Metabolic Evidence Rather Than Amounts of Red or Processed Meat as a Risk on Korean Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Characteristics of Subjects
2.2. Measurement of Exposure and Response Biomarkers
2.3. Epigenetic and Genetic Alterations
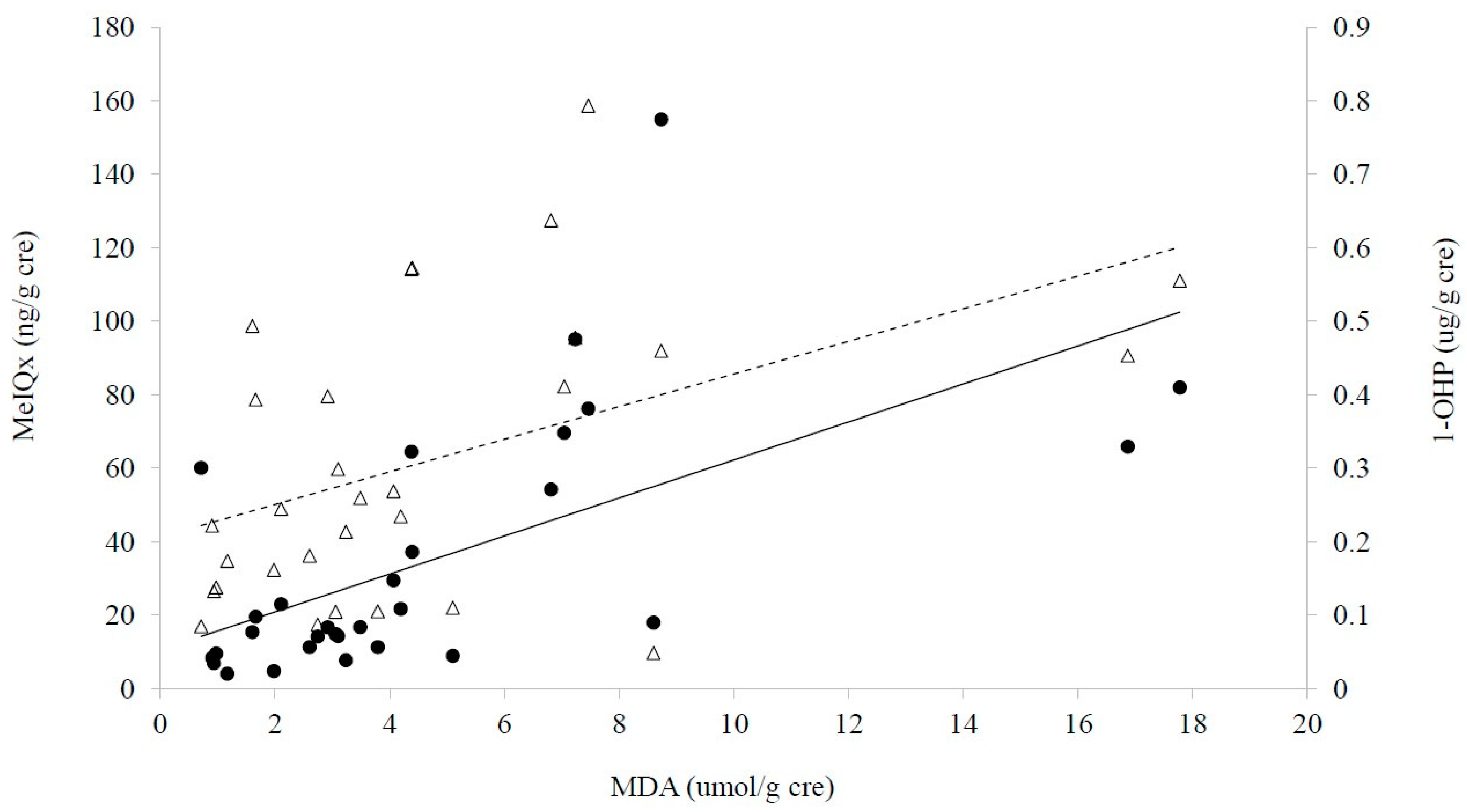
2.4. Alterations of Urinary Fatty Acid
2.5. Associations among CRC, Diet and Biomarkers
3. Discussion
3.1. Meat Consumption and CRC in Koreans
3.2. Exposure Levels
3.3. Multiple Evidences for Effects of Red or Processed Meat on CRC
4. Materials and Methods
4.1. Materials
4.2. Subjects
4.3. Collection of Data and Biospecimens
4.4. Analyses of Urinary 1-OHP
4.5. Analyses of Urinary HCAs and of HCA-DNA Adducts
4.6. Analyses of Urinary MDA
4.7. Analyses of Global DNA Methylation
4.8. Quantification of Gene Expression
4.9. Analyses of Urinary Fatty Acids
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- International Agency for Research on Cancer/ World Health Organization. In Red Meat and Processed Meat; IARC Working Group: Lyon, France, 2015.
- Mattiuzzi, C.; Lippi, G. Epidemiologic Burden of Red and Processed Meat Intake on Colorectal Cancer Mortality. Nutr. Cancer 2020, 73, 562–567. [Google Scholar] [CrossRef]
- Kweon, S. Updates on Cancer Epidemiology in Korea, 2018. Chonnam Med. J. 2018, 54, 90–100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, S.; Moon, H.; Kwak, J.; Kim, J.; Min, B.; Um, J.; Kim, S. Relationship between meat and cereal consumption and colorectal cancer in Korea and Japan. J. Gastroenterol. Hepatol. 2008, 23, 138–140. [Google Scholar] [CrossRef]
- Wong, M.C.; Ding, H.; Wang, J.; Chan, P.S.; Huang, J. Prevalence and risk factors of colorectal cancer in Asia. Intest. Res. 2019, 17, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89. [Google Scholar] [CrossRef] [PubMed]
- Kruger, C.; Zhou, Y. Red meat and colon cancer: A review of mechanistic evidence for heme in the context of risk assessment methodology. Food Chem. Toxicol. 2018, 118, 131–153. [Google Scholar] [CrossRef]
- Bouvard, V.; Loomis, D.; Guyton, K.Z.; Grosse, Y.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Mattock, H.; Straif, K.; Corpet, D. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015, 16, 1599–1600. [Google Scholar] [CrossRef]
- Turesky, R.J. Mechanistic evidence for red meat and processed meat intake and cancer risk: A follow-up on the international agency for research on cancer evaluation of 2015. CHIMIA Int. J. Chem. 2018, 72, 718–724. [Google Scholar] [CrossRef]
- Kameyama, H.; Nagahashi, M.; Shimada, Y.; Tajima, Y.; Ichikawa, H.; Nakano, M.; Sakata, J.; Kobayashi, T.; Narayanan, S.; Takabe, K. Genomic characterization of colitis-associated colorectal cancer. World J. Surg. Oncol. 2018, 16, 121. [Google Scholar] [CrossRef]
- Khare, S.; Verma, M. Epigenetics of colon cancer. Cancer Epigenetics 2012, 177–185. [Google Scholar]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.M.A.; Messias, M.C.; Duarte, G.H.; de Santis, G.K.; Mecatti, G.C.; Porcari, A.M.; Murgu, M.; Simionato, A.V.C.; Rocha, T.; Martinez, C.A. Plasma Lipid Profile Reveals Plasmalogens as Potential Biomarkers for Colon Cancer Screening. Metabolites 2020, 10, 262. [Google Scholar] [CrossRef] [PubMed]
- Christie, W.W.; Harwood, J.L. Oxidation of polyunsaturated fatty acids to produce lipid mediators. Essays Biochem. 2020, 64, 401–421. [Google Scholar] [PubMed]
- Hur, S.J.; Jo, C.; Yoon, Y.; Jeong, J.Y.; Lee, K.T. Controversy on the correlation of red and processed meat consumption with colorectal cancer risk: An Asian perspective. Crit. Rev. Food Sci. Nutr. 2019, 59, 3526–3537. [Google Scholar] [CrossRef]
- Yang, M.; Koga, M.; Katoh, T.; Kawamoto, T. A study for the proper application of urinary naphthols, new biomarkers for airborne polycyclic aromatic hydrocarbons. Arch. Environ. Contam. Toxicol. 1999, 36, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yun, B.; Turesky, R. Biomonitoring of DNA Damage in Humans. In DNA Damage, DNA Repair and Disease: Volume 1; RSC Publishing: London, UK, 2020; pp. 1–26. [Google Scholar]
- Hidalgo-Estévez, A.M.; Stamatakis, K.; Jiménez-Martínez, M.; López-Pérez, R.; Fresno, M. Cyclooxygenase 2-regulated genes an alternative avenue to the development of new therapeutic drugs for colorectal cancer. Front. Pharmacol. 2020, 11, 533. [Google Scholar] [CrossRef] [PubMed]
- Beyerle, J.; Frei, E.; Stiborova, M.; Habermann, N.; Ulrich, C.M. Biotransformation of xenobiotics in the human colon and rectum and its association with colorectal cancer. Drug Metab. Rev. 2015, 47, 199–221. [Google Scholar] [CrossRef]
- Caramujo-Balseiro, S.; Faro, C.; Carvalho, L. Metabolic pathways in sporadic colorectal carcinogenesis: A new proposal. Med. Hypotheses 2021, 148, 110512. [Google Scholar] [CrossRef]
- Chen, C.; Wang, L.; Liao, Q.; Xu, L.; Huang, Y.; Zhang, C.; Ye, H.; Xu, X.; Ye, M.; Duan, S. Association between six genetic polymorphisms and colorectal cancer: A meta-analysis. Genet. Test. Mol. Biomark. 2014, 18, 187–195. [Google Scholar] [CrossRef]
- Sunami, E.; De Maat, M.; Vu, A.; Turner, R.R.; Hoon, D.S. LINE-1 hypomethylation during primary colon cancer progression. PLoS One 2011, 6, e18884. [Google Scholar] [CrossRef]
- Fonteh, A.N.; Cipolla, M.; Chiang, J.; Arakaki, X.; Harrington, M.G. Human cerebrospinal fluid fatty acid levels differ between supernatant fluid and brain-derived nanoparticle fractions, and are altered in Alzheimer’s disease. PLoS ONE 2014, 9, e100519. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, J.Y.; Yang, M. Chemopreventive Effects of Korean Red Ginseng (Panax ginseng Meyer) on Exposure to Polycyclic Aromatic Hydrocarbons. J. Ginseng Res. 2011, 35, 339–343. [Google Scholar] [CrossRef]
- Shiao, S.P.K.; Lie, A.; Yu, C.H. Meta-analysis of homocysteine-related factors on the risk of colorectal cancer. Oncotarget 2018, 9, 25681–25697. [Google Scholar] [CrossRef]
- Holm, M.; Saraswat, M.; Joenväärä, S.; Ristimäki, A.; Haglund, C.; Renkonen, R. Colorectal cancer patients with different C-reactive protein levels and 5-year survival times can be differentiated with quantitative serum proteomics. PLoS ONE 2018, 13, e0195354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xia, Y.; Xia, B.; Nicodemus, K.J.; McGuffey, J.; McGahee, E.; Blount, B.; Wang, L. High-throughput and sensitive analysis of urinary heterocyclic aromatic amines using isotope-dilution liquid chromatography–tandem mass spectrometry and robotic sample preparation system. Anal. Bioanal. Chem. 2016, 408, 8149–8161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aglago, E.K.; Huybrechts, I.; Murphy, N.; Casagrande, C.; Nicolas, G.; Pischon, T.; Fedirko, V.; Severi, G.; Boutron-Ruault, M.; Fournier, A. Consumption of fish and long-chain n-3 polyunsaturated fatty acids is associated with reduced risk of colorectal cancer in a large European cohort. Clin. Gastroenterol. Hepatol. 2020, 18, 654–666. e6. [Google Scholar] [CrossRef] [PubMed]
- GeneCards. Available online: https://www.genecards.org (accessed on 17 June 2021).
- Hammerling, U.; Bergman Laurila, J.; Grafström, R.; Ilbäck, N. Consumption of red/processed meat and colorectal carcinoma: Possible mechanisms underlying the significant association. Crit. Rev. Food Sci. Nutr. 2016, 56, 614–634. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture, Food and Rural Affairs, Korea. Available online: https://lib.mafra.go.kr/skyblueimage/29722.pdf (accessed on 17 June 2021).
- National Institute of Food and Drug Safety Evaluation. Available online: http://nifds.go.kr/brd/m_21 (accessed on 17 June 2021).
- Leem, D.; Yoon, S.; Oh, K. Trends in dietary risk factors contributing to burden of chronic disease in Korean adults: Findings in Korea National Health and Nutrition Examination Survey, 2007-2015. Public Health Wkly. Rep. KCDC 2018, 11, 27–33. [Google Scholar]
- Kim, M.J.; Kim, S.; Choi, S.; Lee, I.; Moon, M.K.; Choi, K.; Park, Y.J.; Cho, Y.H.; Kwon, Y.M.; Yoo, J. Association of exposure to polycyclic aromatic hydrocarbons and heavy metals with thyroid hormones in general adult population and potential mechanisms. Sci. Total Environ. 2021, 762, 144227. [Google Scholar] [CrossRef]
- Lee, I.; Tran, M.; Evans-Nguyen, T.; Stickle, D.; Kim, S.; Han, J.; Park, J.Y.; Yang, M. Detoxification of chlorella supplement on heterocyclic amines in Korean young adults. Environ. Toxicol. Pharmacol. 2015, 39, 441–446. [Google Scholar] [CrossRef]
- Konorev, D.; Koopmeiners, J.S.; Tang, Y. Measurement of the heterocyclic amines 2-amino-9 H-pyrido [2,3-b] indole and 2-amino-1-methyl-6-phenylimidazo [4, 5-b] pyridine in urine: Effects of cigarette smoking. Chem. Res. Toxicol. 2015, 28, 2390–2399. [Google Scholar] [CrossRef]
- Pouzou, J.G.; Costard, S.; Zagmutt, F.J. Probabilistic assessment of dietary exposure to heterocyclic amines and polycyclic aromatic hydrocarbons from consumption of meats and breads in the United States. Food Chem. Toxicol. 2018, 114, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Kidd, L.C.; Stillwell, W.G.; Yu, M.C.; Wishnok, J.S.; Skipper, P.L. Urinary Excretion of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in White, African-American, and Asian-American Men in Los Angeles County. Cancer Epidemiol. Prev. Biomark. 1999, 8, 439–445. [Google Scholar]
- Cuparencu, C.; Praticó, G.; Hemeryck, L.Y.; Harsha, P.S.S.; Noerman, S.; Rombouts, C.; Xi, M.; Vanhaecke, L.; Hanhineva, K.; Brennan, L. Biomarkers of meat and seafood intake: An extensive literature review. Genes Nutr. 2019, 14, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Ruan, M.; Liu, J.; Wilde, P.; Naumova, E.N.; Mozaffarian, D.; Zhang, F.F. Trends in Processed Meat, Unprocessed Red Meat, Poultry, and Fish Consumption in the United States, 1999–2016. J. Acad. Nutr. Diet. 2019, 119, 1085–1098.e12. [Google Scholar] [CrossRef] [PubMed]
- May-Wilson, S.; Sud, A.; Law, P.J.; Palin, K.; Tuupanen, S.; Gylfe, A.; Hänninen, U.A.; Cajuso, T.; Tanskanen, T.; Kondelin, J. Pro-inflammatory fatty acid profile and colorectal cancer risk: A Mendelian randomisation analysis. Eur. J. Cancer 2017, 84, 228–238. [Google Scholar] [CrossRef]
- Parisi, L.R.; Li, N.; Atilla-Gokcumen, G.E. Very long chain fatty acids are functionally involved in necroptosis. Cell Chem. Biol. 2017, 24, 1445–1454.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, B.; Zhang, X.; Chen, X.; Zhu, J.; Zou, Y.; Li, J. Alpha-linolenic acid protects against lipopolysaccharide-induced acute lung injury through anti-inflammatory and anti-oxidative pathways. Microb. Pathog. 2020, 142, 104077. [Google Scholar] [CrossRef]
- Bellamri, M.; Xiao, S.; Murugan, P.; Weight, C.J.; Turesky, R.J. Metabolic activation of the cooked meat carcinogen 2-amino-1-methyl-6-phenylimidazo [4, 5-b] pyridine in human prostate. Toxicol. Sci. 2018, 163, 543–556. [Google Scholar] [CrossRef]
- Park, M.; Noh, H.; Song, N.; Paik, H.; Park, S.; Joung, H.; Song, W.; Kim, J. Validity and reliability of a dish-based, semi-quantitative food frequency questionnaire for Korean diet and cancer research. Asian Pac. J. Cancer Prev. 2012, 13, 545–552. [Google Scholar] [CrossRef]
- Quehenberger, O.; Armando, A.M.; Dennis, E.A. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography–mass spectrometry. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2011, 1811, 648–656. [Google Scholar]

| Variables | Control (n = 15) | CRC (n = 15) | p-Value | ||
|---|---|---|---|---|---|
| Mean | STD c | Mean | STD | ||
| Age (years) | 56.40 | 13.12 | 63.07 | 10.74 | 0.14 |
| Height (cm) | 164.21 | 10.60 | 160.25 | 8.22 | 0.26 |
| Sex (n of men) | 8 | 7 | 0.72 | ||
| Tobacco smoking (ppd) a | 8.00 | 16.67 | 7.67 | 16.13 | 0.56 |
| Body mass index(kg/m2) | 25.53 | 2.49 | 22.73 | 3.49 | 0.02 * |
| Energy intake (kcal/day) | 1730.16 | 826.63 | 1500.30 | 638.85 | 0.43 |
| Meat and lipid intake b (g/day) | |||||
| Red meat | 57.08 | 39.53 | 50.92 | 33.22 | 0.71 |
| Processed meat | 4.79 | 6.13 | 0.31 | 0.87 | <0.01 ** |
| Vegetable lipid | 17.02 | 11.64 | 15.63 | 10.07 | 0.73 |
| Animal lipid | 22.08 | 13.46 | 15.88 | 8.29 | 0.14 |
| Total lipid | 39.10 | 22.63 | 31.51 | 13.38 | 0.73 |
| Frequency of fish intake (n) | 0.47 d | ||||
| Rare | 6 | 9 | |||
| a piece/2 days | 9 | 6 | |||
| a piece/day | 0 | 0 | |||
| Biomarkers | Controls (n = 15) | CRC (n = 15) | p-Value | Adjusted p-Value a | Normal Range | ||
|---|---|---|---|---|---|---|---|
| Mean | STD | Mean | STD | ||||
| Hematological biomarkers | |||||||
| AST (U) | 25.53 | 9.11 | 24.2 | 8.28 | 0.79 | 0.48 | 10–40 |
| ALT (U) | 27.67 | 12.92 | 21.73 | 9.41 | 0.14 | 0.19 | 7–56 |
| CRP (mg/dL) | 0.92 | 0.81 | 0.32 | 0.26 | 0.03 * | <0.01 ** | 0.5–1.0 |
| TC (mg/dL) | 170 | 44.93 | 182.33 | 30.27 | 0.39 | 0.45 | <200 |
| TG (mg/dL) | 160.6 | 160.54 | 121.33 | 47.76 | 0.92 | 0.62 | <150 |
| LDL-C (mg/dL) | 90.11 | 35.19 | 114.67 | 24.54 | 0.04 * | 0.06† | <130 |
| HDL-C (mg/dL) | 43.4 | 14.28 | 43.27 | 11.27 | 0.98 | 0.82 | ≥40 |
| Homocysteine (μM) | 6.87 | 2.13 | 8.8 | 3.93 | 0.09 | 0.10 | <15 |
| Exposure or response biomarkers | |||||||
| MDA (μM) | 2.23 | 2.13 | 2.18 | 1.21 | 0.28 | 0.65 | |
| 1-OHP (μg/L) | 0.12 | 0.04 | 0.16 | 0.06 | 0.06 | 0.09 † | |
| MeIQx (ng/L) | 17.07 | 23.19 | 12.21 | 5.91 | 0.44 | 0.80 | |
| PhIP (ng/L) | 7.84 | 2.65 | 11.96 | 2.65 | 0.28 | 0.44 | |
| dG-C8 MeIQx/1.766ug of DNA | 5.17 | 0.07 | 5.29 | 0.11 | <0.01 ** | <0.01 ** | |
| Global DNA methylation (%) | 4.96 | 1.39 | 5.64 | 1.00 | 0.17 | 0.15 | |
| Contents | Control (n = 15) | CRC (n = 15) | p-Value a | |||
|---|---|---|---|---|---|---|
| Mean (ng/mL) | STD | Mean (ng/mL) | STD | |||
| C14:0 | Myristic acid | 73.12 | 50.25 | 147.04 | 129.71 | 0.04 * |
| C14:1 | Myristoleic acid | 4.67 | 4.4 | 7.08 | 4.61 | 0.11 |
| C15:0 | Pentadecanoic acid | 14.41 | 6.3 | 30.21 | 25.04 | 0.02 * |
| C15:1 | Pentadecenoic acid | 0.58 | 0.73 | 0.7 | 0.45 | 0.10 |
| C16:0 | Palmitic acid | 452.35 | 359.51 | 843.96 | 659.54 | 0.03 * |
| C16:1 | Palmitoleic acid | 11.03 | 15.36 | 28.7 | 26.43 | 0.01 * |
| C16:1T | Palmitelaidic acid | 2.73 | 3.81 | 7.12 | 6.56 | 0.01 * |
| C17:0 | Heptadecanoic acid | 12.66 | 12.92 | 51.69 | 76.51 | 0.03 * |
| C18:0 | Stearic acid | 384.38 | 233.25 | 819.2 | 671.79 | 0.03 * |
| C18:1 Mix | Vaccenic acid, Oleic acid, Elaidic acid | 147.57 | 203.45 | 411.62 | 306.6 | <0.01 ** |
| C18:2 Mix | Linoleic acid, Linolelaidic acid | 23.41 | 26.84 | 54.38 | 30.27 | <0.01 ** |
| C18:3 | α-Linolenic acid | 0.84 | 1.01 | 0.95 | 0.56 | 0.04 * |
| C18:3 | γ- Linolenic acid | 26.19 | 27.54 | 58.21 | 25.79 | <0.001 *** |
| C19:0 | Nonadecylic acid | 1.05 | 0.6 | 5.34 | 7.28 | <0.01 ** |
| C19:1 | 7- Nonadecylic acid | 0.88 | 0.2 | 1.79 | 0.2 | <0.01 ** |
| C20:0 | Arachidic acid | 5.03 | 2.15 | 20.34 | 24.01 | <0.01 ** |
| C20:1 Mix | 11- Eicosenoic acid | 3.3 | 3.92 | 6.47 | 7.25 | 0.06 |
| C20:2 | 11-14- Eicosadienoic acid | 1.1 | 0.87 | 1.36 | 1.33 | 0.60 |
| C20:3 | 11-14-17 Eicosatrienoic acid | 1.07 | 1.47 | 2.44 | 4.33 | 0.09 |
| C20:3 | Homogamma linolenic acid | 3.33 | 3.88 | 8.2 | 6.26 | <0.01 ** |
| C20:4 | Arachidonic acid | 11.37 | 2.23 | 14.95 | 2.23 | 0.27 |
| C20:5 | Eicosapentaenoic acid | 4.89 | 5.33 | 11.9 | 9.23 | <0.01 ** |
| C22:0 | Behenic acid | 8.91 | 3.52 | 15.93 | 11.2 | 0.07 |
| C22:1 | Erucic acid | 3.28 | 1.17 | 3.02 | 2.11 | 0.08 |
| C22:2 | Docosadienoic acid | 0.47 | 0.45 | 0.51 | 0.36 | 0.47 |
| C22:3 | Docosatrienoic acid | 0.19 | 0.23 | 0.22 | 0.17 | 0.09 |
| C22:4 | Docosatetraenoic acid | 0.15 | 0.09 | 0.2 | 0.12 | 0.10 |
| C22:5 | ω-3 Docosapentaenoic acid | 0.22 | 0.2 | 0.3 | 0.18 | 0.13 |
| C22:6 | Docosahexaenoic acid | 2.43 | 1.42 | 3.11 | 1.54 | 0.14 |
| C24:0 | Lignoceric acid | 4.34 | 1.32 | 7.85 | 7.8 | 0.05 |
| C24:1 | Nervonic acid | 1.29 | 0.68 | 1.54 | 1 | 0.60 |
| Total | 1207.26 | 848.67 | 2566.32 | 1883.83 | <0.01 ** | |
| SAFA | 956.25 | 653.39 | 1941.54 | 1533.67 | 0.01 * | |
| MUFA | 175.34 | 230.12 | 468.04 | 346.87 | <0.01 ** | |
| PUFA | 75.66 | 72.43 | 156.74 | 68.06 | <0.01 ** | |
| Fatty Acids | by Variable | Correlation (r) | p-Value |
|---|---|---|---|
| C18:0 Stearic acid (μg/g cre) | 1-OHP (μg/g cre) | 0.563 | 0.001 ** |
| MeIQx (ng/g cre) | 0.391 | 0.033 * | |
| PhIP (ng/g cre) | 0.429 | 0.018 * | |
| C18:3 α-Linolenic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.373 | 0.042 * |
| MeIQx (ng/g cre) | 0.372 | 0.043 * | |
| PhIP (ng/g cre) | 0.386 | 0.035 * | |
| C20:4 Arachidonic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.491 | 0.006 ** |
| MeIQx (ng/g cre) | 0.537 | 0.002 ** | |
| PhIP (ng/g cre) | 0.585 | <0.001 *** | |
| C22:0 Behenic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.575 | <0.001 *** |
| MeIQx (ng/g cre) | 0.512 | 0.004 ** | |
| PhIP (ng/g cre) | 0.506 | 0.004 ** | |
| C22:1 Erucic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.742 | <0.001 *** |
| MeIQx (ng/g cre) | 0.646 | <0.001 *** | |
| PhIP (ng/g cre) | 0.446 | 0.013 * | |
| C22:4 Docosatetraenoic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.537 | 0.002 ** |
| MeIQx (ng/g cre) | 0.572 | <0.001 *** | |
| PhIP (ng/g cre) | 0.570 | <0.001 *** | |
| C22:5 ω-3 Docosapentaenoic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.492 | 0.006 ** |
| MeIQx (ng/g cre) | 0.584 | <0.001 *** | |
| PhIP (ng/g cre) | 0.629 | <0.001 *** | |
| C22:6 Docosahexaenoic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.579 | <0.001 *** |
| MeIQx (ng/g cre) | 0.741 | <0.001 *** | |
| PhIP (ng/g cre) | 0.750 | <0.001 *** | |
| C24:0 Lignoceric acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.431 | 0.017 * |
| MeIQx (ng/g cre) | 0.462 | 0.010 * | |
| PhIP (ng/g cre) | 0.436 | 0.016 * | |
| C24:1 Nervonic acid (μg/g cre) | 1-OHP (μg/g Cre) | 0.566 | 0.001 ** |
| MeIQx (ng/g cre) | 0.607 | <0.001 ** | |
| PhIP (ng/g cre) | 0.595 | <0.001 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.; Lee, J.S.; Kim, E.; Lee, M.-A.; Fonteh, A.N.; Kwong, M.; Cho, Y.H.; Lee, U.J.; Yang, M. Metabolic Evidence Rather Than Amounts of Red or Processed Meat as a Risk on Korean Colorectal Cancer. Metabolites 2021, 11, 462. https://doi.org/10.3390/metabo11070462
Kim E, Lee JS, Kim E, Lee M-A, Fonteh AN, Kwong M, Cho YH, Lee UJ, Yang M. Metabolic Evidence Rather Than Amounts of Red or Processed Meat as a Risk on Korean Colorectal Cancer. Metabolites. 2021; 11(7):462. https://doi.org/10.3390/metabo11070462
Chicago/Turabian StyleKim, Eunbee, Joon Seok Lee, Eunjae Kim, Myung-Ah Lee, Alfred N. Fonteh, Michael Kwong, Yoon Hee Cho, Un Jae Lee, and Mihi Yang. 2021. "Metabolic Evidence Rather Than Amounts of Red or Processed Meat as a Risk on Korean Colorectal Cancer" Metabolites 11, no. 7: 462. https://doi.org/10.3390/metabo11070462
APA StyleKim, E., Lee, J. S., Kim, E., Lee, M.-A., Fonteh, A. N., Kwong, M., Cho, Y. H., Lee, U. J., & Yang, M. (2021). Metabolic Evidence Rather Than Amounts of Red or Processed Meat as a Risk on Korean Colorectal Cancer. Metabolites, 11(7), 462. https://doi.org/10.3390/metabo11070462







