatory subtypes in asthma: Assessment and identification using induced sputum. Respirology 2006, 11, 54–61. [Google Scholar] [CrossRef]
- Tanaka, A.; Sato, H.; Akimoto, K.; Matsunaga, T.; Sagara, H. Spontaneous sputum discriminates inflammatory phenotypes in patients with asthma. Ann. Allergy Asthma Immunol. 2021, 126, 54–60. [Google Scholar] [CrossRef]
- Carpagnano, G.E.; Scioscia, G.; Lacedonia, D.; Soccio, P.; Lepore, G.; Saetta, M.; Barbaro, M.P.F.; Barnes, P.J. Looking for airways periostin in severe asthma: Could it be useful for clustering type 2 endotype? Chest 2018, 154, 1083–1090. [Google Scholar] [CrossRef]
- Gao, J.; Wu, F.; Wu, S.; Yang, X. Inflammatory subtypes in classic asthma and cough variant asthma. J. Inflamm. Res. 2020, 13, 1167–1173. [Google Scholar] [CrossRef]
- Miranda, C.; Busacker, A.; Balzar, S.; Trudeau, J.; Wenzel, S.E. Distinguishing severe asthma phenotypes: Role of age at onset and eosinophilic inflammation. J. Allergy Clin. Immunol. 2004, 113, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ye, L.; She, J.; Song, Y. Clinical differences between early- and late-onset asthma: A population-based cross-sectional study. Can. Respir. J. 2021, 2021, 8886520. [Google Scholar] [CrossRef]
- Nieves, A.; Magan, A.; Boniface, S.; Proudhon, H.; Lanteaume, A.; Romanet, S.; Vervloet, D.; Godard, P.; ARIA. Phenotypes of asthma revisited upon the presence of atopy. Respir. Med. 2005, 99, 347–354. [Google Scholar] [CrossRef] [PubMed]
- de Nijs, S.B.; Venekamp, L.N.; Bel, E.H. Adult-onset asthma: Is it really different? Eur. Respir. Rev. 2013, 22, 44–52. [Google Scholar] [CrossRef]
- Kim, S.H.; Uuganbayar, U.; Trinh, H.K.T.; Le Pham, D.; Kim, N.; Kim, M.; Sohn, H.; Park, H.S. Evaluation of neutrophil activation status according to the phenotypes of adult asthma. Allergy Asthma Immunol. Res. 2019, 11, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Chupp, G. Phenotypes and endotypes of adult asthma: Moving toward precision medicine. J. Allergy Clin. Immunol. 2019, 144, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Loza, M.J.; Adcock, I.; Auffray, C.; Chung, K.F.; Djukanovic, R.; Sterk, P.J.; Susulic, V.S.; Barnathan, E.S.; Baribaud, F.; Silkoff, P.E.; et al. Longitudinally stable, clinically defined clusters of patients with asthma independently in the ADEPT and U-BIOPRED Asthma studies. Ann. Am. Thorac. Soc. 2016, 13, S102–S103. [Google Scholar] [CrossRef]
- Loza, M.; Djukanovic, R.; Chung, K.F.; Horowitz, D.; Ma, K.; Branigan, P.; Barnathan, E.S.; Susulic, V.S.; Silkoff, P.E.; Sterk, P.J.; et al. Validated and longitudinally stable asthma phenotypes based on cluster analysis of the ADEPT study. Respir. Res. 2016, 17, 165. [Google Scholar] [CrossRef]
- Xie, M.; Wenzel, S.E. A global perspective in asthma: From phenotype to endotype. Chin. Med. J. 2013, 126, 166–173. [Google Scholar]
- Popovic-Grle, S.; Stajduhar, A.; Lampalo, M.; Rnjak, D. Biomarkers in different asthma phenotypes. Genes 2021, 12, 801. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Rocha, C.; Beltran, J.; Song, Y.; Posso, M.; Solà, I.; Alonso-Coello, P.; Akdis, C.; Akdis, M.; Canonica, G.W.; et al. Efficacy and safety of treatment with biologicals (benralizumab, dupilumab and omalizumab) for severe allergic asthma: A systematic review for the EAACI guidelines—Recommendations on the use of biologicals in severe asthma. Allergy 2020, 75, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Pité, H.; Aguiar, L.; Morello, J.; Monteiro, E.C.; Alves, A.C.; Bourbon, M.; Morais-Almeida, M. Metabolic dysfunction and asthma: Current perspectives. J. Asthma Allergy. 2020, 13, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Sarandi, E.; Thanasoula, M.; Anamaterou, C.; Papakonstantinou, E.; Geraci, F.; Papamichael, M.M.; Itsiopoulos, C.; Tsoukalas, D. Metabolic profiling of organic and fatty acids in chronic and autoimmune diseases. Adv. Clin. Chem. 2021, 101, 169–229. [Google Scholar] [CrossRef] [PubMed]
- Ravi, A.; Goorsenberg, A.W.M.; Dijkhuis, A.; Dierdorp, B.S.; Dekker, T.; van Weeghel, M.; Piñeros, Y.S.S.; Shah, P.L.; Ten Hacken, N.H.T.; Annema, J.T.; et al. Metabolic differences between bronchial epithelium from healthy individuals and patients with asthma and the effect of bronchial thermoplasty. J. Allergy Clin. Immunol. 2021. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Wang, G.; Wang, C.; Zhang, W.; Liu, J.; Wang, F. Serum metabolomics analysis of asthma in different inflammatory phenotypes: A cross-sectional study in Northeast China. BioMed Res. Int. 2018, 2018, 2860521. [Google Scholar] [CrossRef] [PubMed]
- Sparvero, L.J.; Tian, H.; Amoscato, A.A.; Sun, W.-Y.; Anthonymuthu, T.S.; Tyurina, Y.Y.; Kapralov, O.; Javadov, S.; He, R.-R.; Watkins, S.C.; et al. Direct mapping of phospholipid ferroptotic death signals in cells and tissues by Gas Cluster Ion Beam Secondary Ion Mass Spectrometry (GCIB-SIMS). Angew. Chem. Int. Ed. Engl. 2021, 60, 11784–11788. [Google Scholar] [CrossRef]
- Pité, H.; Morais-Almeida, M.; Rocha, S.M. Metabolomics in asthma: Where do we stand? Curr. Opin. Pulm. Med. 2018, 24, 94–103. [Google Scholar] [CrossRef]
- Farraia, M.; Cavaleiro Rufo, J.; Paciência, I.; Castro Mendes, F.; Delgado, L.; Laerte Boechat, J.; Moreira, A. Metabolic interactions in asthma. Eur. Ann. Allergy Clin. Immunol. 2019, 51, 196–205. [Google Scholar] [CrossRef]
- Plaza, V.; Crespo, A.; Giner, J.; Merino, J.L.; Ramos-Barbón, D.; Mateus, E.F.; Torrego, A.; Cosio, B.G.; Agustí, A.; Sibila, O. Inflammatory asthma phenotype discrimination using an electronic nose breath analyzer. J. Investig. Allergol. Clin. Immunol. 2015, 25, 431–437. [Google Scholar]
- Fens, N.; van der Sluijs, K.F.; van de Pol, M.A.; Dijkhuis, A.; Smids, B.S.; van der Zee, J.S.; Lutter, R.; Zwinderman, A.H.; Sterk, P.J. RESOLVE Research Team. Electronic nose identifies bronchoalveolar lavage fluid eosinophils in asthma. Am. J. Respir. Crit. Care Med. 2015, 191, 1086–1088. [Google Scholar] [CrossRef]
- Brinkman, P.; Wagener, A.H.; Hekking, P.-P.; Bansal, A.T.; Maitland-van der Zee, A.-H.; Wang, Y.; Weda, H.; Knobel, H.H.; Vink, T.J.; Rattray, N.J.; et al. Identification and prospective stability of electronic nose (eNose)-derived inflammatory phenotypes in patients with severe asthma. J. Allergy Clin. Immunol. 2019, 143, 1811–1820. [Google Scholar] [CrossRef]
- Azim, A.; Barber, C.; Dennison, P.; Riley, J.; Howarth, P. Exhaled volatile organic compounds in adult asthma: A systematic review. Eur. Respir. J. 2019, 54, 1900056. [Google Scholar] [CrossRef]
- Riise, G.C.; Torén, K.; Olin, A.C. Subjects in a population study with high levels of FeNO have associated eosinophil airway inflammation. ISRN Allergy 2011, 2011, 792613. [Google Scholar] [CrossRef]
- Berry, M.A.; Hargadon, B.; Shelley, M.; Parker, D.; Shaw, D.E.; Green, R.H.; Bradding, P.; Brightling, C.E.; Wardlaw, A.J.; Pavord, I.D. Evidence of a role of tumor necrosis factor alpha in refractory asthma. N. Engl. J. Med. 2006, 354, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Brightling, C.E.; Symon, F.A.; Birring, S.S.; Bradding, P.; Wardlaw, A.J.; Pavord, I.D. Comparison of airway immunopathology of eosinophilic bronchitis and asthma. Thorax 2003, 58, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Hanagama, M.; Ishida, M.; Ono, M.; Sato, H.; Yanai, M. Exhaled nitric oxide: A biomarker for chronic obstructive pulmonary disease. Respir. Investig. 2021, 59, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Denton, E.; Price, D.B.; Tran, T.N.; Canonica, G.W.; Menzies-Gow, A.; FitzGerald, J.M.; Sadatsafavi, M.; Perez de Llano, L.; Christoff, G.; Quinton, A.; et al. Cluster analysis of inflammatory biomarker expression in the International Severe Asthma Registry. J. Allergy. Clin. Immunol. Pract. 2021, 9, 2680–2688. [Google Scholar] [CrossRef] [PubMed]
- American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: Current understanding, recommendations, and answered questions. Am. J. Respir. Crit. Care Med. 2000, 162, 2341–2351. [CrossRef] [PubMed]
- Takamura, K.; Nasuhara, Y.; Kobayashi, M.; Betsuyaku, T.; Tanino, Y.; Kinoshita, I.; Yamaguchi, E.; Matsukura, S.; Schleimer, R.P.; Nishimura, M. Retinoic acid inhibits interleukin-4-induced eotaxin production in a human bronchial epithelial cell line. Am. J. Physiol. Lung Cell Mol. Physiol. 2004, 286, L777–L785. [Google Scholar] [CrossRef]
- Upham, J.W.; Sehmi, R.; Hayes, L.M.; Howie, K.; Lundahl, J.; Denburg, J.A. Retinoic acid modulates IL-5 receptor expression and selectively inhibitis eosinophil-basophil differentiation of hemopoietic progenitor cells. J. Allergy Clin. Immunol. 2002, 109, 307–313. [Google Scholar] [CrossRef]
- Wang, S.; Tang, K.; Lu, Y.; Tian, Z.; Huang, Z.; Wang, M.; Zhao, J.; Xie, J. Revealing the role of glycerophospholipid metabolism in asthma through plasma lipidomics. Clin. Chim. Acta. 2021, 513, 34–42. [Google Scholar] [CrossRef]
- Guo, C.; Sun, L.; Zhang, L.; Dong, F.; Zhang, X.; Yao, L.; Chang, C. Serum sphingolipid profile in asthma. J. Leukoc. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kowal, K.; Zebrowska, E.; Chabowski, A. Altered sphingolipid metabolism is associated with asthma phenotype in house dust mite-allergic patients. Allergy Asthma Immunol. Res. 2019, 11, 330–342. [Google Scholar] [CrossRef]
- Rago, D.; Pedersen, C.-E.; Huang, M.; Kelly, R.S.; Gürdeniz, G.; Brustad, N.; Knihtilä, H.; Lee-Sarwar, K.A.; Morin, A.; Rasmussen, M.A.; et al. Characteristics and mechanisms of a sphingolipd-associated childhood asthma endotype. Am. J. Respir. Crit. Care Med. 2021, 203, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Jung, H.-W.; Kim, M.; Moon, J.-Y.; Ban, G.-Y.; Kim, S.J.; Yoo, H.-J.; Park, H.-S. Ceramide/sphingosine-1-phosphate imbalance is associated with distinct inflammatory phenotypes of uncontrolled asthma. Allergy 2020, 75, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Schleich, F.N.; Zanella, D.; Stefanuto, P.-H.; Bessonov, K.; Smolinska, A.; Dallinga, J.W.; Henket, M.; Paulus, V.; Guissard, F.; Graff, S.; et al. Exhaled volatile organic compounds are able to discriminate between neutrophilic and eosinophilic asthma. Am. J. Respir. Crit. Care Med. 2019, 200, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, M.I.; Brinkman, P.; Vijverberg, S.J.H.; Neerincx, A.H.; de Vries, R.; Dagelet, Y.W.F.; Riley, J.H.; Hashimoto, S.; Montuschi, P.; Chung, K.F.; et al. eNose breath prints as a surrogate biomarker for classifying patients with asthma by atopy. J. Allergy Clin. Immunol. 2020, 146, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, J.; Zhang, H.P.; Zhang, X.; Wang, L.; Wood, L.; Wang, G. Obesity-associated metabolic signatures correlate to clinical and inflammatory profiles of asthma: A pilot study. Allergy Asthma Immunol. Res. 2018, 10, 628–647. [Google Scholar] [CrossRef]
- Miethe, S.; Guarino, M.; Alhamdan, F.; Simon, H.U.; Renz, H.; Dufour, J.-F.; Potaczek, D.P.; Garn, H. Effects of obesity on asthma: Immunometabolic links. Pol. Arch. Intern. Med. 2018, 128, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, V.A.; Carraro, S.; Pirillo, P.; Gucciardi, A.; Poloniato, G.; Stocchero, M.; Giordano, G.; Zanconato, S.; Baraldi, E. Breathomics in asthmatic children treated with inhaled corticosteroids. Metabolites 2020, 10, 390. [Google Scholar] [CrossRef] [PubMed]
- Nabe, T. Steroid-resistant asthma and neutrophils. Biol. Pharm. Bull. 2020, 43, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.H.; Fitzpatrick, A.M.; Medriano, C.A.; Jones, D.P. High-resolution metabolomics to identify urine biomarkers in corticosteroid-resistant asthmatic children. J. Allergy Clin. Immunol. 2017, 139, 1518–1524. [Google Scholar] [CrossRef][Green Version]
- Orie, N.G.M.; Sluiter, H.J.; De Vries, K.; Tammeling, G.J.; Witkop, J. The host factor in bronchitis. In Bronchitis; Orie, N.G.M., Sluiter, H.J., Eds.; Royal Vangorcum: Assen, The Netherlands, 1961; pp. 43–59. [Google Scholar]
- Bateman, E.D.; Reddel, H.K.; van Zyl-Smit, R.N.; Agusti, A. The asthma-COPD overlap syndrome: Towards a revised taxonomy of chronic airways diseases? Lancet Respir. Med. 2015, 3, 719–728. [Google Scholar] [CrossRef]
- Leung, J.M.; Sin, D.D. Asthma-COPD overlap syndrome: Pathogenesis, clinical features, and therapeutic targets. BMJ 2017, 358, j3772. [Google Scholar] [CrossRef]
- Loureiro, C.C. Blurred lines. Eosinophilic COPD: ACOS or COPD phenotype? Rev. Port. Pneumol. 2006, 22, 279–282. [Google Scholar] [CrossRef]
- Durham, A.L.; Adcock, I.M. The relationship between COPD and lung cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef]
- Greenwald, R.; Johnson, B.A.; Hoskins, A.; Dworski, R. Exhaled breath condensate formate after inhaled allergen provocation in atopic asthmatics in vivo. J. Asthma 2013, 50, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, R.; Fitzpatrick, A.M.; Gaston, B.; Marozkina, N.V.; Erzurum, S.; Teague, G. Breath formate is a marker of airwat S-nitrosothiol depletion in severe asthma. PLoS ONE 2010, 5, e11919. [Google Scholar] [CrossRef] [PubMed]
- Fens, N.; Roldaan, A.C.; van der Schee, M.P.; Boksem, R.J.; Zwinderman, A.H.; Bel, E.H.; Sterk, P.J. External validation of exhaled breath profiling using an electronic nose in the discrimination of asthma with fixed airways obstruction and chronic obstructive pulmonary disease. Clin. Exp. Allergy 2011, 41, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gai, X.Y.; Chang, C.; Zhang, X.; Wang, J.; Li, T.T. Metabolomic profiling differences among asthma, COPD and healthy subjects: A LC-MS-based metabolomic analysis. Biomed. Environ. Sci. 2019, 32, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, N.; Choudhury, P.; Kaushik, S.R.; Arya, R.; Nanda, R.; Bhattacharyya, P.; Roychowdhury, S.; Banerjee, R.; Chaudhury, K. Metabolomic fingerprinting and systemic inflammatory profiling of asthma COPD overlap (ACO). Respir. Res. 2020, 21, 126. [Google Scholar] [CrossRef]
- Kikuchi, S.; Kikuchi, I.; Hagiwara, K.; Kanazawa, M.; Nagata, M. Association of Tumor Necrosis Factor-a and neutrophilic inflammation in severe asthma. Allergol. Int. 2005, 54, 621–625. [Google Scholar] [CrossRef]
- Bar, N.; Korem, T.; Weissbrod, O.; Zeevi, D.; Rotschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weiberger, A.; Le Roy, C.I.; et al. A reference map of potential determinants for the human serum metabolome. Nature 2020, 588, 135–140. [Google Scholar] [CrossRef]
- Ross, A.B.; Barman, M.; Hartvigsson, O.; Lundell, A.-C.; Vaolainen, O.; Hesselmar, B.; Wold, A.E.; Sandberg, A.-S. Umbilical cord metabolome differs in relation to delivery mode, birth order and sex, maternal diet, and possibly future allergy development in rural children. PLoS ONE 2021, 16, e0242978. [Google Scholar] [CrossRef]
- Brandsma, J.; Goss, V.M.; Yang, X.; Bakke, P.S.; Caruso, M.; Chanez, P.; Dahlén, S.-E.; Fowler, S.J.; Horvath, I.; Krug, N.; et al. Lipid phenotyping of lung epithelial fluid in healthy human volunteers. Metabolomics 2018, 14, 123. [Google Scholar] [CrossRef]
- Gai, X.Y.; Zhang, L.J.; Liang, Y.; Guo, C.L.; Mairitifei, A.; Li, W.X.; Chang, C.; Chen, Y.H.; Yao, W.Z.; Zhang, X. Metabolomics analysis identifies serum glycerophospholipid expression: A comparison between men and women with asthma. Zhanghua Yi Xue Za Zhi 2018, 98, 3568–3575. [Google Scholar] [CrossRef]
- Zhang, P.; Zein, J. Novel insights on sex-related differences in asthma. Curr. Allergy Asthma Rep. 2019, 19, 44. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef]
- Smolinska, A.; Klaassen, E.M.; Dallinga, J.W.; van de Kant, K.D.; Jobsis, Q.; Moonen, E.J.; Van Schooten, F.J. Profiling of volatile organic compounds in exhaled breath as a strategy to find early predictive signatures of asthma in children. PLoS ONE 2014, 9, e95668. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, D.; Smolinska, A.; Jöbsis, Q.; Rosias, P.; Muris, J.; Dallinga, J.; Dompeling, E.; van Schooten, F.-J. Can exhaled volatile organic compounds predict asthma exacerbations in children? J. Breath. Res. 2017, 11, 016016. [Google Scholar] [CrossRef]
- Loureiro, C.C.; Duarte, I.F.; Gomes, J.; Carrola, J.; Barros, A.S.; Gil, A.M.; Bousquet, J.; Todo Bom, A.; Rocha, S.M. Urinary metabolomic changes as a predictive biomarker of asthma exacerbation. J. Allergy Clin. Immunol. 2014, 133, 261–263. [Google Scholar] [CrossRef]
- Olopade, C.O.; Zakkar, M.; Swedler, W.I.; Rubinstein, I. Exhaled pentane levels in acute asthma. Chest 1997, 111, 862–865. [Google Scholar] [CrossRef]
- Brinkman, P.; van de Pol, M.A.; Gerritsen, M.G.; Bos, L.D.; Dekker, T.; Smids, B.S.; Sinha, A.; Majoor, C.J.; Sneeboer, M.M.; Knobel, H.H.; et al. Exhaled breath profiles in the monitor and clinical recovery in asthma. Clin. Exp. Allergy. 2017, 47, 1159–1169. [Google Scholar] [CrossRef]
- Bos, L.D.; Sterk, P.J.; Schultz, M.J. Volatile metabolites of pathogens: A systematic review. PLoS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, N.; Alhamwe, B.A.; Caraballo, L.; Ding, M.; Ferrante, A.; Garn, H.; Garssen, J.; Hii, C.S.; Irvine, J.; Llinás-Caballero, K.; et al. Perinatal and early-life nutrition, epigenetics and allergy. Nutrients 2021, 13, 724. [Google Scholar] [CrossRef] [PubMed]
- Lee-Sarwar, K.; Lasky-Su, J.; Kelly, R.S.; Litonjua, A.A.; Weiss, S.T. Gut microbial-derived metabolomics of asthma. Metabolites 2020, 10, 97. [Google Scholar] [CrossRef]
- Nassan, F.L.; Kelly, R.S.; Kosheleva, A.; Koutrakis, P.; Vokonas, P.S.; Lasky-Su, J.A.; Schwartz, J.D. Metabolomic signatures if the long-term exposure to air pollution and temperature. Environ. Health. 2021, 20, 3. [Google Scholar] [CrossRef] [PubMed]
- Jeong, A.; Fiorito, G.; Keski-Rahkonen, P.; Imboden, M.; Kiss, A.; Robinot, N.; Gmuender, H.; Vlaanderen, J.; Vermeulen, R.; Kyrtopoulos, S.; et al. Perturbation of metabolic pathways mediates the association of air pollutants with asthma and cardiovascular diseases. Environ. Int. 2018, 119, 334–345. [Google Scholar] [CrossRef] [PubMed]
| Metabolomics Studies in Relation to Asthma Inflammatory Phenotypes | |||||
|---|---|---|---|---|---|
| Age/Sample Size/Ref. | Sample Biology/ Technique Used | Clinical Characteristics | Main Metabolites Identified | Main Metabolic Pathways Involved | Observations |
| 36.4 years N = 20 (BA) + 10 (HC) [54] | Peripheral blood; 3 detection platforms (UHPLC- MS/MS, optimised for basic species; UHPLC/MS/MS optimised for acidic species; GC/MS. | Patients with controlled severe asthma, patients with non-severe asthma and a healthy group (HC) | Taurine, Aspartic Acid, Glutamic Acid, Asparagine, Serine, Glutamine, Histidine, Glycine, Citrulline, Threonine, Alanine, Arginine, Tyrosine, Amino, Butyric Acid, Methionine, Valine, Tryptophan, Phenylalanine, Isoleucine, Leucine, Ornithine, Lysine 7-α-hydroxy-3-oxo-4- cholestenoate, Androsterone sulfate, Epiandrosterone sulfate, Glycerophosphorylcholine (GPC), Phosphoethanolamine, arachidonate, Oleamide Sphingosine, Glycodeoxycholate, Taurocholate, Lathosterol Adenosine 5-monophosphate | Amino acid, Carbohydrate, Lipid (Fatty acid, Sphingolipid), Bile acid, Cholesterol, Nucleotides | Biochemical differences were found between asthmatics and non-asthmatics, and also between severe and non-severe asthma; in addition, FeNo-high, possibly T2-type asthma phenotype patients had higher levels of branched amino acids and bile acids (glycholate and cholate) |
| 57.7 years N = 82 (BA) + 35 (HC) [59] | Exhaled breath condensate (EBC); Nuclear magnetic resonance (NMR) spectroscopy | Patients with asthma-EA, NA, and a healthy control group (HC) | NMR spectral regions | Not applicable | NMR spectral regions showed potential to discriminate asthmatics from healthy controls but poorly discriminated asthma phenotypes (only NA, but not EA, could be identified) |
| 38 years N = 13 (EA) + 16 (NEA) + 15 (HC) [91] | Peripheral blood and serum; UPLC-MS/MS | Mild and moderate asthma: 2 subgroups—EA and NEA, and a healthy control group (HC) | Glycerolphosphocholine, Monosaccharides, Phosphatidylserine (PS), Cholesterol glucuronide, Lactosylceramide, Phytosphingosine, Lysophosphatidylcholine (LPC), Retinyl ester, Retinols, Phosphatidylcholine (PC), Arachidonic acid (AA), Phosphatidylethanolamine (PE) | Glycerophospholipid, Retinol, Sphingolipid, Lipid ether, Galactose, AA, Inosite phosphate, Starch and Sucrose, Linoleic acid, Glycolysis, Gluconeogenesis | Lipid metabolism is affected in asthmatics; higher levels of monosaccharides, PC (18:1/2:0), PS (18:0/20:0) and arachidonic acid in NEA; higher levels of PC (16:0/18:1), PE (18:3/14:0), LPC (18:1) and lactosylceramide (d18:1/12:0) in EA |
| 48.5 years N = 52 [95] | Exhaled breath; Cyranose 320 eNose | Patients with persistent bronchial asthma (BA)-eosinophilic asthma (EA), various forms of non-eosinophilic asthma (NEA)—neutrophilic asthma (NA) and paucigranulocytic asthma (PGA) phenotypes | VOC breath-prints | Not applicable | Electronic nose can discriminate EA, NA and PGA inflammatory phenotypes in patients with persistent asthma in a regular clinical setting |
| 35.4 years N = 20 [96] | Bronchoalveolar lavage (BAL) Exhaled breath; eNoses | Patients with mild, allergic eosinophilic asthma (EA), who were non-smokers and not on corticosteroid therapy | eNose breath-print | Not applicable | eNose breath-prints were significantly associated with BALF eosinophil-rich inflammation |
| 55 years N = 78 [97] | Exhaled breath; eNose | Severe asthma patients-EA and NA subgroups (U-BIOPRED cohort) | Metabolomic fingerprints obtained from eNoses | Not applicable | eNose technology adequately discriminated between EA and NA (as classifed according to eosinophil and neutrophil numbers in peripheral blood, but not in induced sputum). |
| Lipid metabolism | |||||
| 46.1 years N = 35 (BA) + 23 (HC) [44] | Exhaled breath VOC; GC–MS | Patients with intermittent or persistent asthma: EA and NA, and a healthy control group (HC) | Alkanes, Aldehydes | Lipid (lipid peroxidation) | Respiratory VOCs can discriminate asthmatics from non-asthmatics and identify inflammation-related disease phenotypes |
| 45.6 Years N = 57 (40 non-obese; 17 obese) [55] | Urine; GC×GC-ToFMS | Patients with severe EA and aspirin hypersensitivity | Alkanes, Aldehydes | Lipid (lipid peroxidation) | Peroxydised lipid metabolites are increased in non-obese asthmatics and may be related to EA and disease severity. |
| 41 years N = 24 (BA) + 20 (HC) [107] | Peripheral blood; HPLC-QTOF | Patients with asthma: 2 subgroups-EA and NEA (airway hyperresponsiveness), and a healthy control group (HC) | Fatty acyls, Glycerolipids, Glycerophospholipids, Sphingolipids, Sterol lipids and Prenol lipids | Lipid | Lipid metabolism is affected in asthmatics; significantly higher levels of phosphatidic acids and phosphatidylglycerols-PG (19:0/22:0), PG (P-18:0/18:4), PG (19:1/20:0) and PG (18:0/20:0) in EA than in NEA |
| Age not indicated N = 51 (BA) + 9 (HC) [108] | Serum; LC–MS | Patients with asthma: EA and NEA, early-onset asthma and late-onset asthma, and a healthy control group (HC) | Sphingomyelin (SM) | Sphingolipids | SM levels were reduced in asthma; SM (SM 34:2; SM 38:1 and SM 40:1) levels were significantly more reduced in NEA than in EA |
| N = 421 (149 EA; 71 GA; 155 NA; 46 PGA) [111] | Peripheral blood; LC–MS/MS | Patients with asthma: EA and various types of NEA—mixed granulocytic (GA), NA and PGA phenotypes | Various ceramides, Sphingosine-1-phosphate (S1P), Sphingolipids, Sphingomyelin | Lipid | Asthmatics with NA had higher sphingosine and C16:0 ceramide levels compared with those without neutrophilia; in contrast, patients with EA had higher S1P levels compared with those without eosinophilia. |
| 54 years N = 245 [112] | Exhaled breath; UHGC/MS; GCxGC-HRTOFMS | Patients with EA, NA and PGC asthma phenotypes | Alkanes, Aldehydes | Lipid (Lipid peroxidation) | VOCs discriminate between EA and NA, with hexane and 2-hexanone better identifying EA, and a combination of nonanal, 1-propanol and hexane better identifying NA |
| Metabolomics studies in relation to atopic asthma phenotypes | |||||
| 55 years N = 96 [113] | Exhaled breath; eNoses | Patients with mild, moderate asthma (from two adult cohorts—U-BIOPRED, BreathCloud); atopy detected by positive skin prick tests and/or allergen-specific IgE | VOC breath-prints | Not applicable | e-Nose technology can accurately and robustly differentiate between asthma patients by atopic status |
| Metabolomics studies in relation to obesity-associated asthma phenotype/endotype | |||||
| 38 years N = 25 (OA) + 30 (LA) + 30 (ONA)/ [61] | Exhaled breath condensate (EBC); NMR | Obese asthmatic patients (OA), lean asthmatic (LA) and obese non-asthmatic controls (ONA) | Glucose, butyrate, acetoin levels, formate, tyrosine, ethanol, ethylene glycol, methanol, acetate, saturated fatty acids, propionate levels acetoin, isovalerate, 1,2-propanediol, methanol, acetone, propionate, lactate | Carbohydrate, Lipid, Amino acid | Patients with obesity and asthma have a specific respiratory metabotype (increased levels of glucose, n-valerate, lactate, and various fatty acids), which is different from that of patients with obesity or asthma alone |
| 49 years N = 11 (OA) + 22 (LA) [114] | Peripheral blood and serum Sputum supernatant; GC–TOF–MS | Obese asthmatic patients (OA), lean asthmatic (LA) | Valine, N-Methyl-DL-alanine, Uric acid, D-Glyceric acid, Asparagine 1, Beta-Glycerophosphoric acid, Benzoic acid, 3-Hydroxybutyric acid, Hydrocinnamic acid, Aspartic acid 2, Xanthine, 4-Aminobutyric acid 1, Glutaric acid, Indole-3-acetic acid, Gly-pro, D Glucoheptose, Gluconic lactone 2, L-Glutamic acid, Phytosphingosine, Shikimic acid, Beta-Glutamic acid 1, Pyrrole-2-Carboxylic, Pyrophosphate 3; 3-Aminopropionitrile 1, 3-Hydroxybutyric acid, 3-Hydroxynorvaline 2, Linolenic acid, Isoleucine | Lipid, Amino acid, Carbohydrate, Fatty acid | Metabolomics based on GC–TOF–MS discriminated between obese asthmatics and lean asthmatics |
| Metabolomics studies in asthma compared with COPD and ACO | |||||
| 48 years N = 31 (BA) + 44 (COPD) [52] | Exhaled breath condensate (EBC) Proton NMR spectra | Patients with newly diagnosed asthma or COPD | Methanol, ethanol, acetone, acetaldehyde | Lipid | Asthmatics had lower levels of ethanol and methanol and significantly higher levels of formate and acetone/acetoin than COPD patients |
| 54 years N = 60 (BA)-21 (FA)) + 39 (CA) + 40 (COPD) [126] | Exhaled air eNose | Patients with asthma (BA) with fixed airway obstruction (FA) or with classic, reversible asthma (CA); patients with COPD | Breath-prints | Not applicable | The molecular profile of exhaled breath shows high accuracy in distinguishing between FAO and COPD, as well as between CA and COPD |
| 60.5 years N = 17 (PA) + 17 (COPD) + 15 (HC) [127] | Peripheral blood and serum; LC–MS | Patients with mild, persistent asthma (PA), COPD patients and healthy controls (HC) | Hypoxanthine; P-chlorophenylalanine; L-Glutamine; Glycerophosphocholine; Inosine; Negative ion mode (ESI-); Hypoxanthine, Succinate; Xanthine; Arachidonic Acid (peroxide free); L-Pyroglutamic acid; Indoxyl sulfate; Theophylline; L-Valine; L-Norleucine; Bilirubin; L-Leucine; Inosine; Palmitic acid; L-Phenylalanine | Lipid, Nucleic acid, Amino acid | Asthma patients have a unique serum metabolome, which can distinguish them from individuals with COPD and healthy individuals; in particular, asthmatics had significantly higher levels of hypoxanthine than COPD patients and HC |
| 52.7 years (Cohort 1) 53.6 years (Cohort 2) N = 34 (BA)+ 30 (COPD)+ 35 (ACO)+ 33 (HC) (Cohort 1) N = 32 (BA) + 32 (COPD) + 40 (ACO) (Cohort 2) [128] | Peripheral blood; GC–MS | Patients with moderate and severe asthma (BA), patients with stage II and III COPD, patients with ACO and a healthy group (HC) | L-Serine, L-threonine, Ethanolamine Glucose, D-mannose, Cholesterol, 2-palmitoylglycerol, Stearic acid, Lactic acid, Linoleic acid, Succinic acid | Carbohydrate Lipid Amino acid | 2-palmatoylglycerol and cholesterol were decreased in BA when compared with ACO and COPD; in contrast, stearic acid expression was increased in BA in comparison with ACO and COPD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, A.; Pité, H.; Chaves-Loureiro, C.; Rocha, S.M.; Taborda-Barata, L. Metabolic Phenotypes in Asthmatic Adults: Relationship with Inflammatory and Clinical Phenotypes and Prognostic Implications. Metabolites 2021, 11, 534. https://doi.org/10.3390/metabo11080534
Santos A, Pité H, Chaves-Loureiro C, Rocha SM, Taborda-Barata L. Metabolic Phenotypes in Asthmatic Adults: Relationship with Inflammatory and Clinical Phenotypes and Prognostic Implications. Metabolites. 2021; 11(8):534. https://doi.org/10.3390/metabo11080534
Chicago/Turabian StyleSantos, Adalberto, Helena Pité, Cláudia Chaves-Loureiro, Sílvia M. Rocha, and Luís Taborda-Barata. 2021. "Metabolic Phenotypes in Asthmatic Adults: Relationship with Inflammatory and Clinical Phenotypes and Prognostic Implications" Metabolites 11, no. 8: 534. https://doi.org/10.3390/metabo11080534
APA StyleSantos, A., Pité, H., Chaves-Loureiro, C., Rocha, S. M., & Taborda-Barata, L. (2021). Metabolic Phenotypes in Asthmatic Adults: Relationship with Inflammatory and Clinical Phenotypes and Prognostic Implications. Metabolites, 11(8), 534. https://doi.org/10.3390/metabo11080534