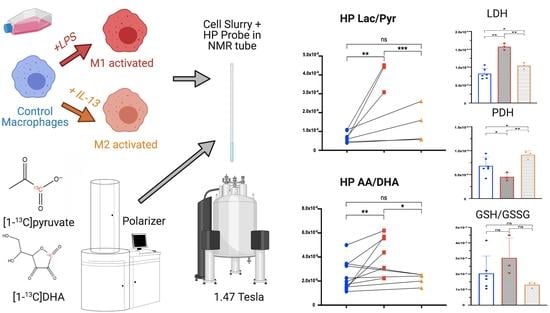
Non-Invasive Differentiation of M1 and M2 Activation in Macrophages Using Hyperpolarized 13C MRS of Pyruvate and DHA at 1.47 Tesla
Abstract
:1. Introduction
2. Results
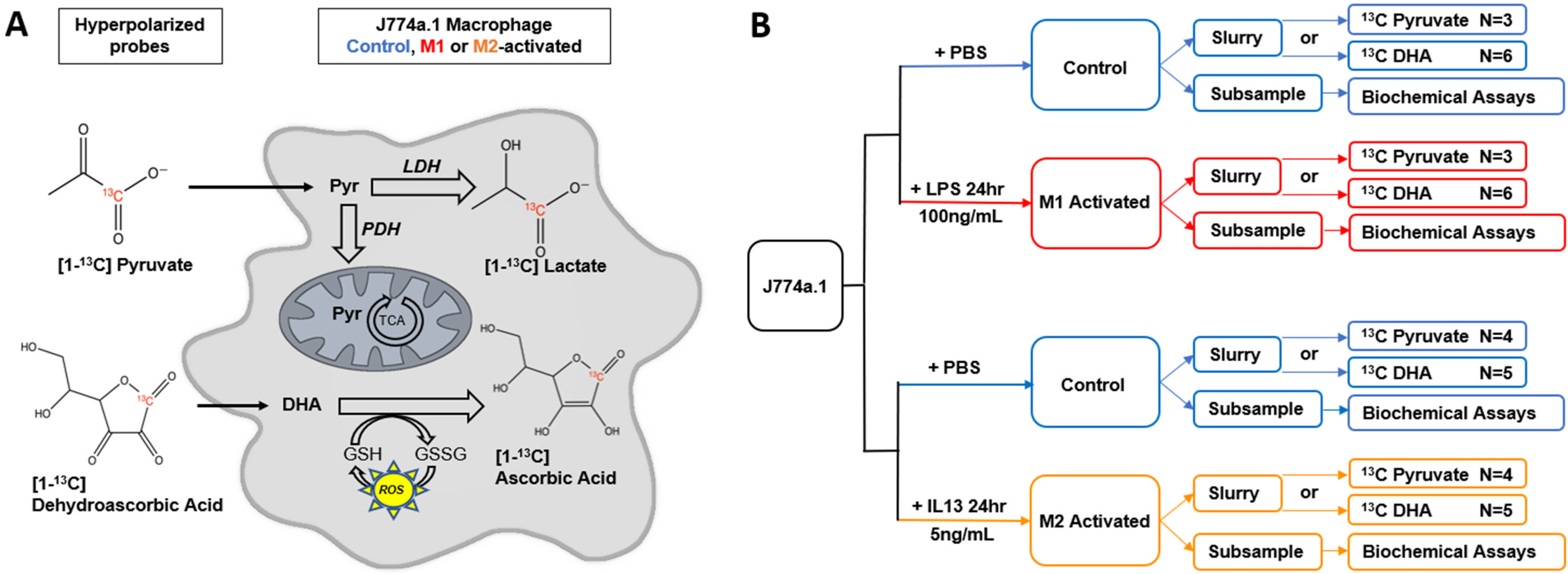
2.1. M1 and M2 Activation of J774a.1 Macrophages
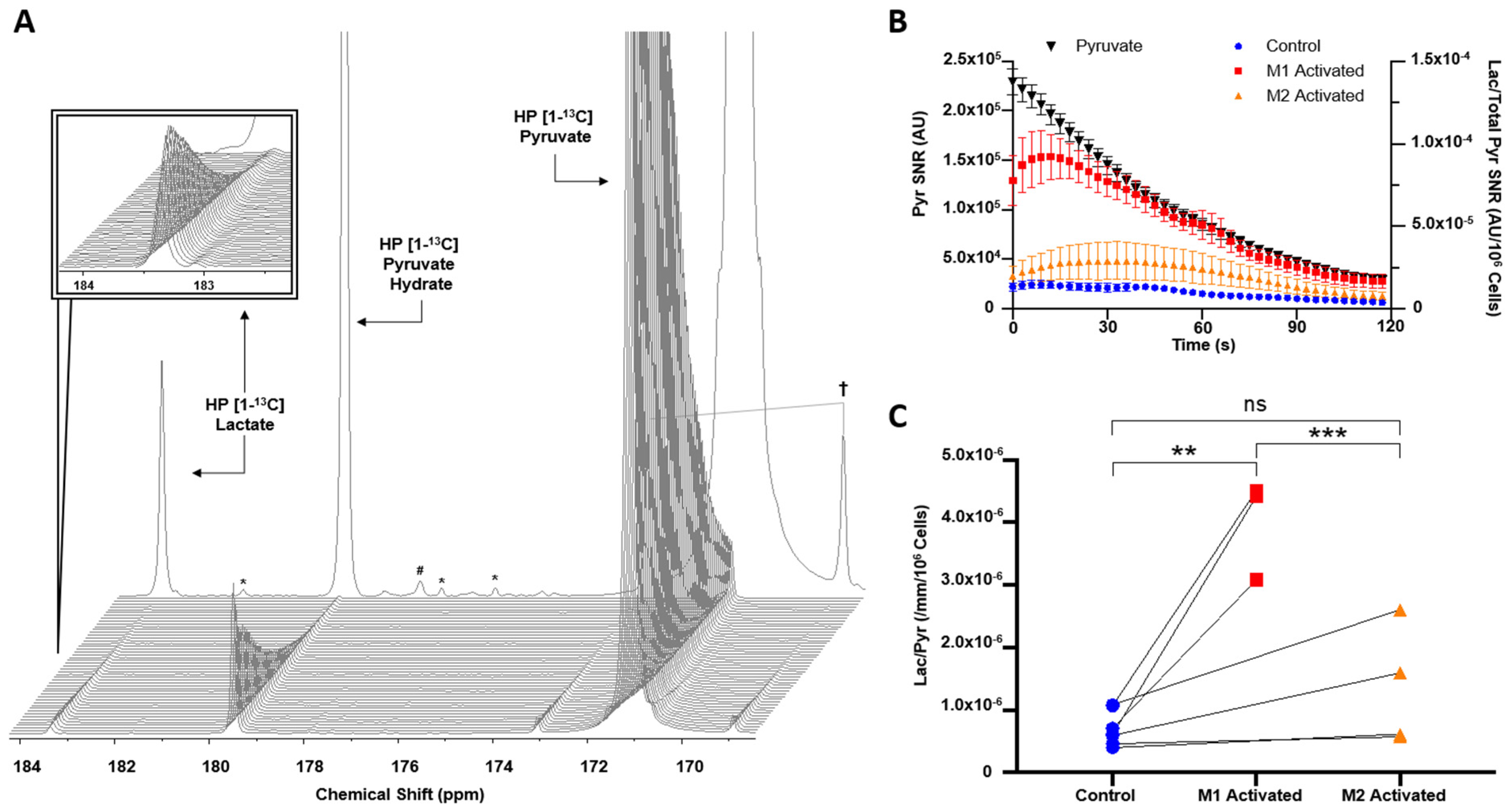
2.2. HP [1-13C]lactate Production Is Differentially Increased by M1/M2 Macrophage Activation
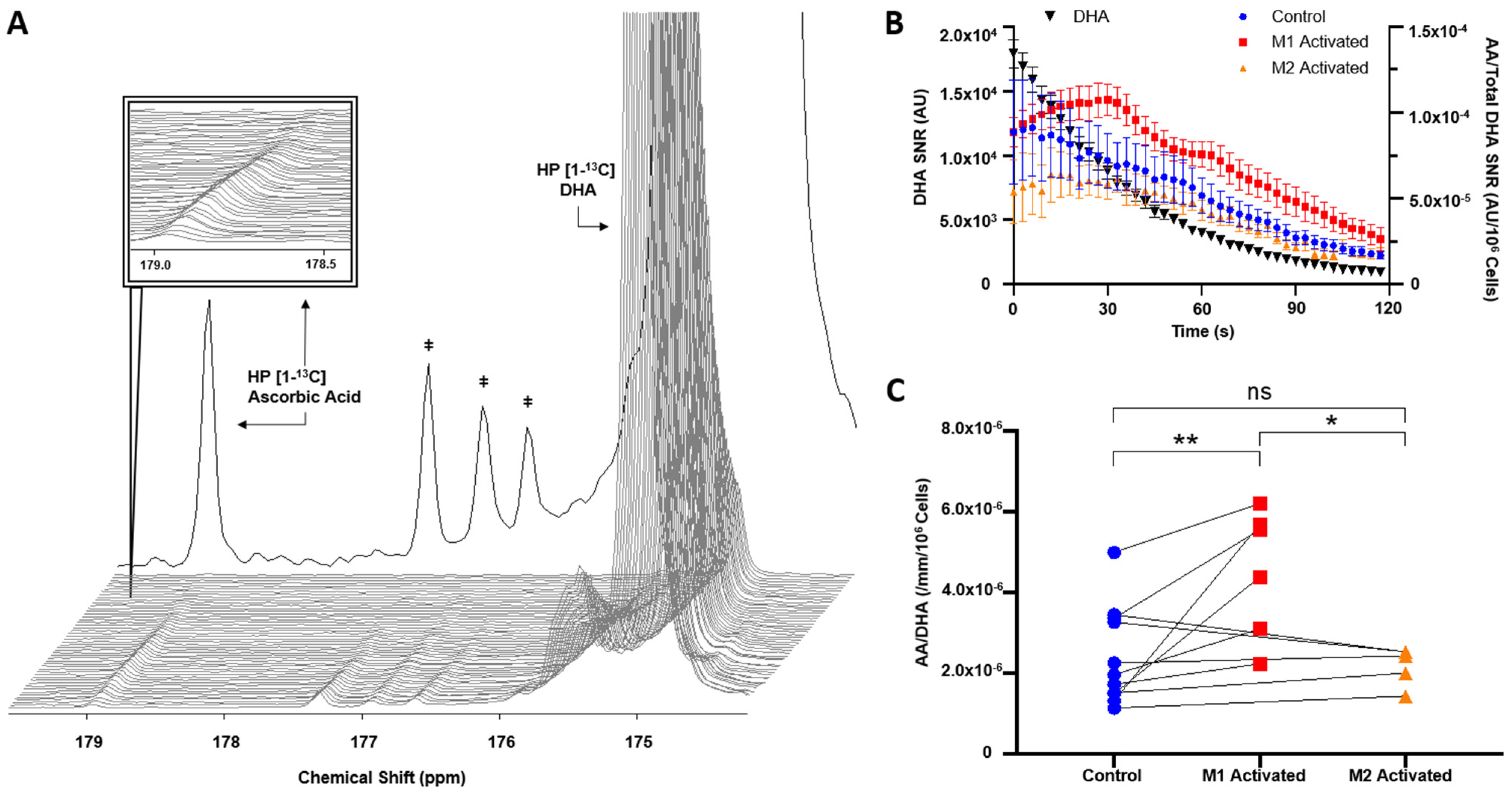
2.3. HP [1-13C]DHA Conversion to HP [1-13C]AA Is Increased with M1 Activation
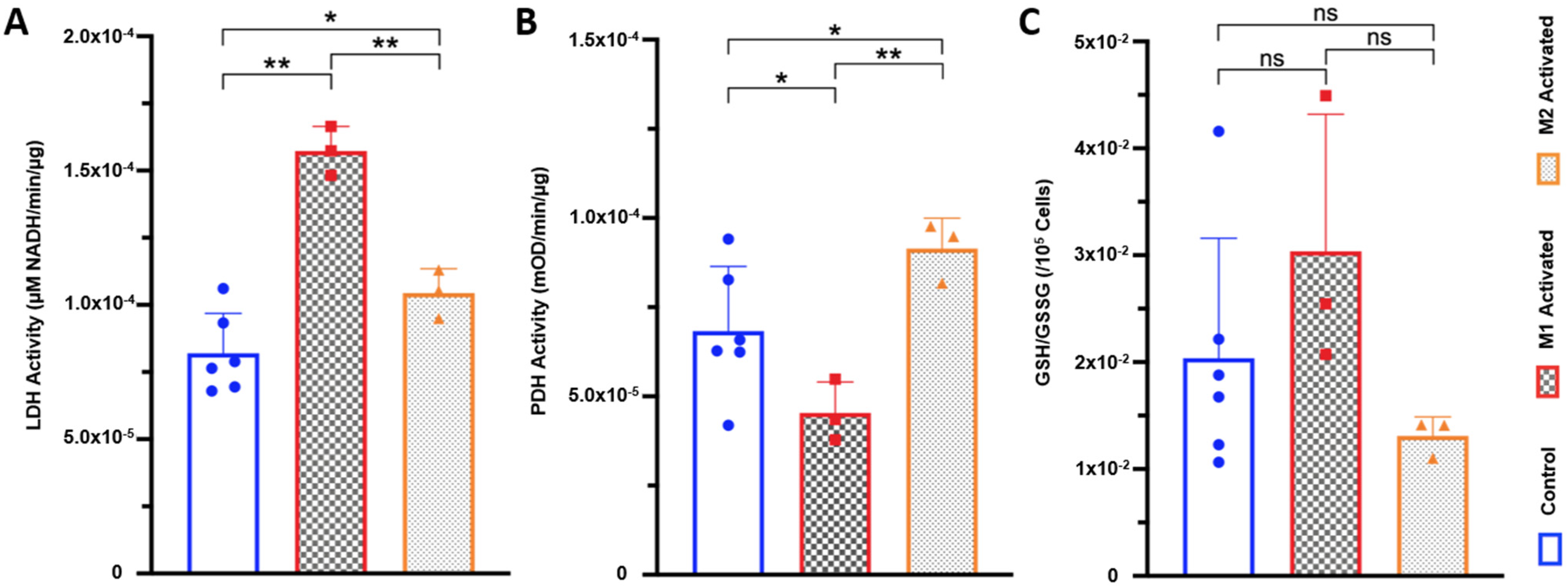
2.4. LDH Activity Is Increased in Both M1- and M2-Activated Macrophages, While PDH Activity Is Decreased in M1 and Increased in M2 Activation
2.5. High Resolution 1H NMR Detects Significant Differences in Extracted Metabolites between Control, M1, and M2 Macrophages
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. MR Acquisitions
4.3. Spectrophotometric Assays
4.4. High Resolution 1H NMR of Extracted Cell Metabolites
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galli, S.J.; Borregaard, N.; Wynn, T.A. Phenotypic and functional plasticity of cells of innate immunity: Macrophages, mast cells and neutrophils. Nat. Immunol. 2011, 12, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- Okabe, Y.; Medzhitov, R. Tissue biology perspective on macrophages. Nat. Immunol. 2016, 17, 9–17. [Google Scholar] [CrossRef]
- Gasteiger, G.; D’Osualdo, A.; Schubert, D.A.; Weber, A.; Bruscia, E.M.; Hartl, D. Cellular Innate Immunity: An Old Game with New Players. J. Innate Immun. 2017, 9, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, A.; Usui, T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediat. Inflamm. 2015, 2015, 568543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraakman, M.J.; Murphy, A.J.; Jandeleit-Dahm, K.; Kammoun, H.L. Macrophage polarization in obesity and type 2 diabetes: Weighing down our understanding of macrophage function? Front. Immunol. 2014, 5, 470. [Google Scholar] [CrossRef]
- Dandekar, R.; Kingaonkar, A.; Dhabekar, G. Role of macrophages in malignancy. Ann. Maxillofac. Surg. 2011, 1, 150–154. [Google Scholar] [CrossRef] [Green Version]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef] [Green Version]
- Stunault, M.I.; Bories, G.; Guinamard, R.R.; Ivanov, S. Metabolism Plays a Key Role during Macrophage Activation. Mediat. Inflamm. 2018, 2018, 2426138. [Google Scholar] [CrossRef]
- Tan, H.-Y.; Wang, N.; Li, S.; Hong, M.; Wang, X.; Feng, Y. The Reactive Oxygen Species in Macrophage Polarization: Reflecting Its Dual Role in Progression and Treatment of Human Diseases. Oxid. Med. Cell. Longev. 2016, 2016, 2795090. [Google Scholar] [CrossRef] [Green Version]
- Raza, H.; John, A.; Shafarin, J. NAC attenuates LPS-induced toxicity in aspirin-sensitized mouse macrophages via suppression of oxidative stress and mitochondrial dysfunction. PLoS ONE 2014, 9, e103379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendra, E.; Riabov, V.; Mossel, D.M.; Sevastyanova, T.; Harmsen, M.C.; Kzhyshkowska, J. Reactive oxygen species (ROS) in macrophage activation and function in diabetes. Immunobiology 2019, 224, 242–253. [Google Scholar] [CrossRef] [PubMed]
- De Santa, F.; Vitiello, L.; Torcinaro, A.; Ferraro, E. The Role of Metabolic Remodeling in Macrophage Polarization and Its Effect on Skeletal Muscle Regeneration. Antioxid. Redox Signal. 2019, 30, 1553–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Deng, Z.; Shi, F.; Zhou, Z.; Sun, F.; Sun, M.H.; Sun, Q.; Chen, L.; Li, D.; Jiang, C.Y.; Zhao, R.Z.; et al. M1 macrophage mediated increased reactive oxygen species (ROS) influence wound healing via the MAPK signaling in vitro and in vivo. Toxicol. Appl. Pharm. 2019, 366, 83–95. [Google Scholar] [CrossRef]
- Mills, E.L.; O’Neill, L.A. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur. J. Immunol. 2016, 46, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.K.; Delaney, C.; Shi, H.; Yung, R. Changes in adipose tissue macrophages and T cells during aging. Crit. Rev. Immunol. 2014, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.S.; Sundareshan, V.; Cory, T.J.; Hayes, D., Jr.; Anstead, M.I.; Feola, D.J. Azithromycin alters macrophage phenotype. J. Antimicrob. Chemother. 2008, 61, 554–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Hu, K. Tissue-type plasminogen activator modulates macrophage M2 to M1 phenotypic change through annexin A2-mediated NF-kappaB pathway. Oncotarget 2017, 8, 88094–88103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Ming, X.F. Functions of arginase isoforms in macrophage inflammatory responses: Impact on cardiovascular diseases and metabolic disorders. Front. Immunol. 2014, 5, 533. [Google Scholar] [CrossRef] [Green Version]
- Rath, M.; Muller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [Green Version]
- Weisser, S.B.; Kozicky, L.K.; Brugger, H.K.; Ngoh, E.N.; Cheung, B.; Jen, R.; Menzies, S.C.; Samarakoon, A.; Murray, P.J.; Lim, C.J.; et al. Arginase activity in alternatively activated macrophages protects PI3Kp110delta deficient mice from dextran sodium sulfate induced intestinal inflammation. Eur. J. Immunol. 2014, 44, 3353–3367. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Ardenkjaer-Larsen, J.H.; Fridlund, B.; Gram, A.; Hansson, G.; Hansson, L.; Lerche, M.H.; Servin, R.; Thaning, M.; Golman, K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. USA 2003, 100, 10158–10163. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, M.M.; Larson, P.E.Z.; Yoshihara, H.A.I.; Danforth, O.M.; Vigneron, D.B.; Nelson, S.J.; Pieper, R.O.; Phillips, J.J.; Ronen, S.M. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat. Commun. 2013, 4, 2429. [Google Scholar] [CrossRef] [Green Version]
- Le Page, L.M.; Guglielmetti, C.; Najac, C.F.; Tiret, B.; Chaumeil, M.M. Hyperpolarized 13 C magnetic resonance spectroscopy detects toxin-induced neuroinflammation in mice. NMR Biomed. 2019, 32, e4164. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, H.A.; Bastiaansen, J.A.; Berthonneche, C.; Comment, A.; Schwitter, J. An intact small animal model of myocardial ischemia-reperfusion: Characterization of metabolic changes by hyperpolarized 13C MR spectroscopy. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H2058–H2066. [Google Scholar] [CrossRef] [Green Version]
- Kurhanewicz, J.; Vigneron, D.B.; Ardenkjaer-Larsen, J.H.; Bankson, J.A.; Brindle, K.; Cunningham, C.H.; Gallagher, F.A.; Keshari, K.R.; Kjaer, A.; Laustsen, C.; et al. Hyperpolarized (13)C MRI: Path to Clinical Translation in Oncology. Neoplasia 2019, 21, 1–16. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Imaging of Traumatic Brain Injury Metabolism Using Hyperpolarized Carbon-13 Pyruvate; U.S. National Library of Medicine: Bethesda, MD, USA, 2018.
- Nelson, S.J.; Kurhanewicz, J.; Vigneron, D.B.; Larson, P.E.Z.; Harzstark, A.L.; Ferrone, M.; van Criekinge, M.; Chang, J.W.; Bok, R.; Park, I.; et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-¹3C]pyruvate. Sci. Transl. Med. 2013, 5, 198ra108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Page, L.M.; Guglielmetti, C.; Taglang, C.; Chaumeil, M.M. Imaging Brain Metabolism Using Hyperpolarized 13C Magnetic Resonance Spectroscopy. Trends Neurosci. 2020, 43, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Grist, J.T.; Miller, J.J.; Zaccagna, F.; McLean, M.A.; Riemer, F.; Matys, T.; Tyler, D.J.; Laustsen, C.; Coles, A.J.; Gallagher, F.A. Hyperpolarized (13)C MRI: A novel approach for probing cerebral metabolism in health and neurological disease. J. Cereb. Blood Flow Metab. 2020, 40, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Najac, C.; Chaumeil, M.M.; Kohanbash, G.; Guglielmetti, C.; Gordon, J.W.; Okada, H.; Ronen, S.M. Detection of inflammatory cell function using 13C magnetic resonance spectroscopy of hyperpolarized [6-13C]-arginine. Sci. Rep. 2016, 6, 31397. [Google Scholar] [CrossRef] [Green Version]
- Sriram, R.; Nguyen, J.; Santos, J.D.; Nguyen, L.; Sun, J.; Vigneron, S.; Van Criekinge, M.; Kurhanewicz, J.; MacKenzie, J.D. Molecular detection of inflammation in cell models using hyperpolarized (13)C-pyruvate. Theranostics 2018, 8, 3400–3407. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.J.M.; Miller, J.J.; Lau, A.Z.; Curtis, M.K.; Rider, O.J.; Choudhury, R.P.; Neubauer, S.; Cunningham, C.H.; Carr, C.A.; Tyler, D.J. Noninvasive Immunometabolic Cardiac Inflammation Imaging Using Hyperpolarized Magnetic Resonance. Circ. Res. 2018, 122, 1084–1093. [Google Scholar] [CrossRef] [Green Version]
- Kohler, S.J.; Yen, Y.; Wolber, J.; Chen, A.P.; Albers, M.J.; Bok, R.; Zhang, V.; Tropp, J.; Nelson, S.; Vigneron, D.B.; et al. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn. Reson. Med. 2007, 58, 65–69. [Google Scholar] [CrossRef]
- Keshari, K.R.; Kurhanewicz, J.; Bok, R.; Larson, P.E.Z.; Vigneron, D.B.; Wilson, D.M. Hyperpolarized 13C dehydroascorbate as an endogenous redox sensor for in vivo metabolic imaging. Proc. Natl. Acad. Sci. USA 2011, 108, 18606–18611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csóka, B.; Selmeczy, Z.; Koscsó, B.; Németh, Z.H.; Pacher, P.; Murray, P.J.; Kepka-Lenhart, D.; Morris, S.M., Jr.; Gause, W.C.; Leibovich, S.J.; et al. Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 2012, 26, 376–386. [Google Scholar] [CrossRef] [Green Version]
- Koo, S.-J.; Chowdhury, I.H.; Szczesny, B.; Wan, X.; Garg, N.J. Macrophages Promote Oxidative Metabolism To Drive Nitric Oxide Generation in Response to Trypanosoma cruzi. Infect. Immun. 2016, 84, 3527–3541. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, E.M.; Gonzalez-Cotto, M.; Baseler, W.A.; Davies, L.C.; Ghesquière, B.; Maio, N.; Rice, C.M.; Rouault, T.A.; Cassel, T.; Higashi, R.M.; et al. Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat. Commun. 2020, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Michelucci, A.; Cordes, T.; Ghelfi, J.; Pailot, A.; Reiling, N.; Goldmann, O.; Binz, T.; Wegner, A.; Tallam, A.; Rausell, A.; et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA 2013, 110, 7820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordes, T.; Wallace, M.; Michelucci, A.; Divakaruni, A.S.; Sapcariu, S.C.; Sousa, C.; Koseki, H.; Cabrales, P.; Murphy, A.N.; Hiller, K.; et al. Immunoresponsive Gene 1 and Itaconate Inhibit Succinate Dehydrogenase to Modulate Intracellular Succinate Levels. J. Biol. Chem. 2016, 291, 14274–14284. [Google Scholar] [CrossRef] [Green Version]
- Tavakoli, S.; Downs, K.; Short, J.D.; Nguyen, H.N.; Lai, Y.; Jerabek, P.A.; Goins, B.; Toczek, J.; Sadeghi, M.M.; Asmis, R. Characterization of Macrophage Polarization States Using Combined Measurement of 2-Deoxyglucose and Glutamine Accumulation: Implications for Imaging of Atherosclerosis. Arterioscl. Thromb. Vasc. Biol. 2017, 37, 1840–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, A.L.; Schiller, S.M.; Keegan, W.J.; Ammons, M.C.B.; Eilers, B.; Tripet, B.; Copié, V. Quantitative (1)H NMR Metabolomics Reveal Distinct Metabolic Adaptations in Human Macrophages Following Differential Activation. Metabolites 2019, 9, 248. [Google Scholar] [CrossRef] [Green Version]
- Andreu, N.; Phelan, J.; de Sessions, P.F.; Cliff, J.M.; Clark, T.G.; Hibberd, M.L. Primary macrophages and J774 cells respond differently to infection with Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 42225. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Balakrishnan, A.; Bok, R.A.; Anderton, B.; Larson, P.E.Z.; Nelson, S.J.; Kurhanewicz, J.; Vigneron, D.B.; Goga, A. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011, 14, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.C.-C.; Smith, A.M.; Everts, B.; Colonna, M.; Pearce, E.L.; Schilling, J.D.; Pearce, E.J. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity 2016, 45, 817–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, B.-K.; Park, S.; Kang, H.-J.; Kim, D.W.; Ham, H.J.; Ha, C.-M.; Choi, B.-J.; Lee, J.Y.; Oh, C.J.; Yoo, E.K.; et al. Pyruvate Dehydrogenase Kinase Is a Metabolic Checkpoint for Polarization of Macrophages to the M1 Phenotype. Front. Immunol. 2019, 10, 944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merritt, M.E.; Harrison, C.; Sherry, A.D.; Malloy, C.R.; Burgess, S.C. Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc. Natl. Acad. Sci. USA 2011, 108, 19084–19089. [Google Scholar] [CrossRef] [Green Version]
- Merritt, M.E.; Harrison, C.; Storey, C.; Jeffrey, F.M.; Sherry, A.D.; Malloy, C.R. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc. Natl. Acad. Sci. USA 2007, 104, 19773–19777. [Google Scholar] [CrossRef] [Green Version]
- Hong, C.S.; Graham, N.A.; Gu, W.; Espindola Camacho, C.; Mah, V.; Maresh, E.L.; Alavi, M.; Bagryanova, L.; Krotee, P.A.L.; Gardner, B.K.; et al. MCT1 Modulates Cancer Cell Pyruvate Export and Growth of Tumors that Co-express MCT1 and MCT4. Cell Rep. 2016, 14, 1590–1601. [Google Scholar] [CrossRef] [Green Version]
- Vincent, S.J.; Zwahlen, C.; Post, C.B.; Burgner, J.W.; Bodenhausen, G. The conformation of NAD+ bound to lactate dehydrogenase determined by nuclear magnetic resonance with suppression of spin diffusion. Proc. Natl. Acad. Sci. USA 1997, 94, 4383–4388. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, M.M.; Najac, C.; Ronen, S.M. Studies of Metabolism Using (13)C MRS of Hyperpolarized Probes. Methods Enzym. 2015, 561, 1–71. [Google Scholar] [CrossRef]
- Armstrong, J.S.; Steinauer, K.K.; Hornung, B.; Irish, J.M.; Lecane, P.; Birrell, G.W.; Peehl, D.M.; Knox, S.J. Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 2002, 9, 252–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linster, C.L.; Van Schaftingen, E. Vitamin C. Biosynthesis, recycling and degradation in mammals. FEBS J. 2007, 274, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Carroll, V.N.; Sriram, R.; Villanueva-Meyer, J.E.; von Morze, C.; Wang, Z.J.; Mutch, C.A.; Keshari, K.R.; Flavell, R.R.; Kurhanewicz, J.; et al. Imaging glutathione depletion in the rat brain using ascorbate-derived hyperpolarized MR and PET probes. Sci. Rep. 2018, 8, 7928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadl, A.; Meher, A.K.; Sharma, P.R.; Lee, M.Y.; Doran, A.C.; Johnstone, S.R.; Elliott, M.R.; Gruber, F.; Han, J.; Chen, W.; et al. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ. Res. 2010, 107, 737–746. [Google Scholar] [CrossRef]
- Vera, J.C.; Rivas, C.I.; Fischbarg, J.; Golde, D.W. Mammalian facilitative hexose transporters mediate the transport of dehydroascorbic acid. Nature 1993, 364, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sanchez-Rodriguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Obaid, M.; Udden, S.M.N.; Alluri, P.; Mandal, S.S. LncRNA HOTAIR regulates glucose transporter Glut1 expression and glucose uptake in macrophages during inflammation. Sci. Rep. 2021, 11, 232. [Google Scholar] [CrossRef]
- Satomi, T.; Ogawa, M.; Mori, I.; Ishino, S.; Kubo, K.; Magata, Y.; Nishimoto, T. Comparison of contrast agents for atherosclerosis imaging using cultured macrophages: FDG versus ultrasmall superparamagnetic iron oxide. J. Nucl. Med. 2013, 54, 999–1004. [Google Scholar] [CrossRef] [Green Version]
- Autry, A.W.; Gordon, J.W.; Carvajal, L.; Mareyam, A.; Chen, H.Y.; Park, I.; Mammoli, D.; Vareth, M.; Chang, S.M.; Wald, L.L.; et al. Comparison between 8- and 32-channel phased-array receive coils for in vivo hyperpolarized (13) C imaging of the human brain. Magn. Reson. Med. 2019, 82, 833–841. [Google Scholar] [CrossRef]
- Chung, B.T.; Chen, H.Y.; Gordon, J.; Mammoli, D.; Sriram, R.; Autry, A.W.; Le Page, L.M.; Chaumeil, M.M.; Shin, P.; Slater, J.; et al. First hyperpolarized [2-(13)C]pyruvate MR studies of human brain metabolism. J. Magn. Reson. 2019, 309, 106617. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.W.; Chen, H.Y.; Autry, A.; Park, I.; Van Criekinge, M.; Mammoli, D.; Milshteyn, E.; Bok, R.; Xu, D.; Li, Y.; et al. Translation of Carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients. Magn. Reson. Med. 2019, 81, 2702–2709. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Soliman, H.; Geraghty, B.J.; Chen, A.P.; Connelly, K.A.; Endre, R.; Perks, W.J.; Heyn, C.; Black, S.E.; Cunningham, C.H. Lactate topography of the human brain using hyperpolarized (13)C-MRI. Neuroimage 2019, 204, 116202. [Google Scholar] [CrossRef] [PubMed]
- Miloushev, V.Z.; Granlund, K.L.; Boltyanskiy, R.; Lyashchenko, S.K.; DeAngelis, L.M.; Mellinghoff, I.K.; Brennan, C.W.; Tabar, V.; Yang, T.J.; Holodny, A.I.; et al. Metabolic Imaging of the Human Brain with Hyperpolarized (13)C Pyruvate Demonstrates (13)C Lactate Production in Brain Tumor Patients. Cancer Res. 2018, 78, 3755–3760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, I.; Larson, P.E.Z.; Gordon, J.W.; Carvajal, L.; Chen, H.Y.; Bok, R.; Van Criekinge, M.; Ferrone, M.; Slater, J.B.; Xu, D.; et al. Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn. Reson. Med. 2018, 80, 864–873. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, F.A.; Woitek, R.; McLean, M.A.; Gill, A.B.; Manzano Garcia, R.; Provenzano, E.; Riemer, F.; Kaggie, J.; Chhabra, A.; Ursprung, S.; et al. Imaging breast cancer using hyperpolarized carbon-13 MRI. Proc. Natl. Acad. Sci. USA 2020, 117, 2092–2098. [Google Scholar] [CrossRef] [Green Version]
- Granlund, K.L.; Tee, S.S.; Vargas, H.A.; Lyashchenko, S.K.; Reznik, E.; Fine, S.; Laudone, V.; Eastham, J.A.; Touijer, K.A.; Reuter, V.E.; et al. Hyperpolarized MRI of Human Prostate Cancer Reveals Increased Lactate with Tumor Grade Driven by Monocarboxylate Transporter 1. Cell Metab. 2020, 31, 105–114.e3. [Google Scholar] [CrossRef]
- Grist, J.T.; Hansen, E.S.S.; Sanchez-Heredia, J.D.; McLean, M.A.; Tougaard, R.; Riemer, F.; Schulte, R.F.; Kaggie, J.D.; Ardenkjaer-Larsen, J.H.; Laustsen, C.; et al. Creating a clinical platform for carbon-13 studies using the sodium-23 and proton resonances. Magn. Reson. Med. 2020, 84, 1817–1827. [Google Scholar] [CrossRef] [Green Version]
- Woitek, R.; McLean, M.A.; Gill, A.B.; Grist, J.T.; Provenzano, E.; Patterson, A.J.; Ursprung, S.; Torheim, T.; Zaccagna, F.; Locke, M.; et al. Hyperpolarized (13)C MRI of Tumor Metabolism Demonstrates Early Metabolic Response to Neoadjuvant Chemotherapy in Breast Cancer. Radiol. Imaging Cancer 2020, 2, e200017. [Google Scholar] [CrossRef]
- Heller, N.M.; Qi, X.; Junttila, I.S.; Shirey, K.A.; Vogel, S.N.; Paul, W.E.; Keegan, A.D. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci. Signal. 2008, 1, ra17. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhang, S.; Jeon, R.; Vuckovic, I.; Jiang, X.; Lerman, A.; Folmes, C.D.; Dzeja, P.D.; Herrmann, J. Interferon Gamma Induces Reversible Metabolic Reprogramming of M1 Macrophages to Sustain Cell Viability and Pro-Inflammatory Activity. EBioMedicine 2018, 30, 303–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larson, P.E.Z.; Bok, R.; Kerr, A.B.; Lustig, M.; Hu, S.; Chen, A.P.; Nelson, S.J.; Pauly, J.M.; Kurhanewicz, J.; Vigneron, D.B. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magn. Reson. Med. 2010, 63, 582–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapcariu, S.C.; Kanashova, T.; Weindl, D.; Ghelfi, J.; Dittmar, G.; Hiller, K. Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX 2014, 1, 74–80. [Google Scholar] [CrossRef] [PubMed]
- de Graaf, R.A.; Brown, P.B.; McIntyre, S.; Nixon, T.W.; Behar, K.L.; Rothman, D.L. High magnetic field water and metabolite proton T1 and T2 relaxation in rat brain in vivo. Magn. Reson. Med. 2006, 56, 386–394. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
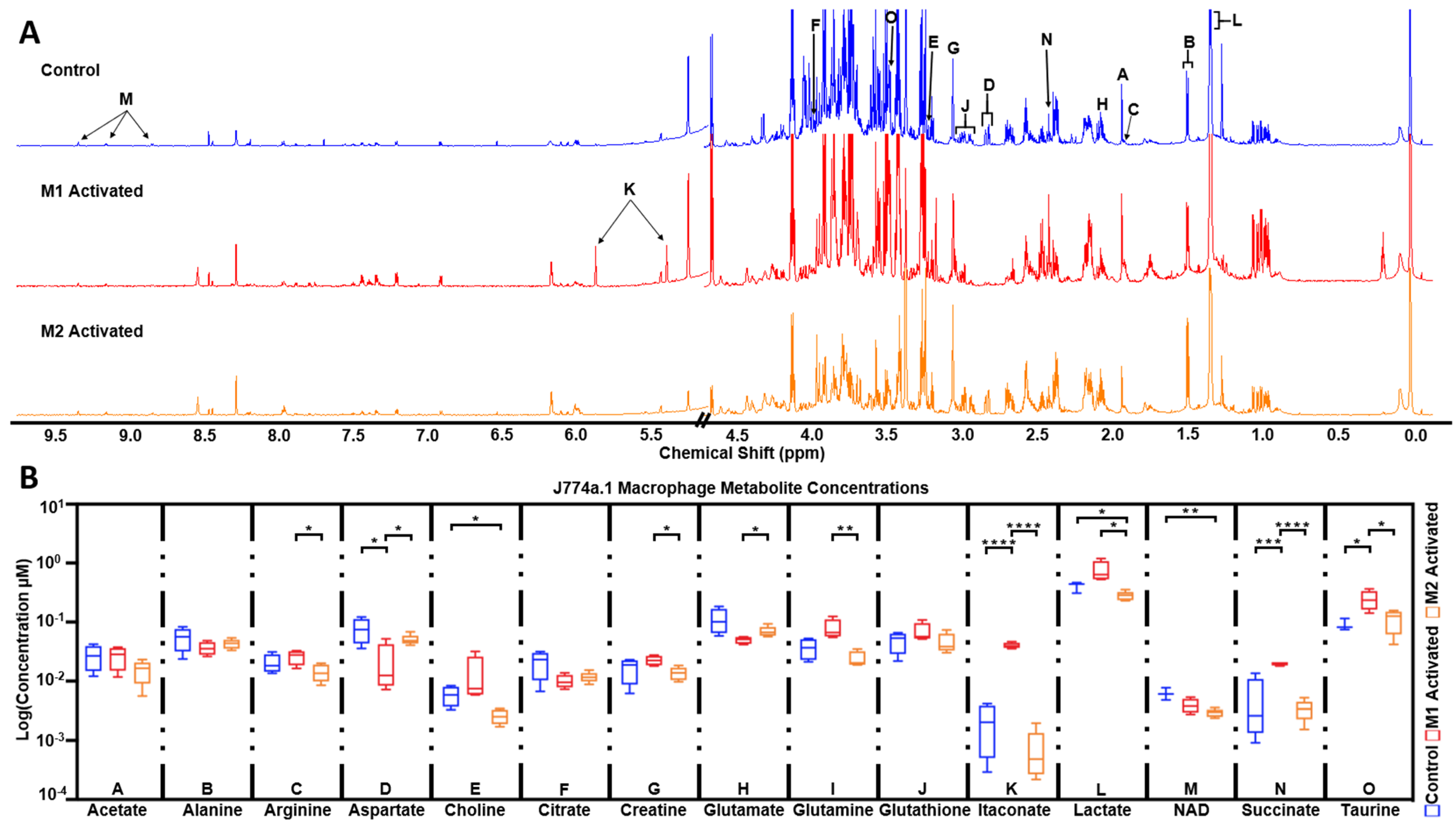
| J774a.1 Non-Activated vs. Activated Metabolite Concentrations (µM) | |||
|---|---|---|---|
| Metabolite | Control | M1-Activated | M2-Activated |
| Acetate | 27 ± 12.7 | 26.52 ± 11.1 | 15.18 ± 6.58 |
| Alanine | 54.91 ± 24.3 | 36.48 ± 9.34 | 43.02 ± 7.98 |
| Arginine | 20.37 ± 7.68 | 26.17 ± 7.24 | 14.28 ± 4.75 |
| Aspartate | 76.93 ± 36.1 | 21.05 ± 20.89 | 50.57 ± 10.53 |
| Choline | 5.86 ± 2.23 | 13.13 ± 12.5 | 2.58 ± 0.72 |
| Citrate | 21.16 ± 10.57 | 10.03 ± 2.69 | 11.76 ± 2.38 |
| Creatine | 16.72 ± 7.54 | 22.59 ± 4.31 | 13.69 ± 3.45 |
| Glutamate | 111.09 ± 54.14 | 49.98 ± 6.89 | 69.95 ± 14.23 |
| Glutamine | 36.97 ± 14.99 | 78.15 ± 31.56 | 24.51 ± 7.28 |
| Glutathione | 48.79 ± 19.16 | 68.46 ± 26.94 | 46.3 ± 17.43 |
| Itaconic acid | 2.13 ± 1.74 | 40.27 ± 4.59 | 0.74 ± 0.71 |
| Lactate | 406.03 ± 83.92 | 748.64 ± 310.05 | 280.91 ± 49.32 |
| NAD | 6.2 ± 1.45 | 3.93 ± 1.09 | 2.94 ± 0.45 |
| Succinate | 4.93 ± 5.81 | 19.86 ± 1.4 | 3.38 ± 1.34 |
| Taurine | 90.85 ± 21.66 | 244.7 ± 92.42 | 113.2 ± 49.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, K.; Le Page, L.M.; Chaumeil, M.M. Non-Invasive Differentiation of M1 and M2 Activation in Macrophages Using Hyperpolarized 13C MRS of Pyruvate and DHA at 1.47 Tesla. Metabolites 2021, 11, 410. https://doi.org/10.3390/metabo11070410
Qiao K, Le Page LM, Chaumeil MM. Non-Invasive Differentiation of M1 and M2 Activation in Macrophages Using Hyperpolarized 13C MRS of Pyruvate and DHA at 1.47 Tesla. Metabolites. 2021; 11(7):410. https://doi.org/10.3390/metabo11070410
Chicago/Turabian StyleQiao, Kai, Lydia M. Le Page, and Myriam M. Chaumeil. 2021. "Non-Invasive Differentiation of M1 and M2 Activation in Macrophages Using Hyperpolarized 13C MRS of Pyruvate and DHA at 1.47 Tesla" Metabolites 11, no. 7: 410. https://doi.org/10.3390/metabo11070410
APA StyleQiao, K., Le Page, L. M., & Chaumeil, M. M. (2021). Non-Invasive Differentiation of M1 and M2 Activation in Macrophages Using Hyperpolarized 13C MRS of Pyruvate and DHA at 1.47 Tesla. Metabolites, 11(7), 410. https://doi.org/10.3390/metabo11070410