Metabolic View on Human Healthspan: A Lipidome-Wide Association Study
Abstract
1. Introduction
2. Results
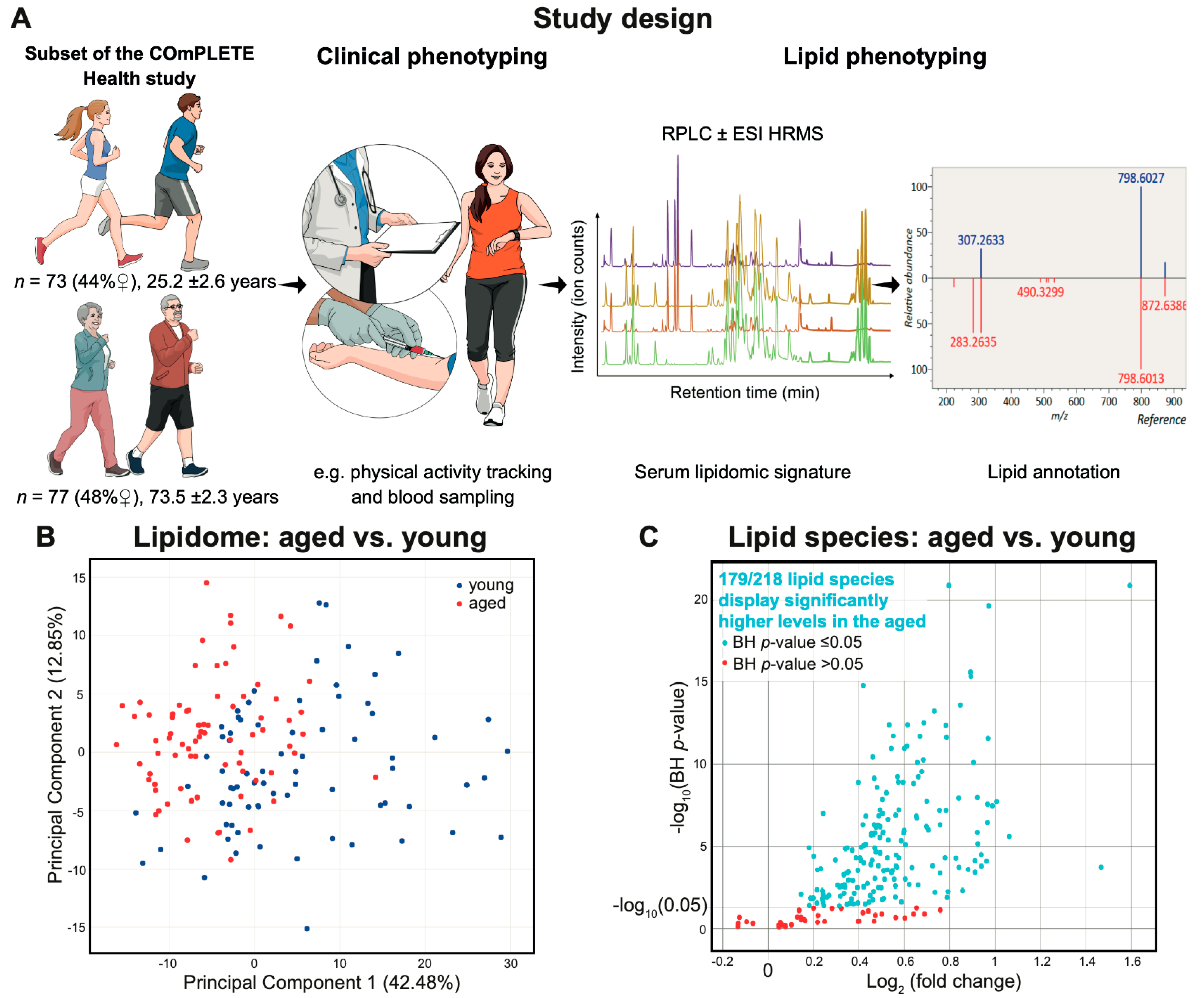
2.1. Characteristics of the Clinically Healthy Participants
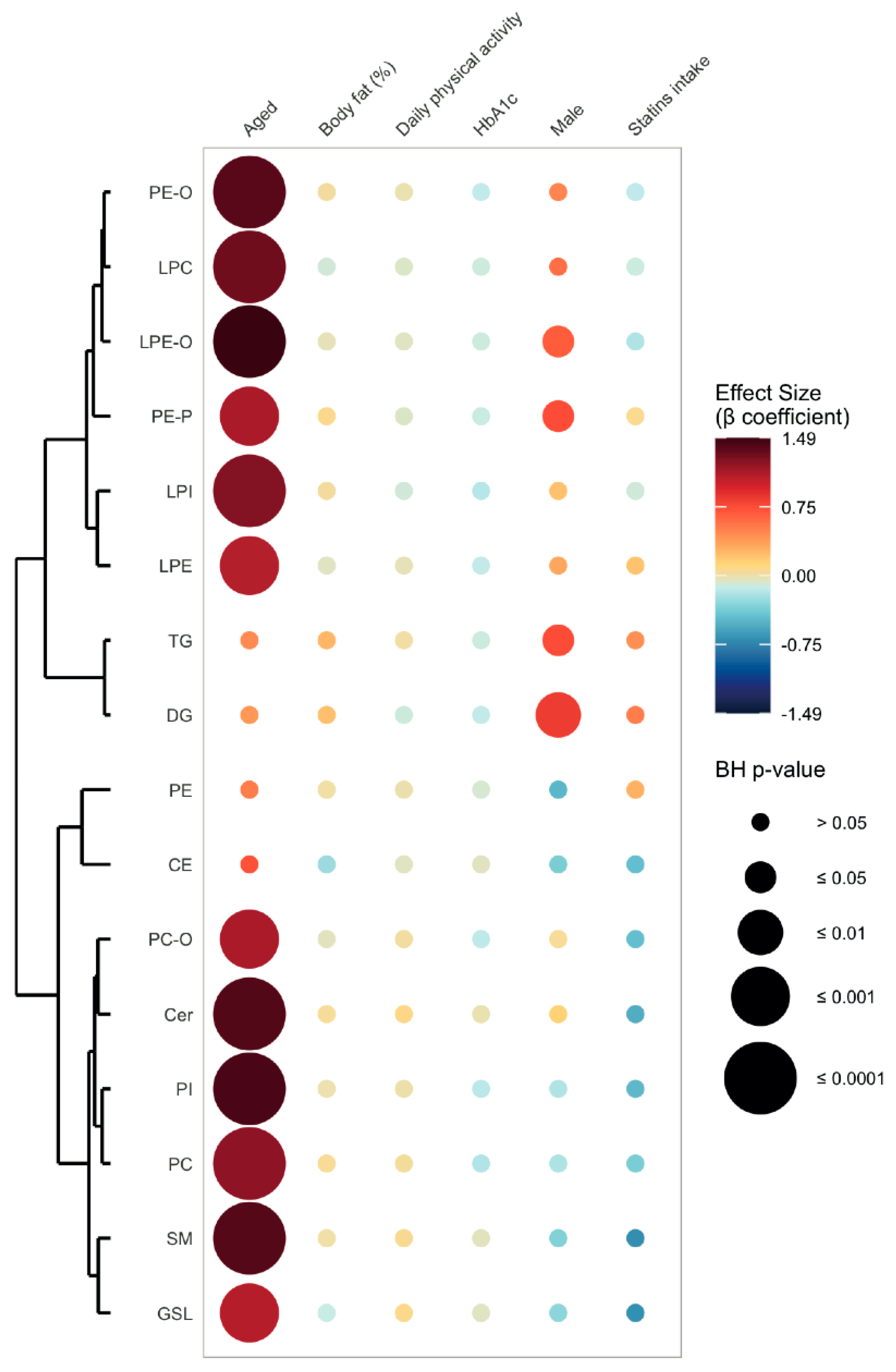
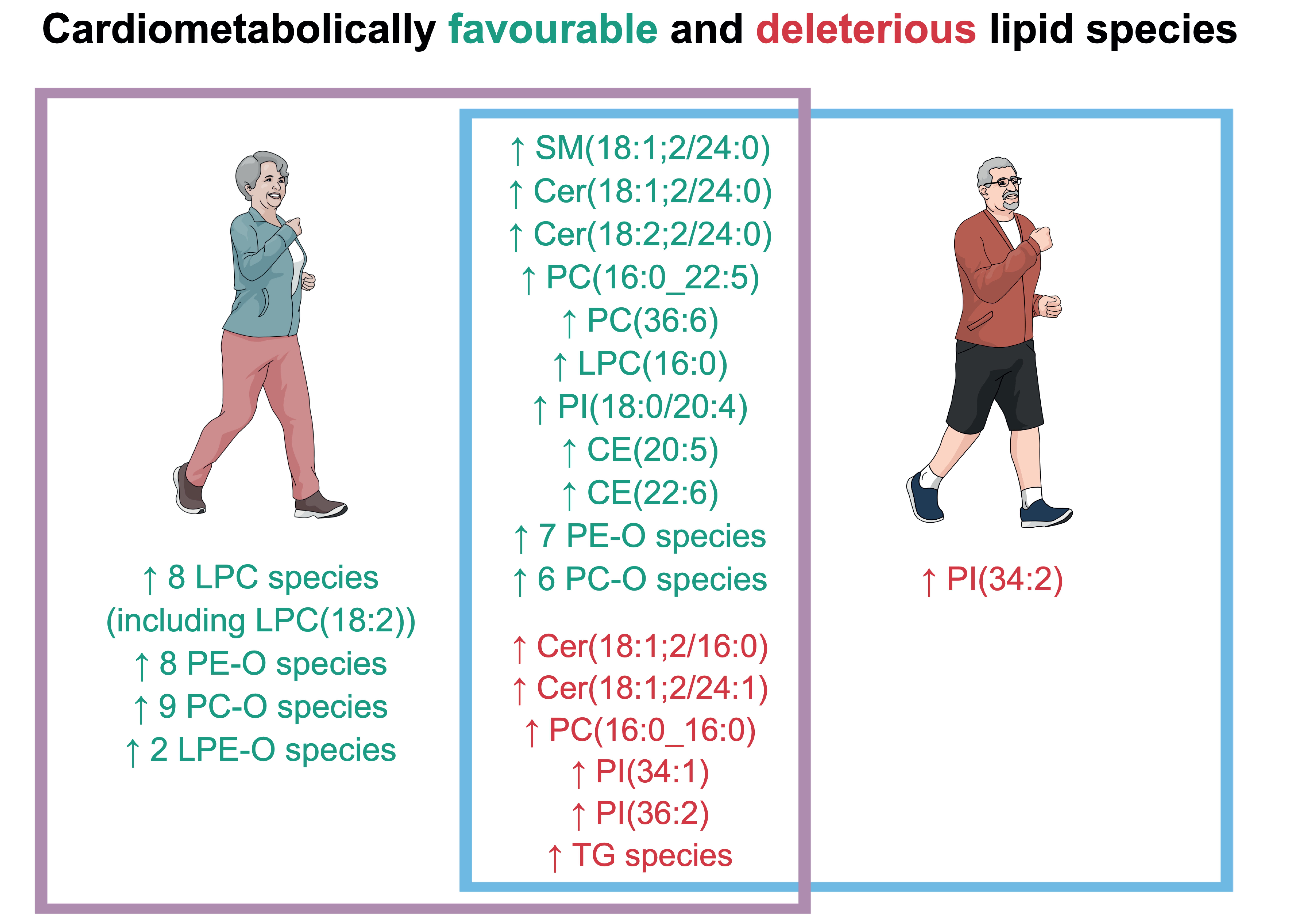
2.2. Lipid Signature of Clinically Healthy Young and Aged Phenotypes
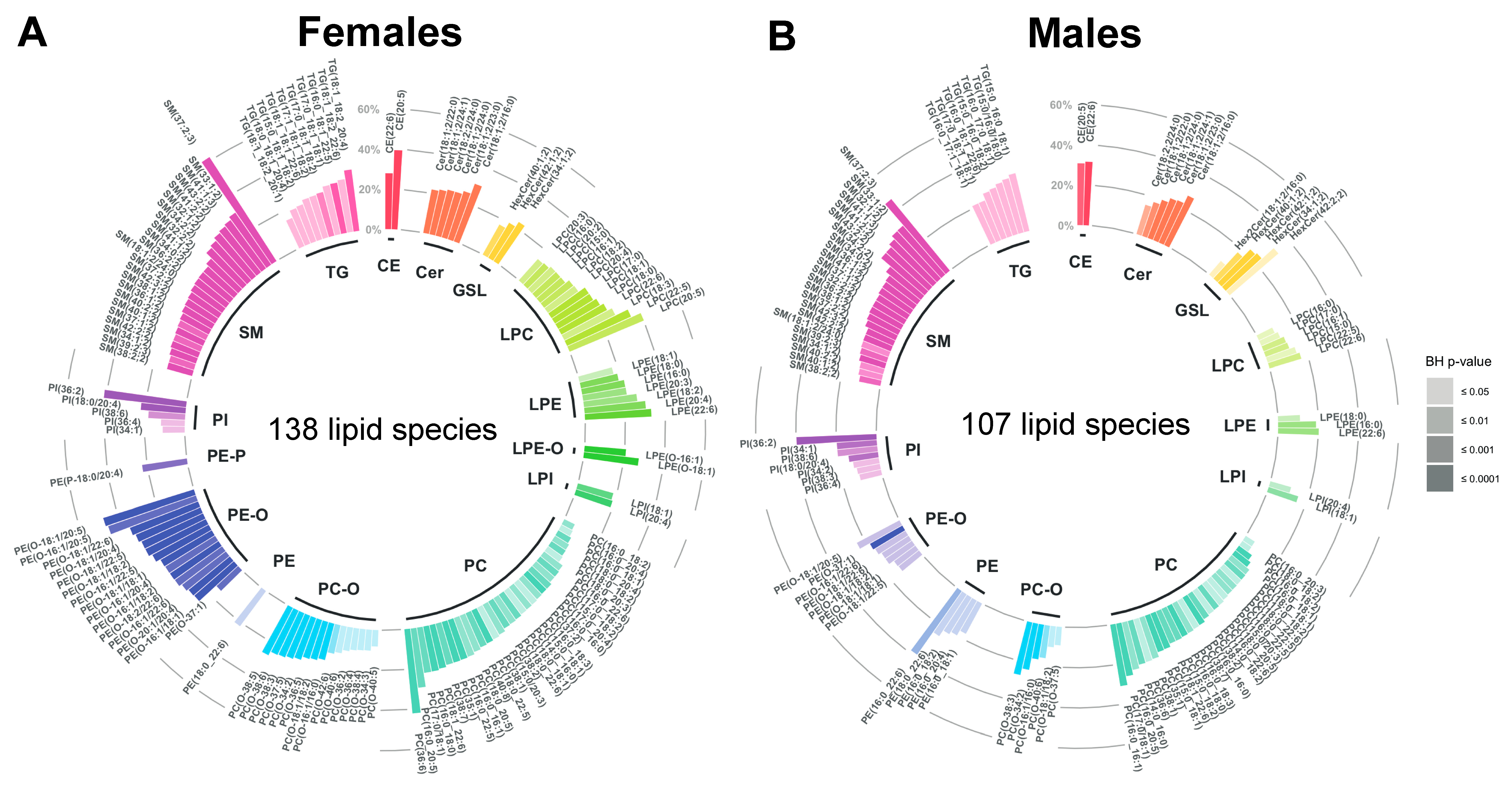
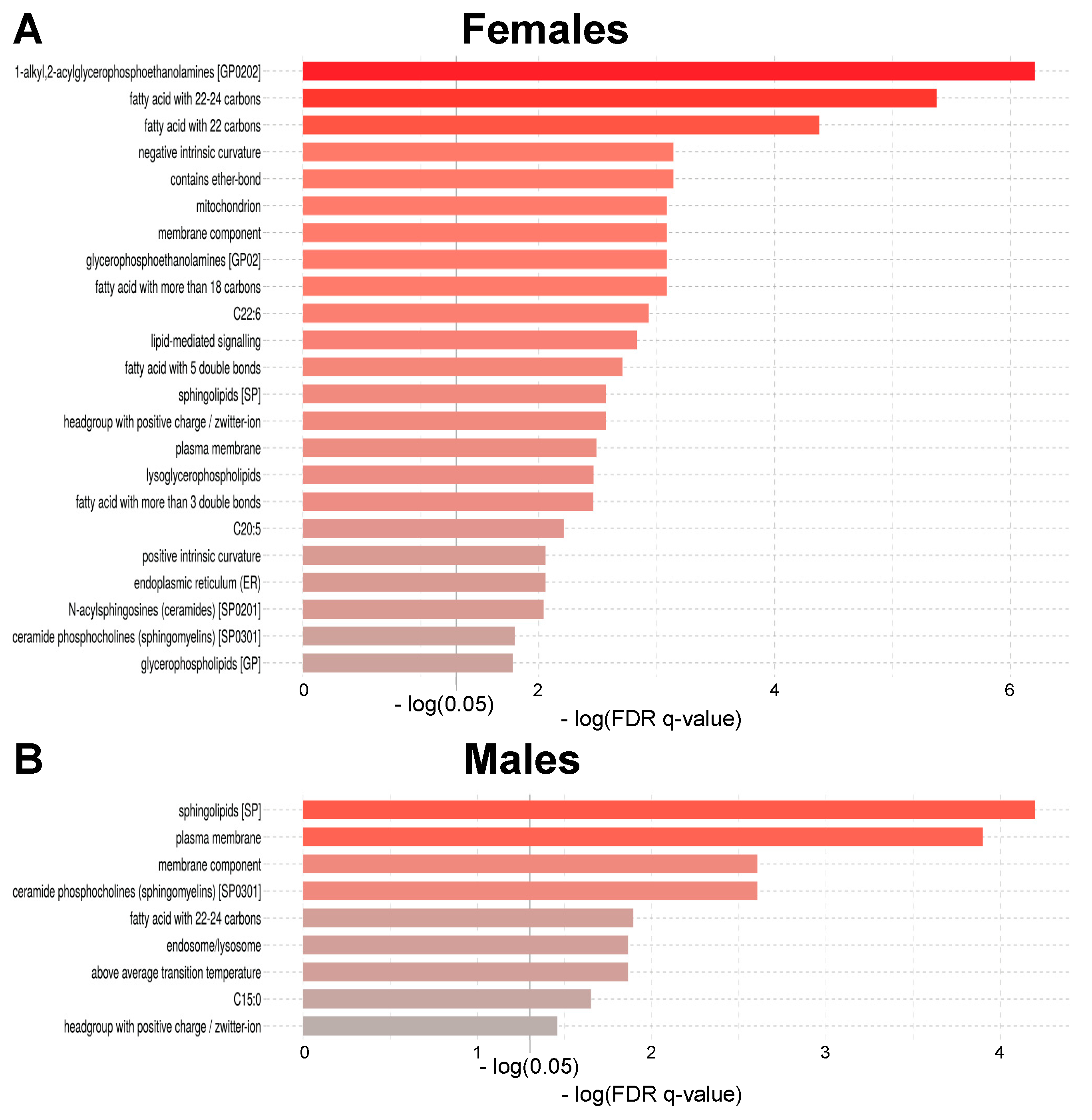
2.3. Age-Related Sex Differences in the Circulatory Lipidome Composition
3. Discussion
3.1. Unravelling Age- and Sex-Associated Lipid Signature
3.2. Cardiometabolic Significance of the Identified Lipid Signature
3.2.1. Sphingolipids
3.2.2. Glycerophospholipids
3.2.3. Cholesterol Esters, Glycerolipids and Saturation Levels
3.3. Moving Away from Subclass towards Species Analysis in Clinical Medicine
3.4. Limitations
4. Materials and Methods
4.1. Participants
4.2. Data Collection
4.3. Biochemical Analysis
4.4. Lipid Extraction
4.5. Untargeted Lipidomics
4.6. Quality Control
4.7. Data Processing and Lipid Annotation
4.8. Lipid Shorthand Notation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beard, J.R.; Officer, A.; de Carvalho, I.A.; Sadana, R.; Pot, A.M.; Michel, J.P.; Lloyd-Sherlock, P.; Epping-Jordan, J.E.; Peeters, G.; Mahanani, W.R.; et al. The World report on ageing and health: A policy framework for healthy ageing. Lancet 2016, 387, 2145–2154. [Google Scholar] [CrossRef]
- Seals, D.R.; Justice, J.N.; LaRocca, T.J. Physiological geroscience: Targeting function to increase healthspan and achieve optimal longevity. J. Physiol. 2016, 594, 2001–2024. [Google Scholar] [CrossRef]
- Nikolich-Žugich, J.; Goldman, D.P.; Cohen, P.R.; Cortese, D.; Fontana, L.; Kennedy, B.K.; Mohler, M.J.; Olshansky, S.J.; Perls, T.; Perry, D.; et al. Preparing for an Aging World: Engaging Biogerontologists, Geriatricians, and the Society. J. Gerontol. Ser. Abiological. Sci. Med. Sci. 2015, 71, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Crimmins, E.M. Lifespan and Healthspan: Past, Present, and Promise. Gerontologist 2015, 55, 901–911. [Google Scholar] [CrossRef]
- Chang, A.Y.; Skirbekk, V.F.; Tyrovolas, S.; Kassebaum, N.J.; Dieleman, J.L. Measuring population ageing: An analysis of the Global Burden of Disease Study 2017. Lancet Public Health 2019, 4, e159–e167. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Rabinovitch, P.S.; Martin, G.M. Healthy aging: The ultimate preventative medicine. Science 2015, 350, 1191–1193. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.E.; Chatterji, S.; Kowal, P.; Lloyd-Sherlock, P.; McKee, M.; Rechel, B.; Rosenberg, L.; Smith, J.P. Macroeconomic implications of population ageing and selected policy responses. Lancet 2015, 385, 649–657. [Google Scholar] [CrossRef]
- Suhre, K.; Shin, S.Y.; Petersen, A.K.; Mohney, R.P.; Meredith, D.; Wägele, B.; Altmaier, E.; Deloukas, P.; Erdmann, J.; Grundberg, E.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Després, J.-P. Predicting longevity using metabolomics: A novel tool for precision lifestyle medicine? Nat. Rev. Cardiol. 2020, 17, 67–68. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk: The Task Force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS). Eur. Heart J. 2019, 41, 111–188. [Google Scholar] [CrossRef]
- Morrison, L.M.; Hall, L.; Chaney, A.L. Cholesterol metabolism and its relationship to atherosclerosis, coronary artery disease, and arteriosclerosis. Am. J. Med. 1948, 4, 616. [Google Scholar] [CrossRef]
- Quehenberger, O.; Dennis, E.A. The Human Plasma Lipidome. N. Engl. J. Med. 2011, 365, 1812–1823. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, C.; Han, R.H.; Han, X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid. Res. 2016, 61, 83–108. [Google Scholar] [CrossRef]
- Yang, K.; Han, X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef] [PubMed]
- Lydic, T.A.; Goo, Y.-H. Lipidomics unveils the complexity of the lipidome in metabolic diseases. Clin. Transl. Med. 2018, 7, e4. [Google Scholar] [CrossRef]
- de Diego, I.; Peleg, S.; Fuchs, B. The role of lipids in aging-related metabolic changes. Chem. Phys. Lipids 2019, 222, 59–69. [Google Scholar] [CrossRef]
- Maxfield, F.R.; Tabas, I. Role of cholesterol and lipid organization in disease. Nature 2005, 438, 612–621. [Google Scholar] [CrossRef]
- Arish, M.; Husein, A.; Kashif, M.; Sandhu, P.; Hasnain, S.E.; Akhter, Y.; Rub, A. Orchestration of membrane receptor signaling by membrane lipids. Biochimie 2015, 113, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Meer, G.v.; Sprong, H. Membrane lipids and vesicular traffic. Curr. Opin. Cell Biol. 2004, 16, 373–378. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Walsh, M.T.; Hammad, S.M.; Hussain, M.M. Sphingolipids and Lipoproteins in Health and Metabolic Disorders. Trends Endocrinol. Metab. 2017, 28, 506–518. [Google Scholar] [CrossRef]
- Li, W.; Yang, X.; Xing, S.; Bian, F.; Yao, W.; Bai, X.; Zheng, T.; Wu, G.; Jin, S. Endogenous ceramide contributes to the transcytosis of oxLDL across endothelial cells and promotes its subendothelial retention in vascular wall. Oxidative. Med. Cell. Longev. 2014, 2014, 823071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Wang, P.; Zhang, S.-Y.; Dong, Y.; Zeng, G.; Yan, Y.; Sun, L.; Wu, Q.; Liu, H.; et al. Adipocyte Hypoxia-Inducible Factor 2α Suppresses Atherosclerosis by Promoting Adipose Ceramide Catabolism. Cell Metab. 2019, 30, 937–951.e935. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, B.; Summers, S.A. Ceramides—Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. 2015, 26, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Laaksonen, R.; Ekroos, K.; Sysi-Aho, M.; Hilvo, M.; Vihervaara, T.; Kauhanen, D.; Suoniemi, M.; Hurme, R.; Marz, W.; Scharnagl, H.; et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur. Heart J. 2016, 37, 1967–1976. [Google Scholar] [CrossRef]
- Havulinna, A.S.; Sysi-Aho, M.; Hilvo, M.; Kauhanen, D.; Hurme, R.; Ekroos, K.; Salomaa, V.; Laaksonen, R. Circulating Ceramides Predict Cardiovascular Outcomes in the Population-Based FINRISK 2002 Cohort. Arter. Thromb. Vasc. Biol. 2016, 36, 2424–2430. [Google Scholar] [CrossRef]
- Meeusen, J.W.; Donato, L.J.; Bryant, S.C.; Baudhuin, L.M.; Berger, P.B.; Jaffe, A.S. Plasma Ceramides. Arter. Thromb. Vasc. Biol. 2018, 38, 1933–1939. [Google Scholar] [CrossRef]
- Hilvo, M.; Meikle, P.J.; Pedersen, E.R.; Tell, G.S.; Dhar, I.; Brenner, H.; Schöttker, B.; Lääperi, M.; Kauhanen, D.; Koistinen, K.M.; et al. Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur. Heart J. 2019, 41, 371–380. [Google Scholar] [CrossRef]
- Summers, S.A. Could Ceramides Become the New Cholesterol? Cell Metab. 2018, 27, 276–280. [Google Scholar] [CrossRef]
- Bustos, V.; Partridge, L. Good Ol’ Fat: Links between Lipid Signaling and Longevity. Trends Biochem. Sci. 2017, 42, 812–823. [Google Scholar] [CrossRef]
- Jove, M.; Naudi, A.; Gambini, J.; Borras, C.; Cabre, R.; Portero-Otin, M.; Vina, J.; Pamplona, R. A Stress-Resistant Lipidomic Signature Confers Extreme Longevity to Humans. J. Gerontol. Ser. Abiological. Sci. Med. Sci. 2017, 72, 30–37. [Google Scholar] [CrossRef]
- Huynh, K.; Barlow, C.K.; Jayawardana, K.S.; Weir, J.M.; Mellett, N.A.; Cinel, M.; Magliano, D.J.; Shaw, J.E.; Drew, B.G.; Meikle, P.J. High-Throughput Plasma Lipidomics: Detailed Mapping of the Associations with Cardiometabolic Risk Factors. Cell Chem. Biol. 2019, 26, 71–84.e74. [Google Scholar] [CrossRef]
- Kawanishi, N.; Kato, Y.; Yokozeki, K.; Sawada, S.; Sakurai, R.; Fujiwara, Y.; Shinkai, S.; Goda, N.; Suzuki, K. Effects of aging on serum levels of lipid molecular species as determined by lipidomics analysis in Japanese men and women. Lipids Health Dis. 2018, 17, 135. [Google Scholar] [CrossRef]
- Gonzalez-Covarrubias, V.; Beekman, M.; Uh, H.W.; Dane, A.; Troost, J.; Paliukhovich, I.; van der Kloet, F.M.; Houwing-Duistermaat, J.; Vreeken, R.J.; Hankemeier, T.; et al. Lipidomics of familial longevity. Aging Cell 2013, 12, 426–434. [Google Scholar] [CrossRef]
- Ishikawa, M.; Maekawa, K.; Saito, K.; Senoo, Y.; Urata, M.; Murayama, M.; Tajima, Y.; Kumagai, Y.; Saito, Y. Plasma and Serum Lipidomics of Healthy White Adults Shows Characteristic Profiles by Subjects’ Gender and Age. PLoS ONE 2014, 9, e91806. [Google Scholar] [CrossRef]
- Wong, M.W.K.; Braidy, N.; Pickford, R.; Vafaee, F.; Crawford, J.; Muenchhoff, J.; Schofield, P.; Attia, J.; Brodaty, H.; Sachdev, P.; et al. Plasma lipidome variation during the second half of the human lifespan is associated with age and sex but minimally with BMI. PLoS ONE 2019, 14, e0214141. [Google Scholar] [CrossRef]
- Maekawa, K.; Okemoto, K.; Ishikawa, M.; Tanaka, R.; Kumagai, Y.; Saito, Y. Plasma Lipidomics of Healthy Japanese Adults Reveals Gender- and Age-Related Differences. J. Pharm. Sci. 2017, 106, 2914–2918. [Google Scholar] [CrossRef]
- Naudí, A.; Jové, M.; Ayala, V.; Portero-Otín, M.; Barja, G.; Pamplona, R. Membrane lipid unsaturation as physiological adaptation to animal longevity. Front. Physiol. 2013, 4, 372. [Google Scholar] [CrossRef] [PubMed]
- Montoliu, I.; Scherer, M.; Beguelin, F.; DaSilva, L.; Mari, D.; Salvioli, S.; Martin, F.P.; Capri, M.; Bucci, L.; Ostan, R.; et al. Serum profiling of healthy aging identifies phospho- and sphingolipid species as markers of human longevity. Aging 2014, 6, 9–25. [Google Scholar] [CrossRef]
- Chaleckis, R.; Murakami, I.; Takada, J.; Kondoh, H.; Yanagida, M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. USA 2016, 113, 4252–4259. [Google Scholar] [CrossRef]
- Darst, B.F.; Koscik, R.L.; Hogan, K.J.; Johnson, S.C.; Engelman, C.D. Longitudinal plasma metabolomics of aging and sex. Aging 2019, 11, 1262–1282. [Google Scholar] [CrossRef]
- Collino, S.; Montoliu, I.; Martin, F.-P.J.; Scherer, M.; Mari, D.; Salvioli, S.; Bucci, L.; Ostan, R.; Monti, D.; Biagi, E.; et al. Metabolic Signatures of Extreme Longevity in Northern Italian Centenarians Reveal a Complex Remodeling of Lipids, Amino Acids, and Gut Microbiota Metabolism. PLoS ONE 2013, 8, e56564. [Google Scholar] [CrossRef]
- Menni, C.; Kastenmüller, G.; Petersen, A.K.; Bell, J.T.; Psatha, M.; Tsai, P.-C.; Gieger, C.; Schulz, H.; Erte, I.; John, S.; et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013, 42, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Larson, M.G.; McCabe, E.L.; Murabito, J.M.; Rhee, E.P.; Ho, J.E.; Jacques, P.F.; Ghorbani, A.; Magnusson, M.; Souza, A.L.; et al. Distinct metabolomic signatures are associated with longevity in humans. Nat. Commun. 2015, 6, 6791. [Google Scholar] [CrossRef] [PubMed]
- Beyene, H.B.; Olshansky, G.; Smith, A.A.; Giles, C.; Huynh, K.; Cinel, M.; Mellett, N.A.; Cadby, G.; Hung, J.; Hui, J.; et al. High-coverage plasma lipidomics reveals novel sex-specific lipidomic fingerprints of age and BMI: Evidence from two large population cohort studies. PLoS Biol. 2020, 18, e3000870. [Google Scholar] [CrossRef]
- Yu, Z.; Zhai, G.; Singmann, P.; He, Y.; Xu, T.; Prehn, C.; Römisch-Margl, W.; Lattka, E.; Gieger, C.; Soranzo, N.; et al. Human serum metabolic profiles are age dependent. Aging Cell 2012, 11, 960–967. [Google Scholar] [CrossRef]
- Rist, M.J.; Roth, A.; Frommherz, L.; Weinert, C.H.; Krüger, R.; Merz, B.; Bunzel, D.; Mack, C.; Egert, B.; Bub, A.; et al. Metabolite patterns predicting sex and age in participants of the Karlsruhe Metabolomics and Nutrition (KarMeN) study. PLoS ONE 2017, 12, e0183228. [Google Scholar] [CrossRef]
- Dunn, W.B.; Lin, W.; Broadhurst, D.; Begley, P.; Brown, M.; Zelena, E.; Vaughan, A.A.; Halsall, A.; Harding, N.; Knowles, J.D.; et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics 2015, 11, 9–26. [Google Scholar] [CrossRef]
- Johnson, L.C.; Parker, K.; Aguirre, B.F.; Nemkov, T.G.; D’Alessandro, A.; Johnson, S.A.; Seals, D.R.; Martens, C.R. The plasma metabolome as a predictor of biological aging in humans. GeroScience 2019, 41, 895–906. [Google Scholar] [CrossRef]
- Kohl, H.W., 3rd; Craig, C.L.; Lambert, E.V.; Inoue, S.; Alkandari, J.R.; Leetongin, G.; Kahlmeier, S. The pandemic of physical inactivity: Global action for public health. Lancet 2012, 380, 294–305. [Google Scholar] [CrossRef]
- Bull, F.C.; Al-Ansari, S.S.; Biddle, S.; Borodulin, K.; Buman, M.P.; Cardon, G.; Carty, C.; Chaput, J.-P.; Chastin, S.; Chou, R.; et al. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br. J. Sports Med. 2020, 54, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.; Blair, S.N.; Arena, R.; Church, T.S.; Despres, J.P.; Franklin, B.A.; Haskell, W.L.; Kaminsky, L.A.; Levine, B.D.; Lavie, C.J.; et al. Importance of Assessing Cardiorespiratory Fitness in Clinical Practice: A Case for Fitness as a Clinical Vital Sign: A Scientific Statement From the American Heart Association. Circulation 2016, 134, e653–e699. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Knaier, R.; Infanger, D.; Arbeev, K.; Briel, M.; Dieterle, T.; Hanssen, H.; Faude, O.; Roth, R.; Hinrichs, T.; et al. Functional aging in health and heart failure: The COmPLETE Study. BMC Cardiovasc. Disord. 2019, 19, 180. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Mancia, G.; Spiering, W.; Agabiti Rosei, E.; Azizi, M.; Burnier, M.; Clement, D.L.; Coca, A.; de Simone, G.; Dominiczak, A.; et al. 2018 ESC/ESH Guidelines for the management of arterial hypertension: The Task Force for the management of arterial hypertension of the European Society of Cardiology (ESC) and the European Society of Hypertension (ESH). Eur. Heart J. 2018, 39, 3021–3104. [Google Scholar] [CrossRef]
- Cosentino, F.; Grant, P.J.; Aboyans, V.; Bailey, C.J.; Ceriello, A.; Delgado, V.; Federici, M.; Filippatos, G.; Grobbee, D.E.; Hansen, T.B.; et al. 2019 ESC Guidelines on diabetes, pre-diabetes, and cardiovascular diseases developed in collaboration with the EASD: The Task Force for diabetes, pre-diabetes, and cardiovascular diseases of the European Society of Cardiology (ESC) and the European Association for the Study of Diabetes (EASD). Eur. Heart J. 2019, 41, 255–323. [Google Scholar] [CrossRef]
- Searle, S.R.; Speed, F.M.; Milliken, G.A. Population Marginal Means in the Linear Model: An Alternative to Least Squares Means. Am. Stat. 1980, 34, 216–221. [Google Scholar] [CrossRef]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; van de Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/web: A web-based ontology enrichment tool for lipidomic data analysis. GigaScience 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Covarrubias, V. Lipidomics in longevity and healthy aging. Biogerontology 2013, 14, 663–672. [Google Scholar] [CrossRef]
- Endapally, S.; Frias, D.; Grzemska, M.; Gay, A.; Tomchick, D.R.; Radhakrishnan, A. Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes. Cell 2019, 176, 1040–1053.e1017. [Google Scholar] [CrossRef]
- Trayssac, M.; Hannun, Y.A.; Obeid, L.M. Role of sphingolipids in senescence: Implication in aging and age-related diseases. J. Clin. Investig. 2018, 128, 2702–2712. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Sigruener, A.; Kleber, M.E.; Heimerl, S.; Liebisch, G.; Schmitz, G.; Maerz, W. Glycerophospholipid and Sphingolipid Species and Mortality: The Ludwigshafen Risk and Cardiovascular Health (LURIC) Study. PLoS ONE 2014, 9, e85724. [Google Scholar] [CrossRef]
- Hilvo, M.; Wallentin, L.; Lakic, T.G.; Held, C.; Kauhanen, D.; Jylhä, A.; Lindbäck, J.; Siegbahn, A.; Granger, C.B.; Koenig, W.; et al. Prediction of Residual Risk by Ceramide-Phospholipid Score in Patients With Stable Coronary Heart Disease on Optimal Medical Therapy. J. Am. Heart Assoc. 2020, 9, e015258. [Google Scholar] [CrossRef] [PubMed]
- Chew, W.S.; Torta, F.; Ji, S.; Choi, H.; Begum, H.; Sim, X.; Khoo, C.M.; Khoo, E.Y.H.; Ong, W.Y.; Van Dam, R.M.; et al. Large-scale lipidomics identifies associations between plasma sphingolipids and T2DM incidence. JCI Insight 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Dugo, C. Ceramides and risk of major adverse cardiovascular events: A meta-analysis of longitudinal studies. J. Clin. Lipidol. 2020, 14, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Fang, Z.; Li, S.; Xu, M.; Zhang, J.; Han, D.; Hu, W.; Yan, L.; Wang, Y.; Fan, L.; et al. Circulating Ceramide: A New Cardiometabolic Biomarker in Patients With Comorbid Acute Coronary Syndrome and Type 2 Diabetes Mellitus. Front. Physiol. 2020, 11, 1104. [Google Scholar] [CrossRef] [PubMed]
- Castro, B.M.; Prieto, M.; Silva, L.C. Ceramide: A simple sphingolipid with unique biophysical properties. Prog. Lipid Res. 2014, 54, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Bolla, J.R.; Agasid, M.T.; Mehmood, S.; Robinson, C.V. Membrane Protein-Lipid Interactions Probed Using Mass Spectrometry. Annu. Rev. Biochem. 2019, 88, 85–111. [Google Scholar] [CrossRef]
- Xu, H.; Valenzuela, N.; Fai, S.; Figeys, D.; Bennett, S.A.L. Targeted lipidomics—Advances in profiling lysophosphocholine and platelet-activating factor second messengers. FEBS J. 2013, 280, 5652–5667. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, O.; Shih, D.M.; Aviram, M. Human Serum Paraoxonase 1 Decreases Macrophage Cholesterol Biosynthesis. Arter. Thromb. Vasc. Biol. 2003, 23, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Sandin, M.; Sampaio, J.L.; Almgren, P.; Narkiewicz, K.; Hoffmann, M.; Hedner, T.; Wahlstrand, B.; Simons, K.; Shevchenko, A.; et al. Plasma Lipid Composition and Risk of Developing Cardiovascular Disease. PLoS ONE 2013, 8, e71846. [Google Scholar] [CrossRef]
- Wang-Sattler, R.; Yu, Z.; Herder, C.; Messias, A.C.; Floegel, A.; He, Y.; Heim, K.; Campillos, M.; Holzapfel, C.; Thorand, B.; et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012, 8, 615. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Et Biophys. Acta (BBA)—Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Calzada, E.; Onguka, O.; Claypool, S.M. Chapter Two—Phosphatidylethanolamine Metabolism in Health and Disease. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 321, pp. 29–88. [Google Scholar]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny Lipids With Giant Impact on Cell Regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef]
- Bozelli, J.C., Jr.; Epand, R.M. Specificity of Acyl Chain Composition of Phosphatidylinositols. Proteomics 2019, 19, 1900138. [Google Scholar] [CrossRef] [PubMed]
- Naguib, A.; Bencze, G.; Engle, D.D.; Chio, I.I.; Herzka, T.; Watrud, K.; Bencze, S.; Tuveson, D.A.; Pappin, D.J.; Trotman, L.C. P53 Mutations Change Phosphatidylinositol Acyl Chain Composition. Cell Rep. 2015, 10, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Soussi, T.; Béroud, C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer 2001, 1, 233–239. [Google Scholar] [CrossRef]
- Dean, J.M.; Lodhi, I.J. Structural and functional roles of ether lipids. Protein Cell 2018, 9, 196–206. [Google Scholar] [CrossRef]
- Watschinger, K.; Werner, E.R. Orphan enzymes in ether lipid metabolism. Biochimie 2013, 95, 59–65. [Google Scholar] [CrossRef]
- Puri, P.; Wiest, M.M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.-K.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H.; et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef]
- Oresic, M.; Simell, S.; Sysi-Aho, M.; Nanto-Salonen, K.; Seppänen-Laakso, T.; Parikka, V.; Katajamaa, M.; Hekkala, A.; Mattila, I.; Keskinen, P.; et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J. Exp. Med. 2008, 205, 2975–2984. [Google Scholar] [CrossRef]
- Tatsuta, T.; Scharwey, M.; Langer, T. Mitochondrial lipid trafficking. Trends Cell Biol. 2014, 24, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Toledo, E.; Wang, D.D.; Ruiz-Canela, M.; Clish, C.B.; Razquin, C.; Zheng, Y.; Guasch-Ferré, M.; Hruby, A.; Corella, D.; Gómez-Gracia, E.; et al. Plasma lipidomic profiles and cardiovascular events in a randomized intervention trial with the Mediterranean diet. Am. J. Clin. Nutr. 2017, 106, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Gerl, M.J.; Vaz, W.L.C.; Domingues, N.; Klose, C.; Surma, M.A.; Sampaio, J.L.; Almeida, M.S.; Rodrigues, G.; Araújo-Gonçalves, P.; Ferreira, J.; et al. Cholesterol is Inefficiently Converted to Cholesteryl Esters in the Blood of Cardiovascular Disease Patients. Sci. Rep. 2018, 8, 14764. [Google Scholar] [CrossRef]
- Del Gobbo, L.C.; Imamura, F.; Aslibekyan, S.; Marklund, M.; Virtanen, J.K.; Wennberg, M.; Yakoob, M.Y.; Chiuve, S.E.; dela Cruz, L.; Frazier-Wood, A.C.; et al. ω-3 Polyunsaturated Fatty Acid Biomarkers and Coronary Heart Disease: Pooling Project of 19 Cohort Studies. JAMA Intern. Med. 2016, 176, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Aimo, L.; Liechti, R.; Hyka-Nouspikel, N.; Niknejad, A.; Gleizes, A.; Götz, L.; Kuznetsov, D.; David, F.P.A.; van der Goot, F.G.; Riezman, H.; et al. The SwissLipids knowledgebase for lipid biology. Bioinformatics 2015, 31, 2860–2866. [Google Scholar] [CrossRef]
- Tully, A.M.; Roche, H.M.; Doyle, R.; Fallon, C.; Bruce, I.; Lawlor, B.; Coakley, D.; Gibney, M.J. Low serum cholesteryl ester-docosahexaenoic acid levels in Alzheimer’s disease: A case-control study. Br. J. Nutr. 2003, 89, 483–489. [Google Scholar] [CrossRef]
- Sugiyama, M.G.; Agellon, L.B. Sex differences in lipid metabolism and metabolic disease risk. Biochem. Cell Biol. 2012, 90, 124–141. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Stone, N.J.; Ballantyne, C.; Bittner, V.; Criqui, M.H.; Ginsberg, H.N.; Goldberg, A.C.; Howard, W.J.; Jacobson, M.S.; Kris-Etherton, P.M.; et al. Triglycerides and Cardiovascular Disease. Circulation 2011, 123, 2292–2333. [Google Scholar] [CrossRef]
- Pamplona, R.; Barja, G.; Portero-Otin, M. Membrane fatty acid unsaturation, protection against oxidative stress, and maximum life span: A homeoviscous-longevity adaptation? Ann. N. Y. Acad. Sci. 2002, 959, 475–490. [Google Scholar] [CrossRef]
- Hulbert, A.J.; Pamplona, R.; Buffenstein, R.; Buttemer, W.A. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol. Rev. 2007, 87, 1175–1213. [Google Scholar] [CrossRef]
- Pamplona, R. Membrane phospholipids, lipoxidative damage and molecular integrity: A causal role in aging and longevity. Biochim. Biophys. Acta (BBA) Bioenerg. 2008, 1777, 1249–1262. [Google Scholar] [CrossRef]
- Pamplona, R. Advanced lipoxidation end-products. Chem. Biol. Interact. 2011, 192, 14–20. [Google Scholar] [CrossRef]
- Chu, S.H.; Huang, M.; Kelly, R.S.; Benedetti, E.; Siddiqui, J.K.; Zeleznik, O.A.; Pereira, A.; Herrington, D.; Wheelock, C.E.; Krumsiek, J.; et al. Integration of Metabolomic and Other Omics Data in Population-Based Study Designs: An Epidemiological Perspective. Metabolites 2019, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Silsirivanit, A.; Phoomak, C.; Teeravirote, K.; Wattanavises, S.; Seubwai, W.; Saengboonmee, C.; Zhan, Z.; Inokuchi, J.I.; Suzuki, A.; Wongkham, S. Overexpression of HexCer and LacCer containing 2-hydroxylated fatty acids in cholangiocarcinoma and the association of the increase of LacCer (d18:1-h23:0) with shorter survival of the patients. Glycoconj. J. 2019, 36, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Tamiya-Koizumi, K.; Yamada, M.; Murate, T.; Kannagi, R.; Kyogashima, M. Hypoxia remodels the composition of the constituent ceramide species of HexCer and Hex2Cer with phytosphingosine and hydroxy fatty acids in human colon cancer LS174T cells. Glycoconj. J. 2015, 32, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Corbacho, M.J.; Jenkins, R.W.; Clarke, C.J.; Hannun, Y.A.; Obeid, L.M.; Snider, A.J.; Siskind, L.J. Accumulation of long-chain glycosphingolipids during aging is prevented by caloric restriction. PLoS ONE 2011, 6, e20411. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro, R.; Falasca, M. Lysophosphatidylinositol signalling: New wine from an old bottle. Biochim. Biophys. Acta 2012, 1821, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Arifin, S.A.; Falasca, M. Lysophosphatidylinositol Signalling and Metabolic Diseases. Metabolites 2016, 6, 6. [Google Scholar] [CrossRef]
- Liebisch, G.; Vizcaíno, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J.O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Esliger, D.W.; Rowlands, A.V.; Hurst, T.L.; Catt, M.; Murray, P.; Eston, R.G. Validation of the GENEA Accelerometer. Med. Sci. Sports Exerc. 2011, 43, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Lofgren, L.; Forsberg, G.B.; Stahlman, M. The BUME method: A new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Sci. Rep. 2016, 6, 27688. [Google Scholar] [CrossRef] [PubMed]
- Lofgren, L.; Stahlman, M.; Forsberg, G.B.; Saarinen, S.; Nilsson, R.; Hansson, G.I. The BUME method: A novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 2012, 53, 1690–1700. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal. Chem. Trac 2014, 61, 192–206. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. LC-MS-Based Lipidomics and Automated Identification of Lipids Using the LipidBlast In-Silico MS/MS Library. Methods Mol. Biol. 2017, 1609, 149–170. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Gagnebin, Y.; Tonoli, D.; Lescuyer, P.; Ponte, B.; de Seigneux, S.; Martin, P.Y.; Schappler, J.; Boccard, J.; Rudaz, S. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Anal. Chim. Acta 2017, 955, 27–35. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Ni, Z.; Fedorova, M. LipidLynxX: Lipid annotations converter for large scale lipidomics and epilipidomics datasets. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Taverna, F.; Goveia, J.; Karakach, T.K.; Khan, S.; Rohlenova, K.; Treps, L.; Subramanian, A.; Schoonjans, L.; Dewerchin, M.; Eelen, G.; et al. BIOMEX: An interactive workflow for (single cell) omics data interpretation and visualization. Nucleic Acids Res. 2020. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Gerl, M.J.; Klose, C.; Surma, M.A.; Fernandez, C.; Melander, O.; Männistö, S.; Borodulin, K.; Havulinna, A.S.; Salomaa, V.; Ikonen, E.; et al. Machine learning of human plasma lipidomes for obesity estimation in a large population cohort. PLoS Biol. 2019, 17, e3000443. [Google Scholar] [CrossRef]
- Chapman, M.J.; Orsoni, A.; Tan, R.; Mellett, N.A.; Nguyen, A.; Robillard, P.; Giral, P.; Thérond, P.; Meikle, P.J. LDL subclass lipidomics in atherogenic dyslipidemia: Effect of statin therapy on bioactive lipids and dense LDL. J. Lipid Res. 2020, 61, 911–932. [Google Scholar] [CrossRef]
- Takeda, H.; Izumi, Y.; Tamura, S.; Koike, T.; Koike, Y.; Shiomi, M.; Bamba, T. Lipid Profiling of Serum and Lipoprotein Fractions in Response to Pitavastatin Using an Animal Model of Familial Hypercholesterolemia. J. Proteome Res. 2020, 19, 1100–1108. [Google Scholar] [CrossRef]
- Lu, J.; Lam, S.M.; Wan, Q.; Shi, L.; Huo, Y.; Chen, L.; Tang, X.; Li, B.; Wu, X.; Peng, K.; et al. High-Coverage Targeted Lipidomics Reveals Novel Serum Lipid Predictors and Lipid Pathway Dysregulation Antecedent to Type 2 Diabetes Onset in Normoglycemic Chinese Adults. Diabetes Care 2019, 42, 2117–2126. [Google Scholar] [CrossRef]
- Pino, M.F.; Stephens, N.A.; Eroshkin, A.M.; Yi, F.; Hodges, A.; Cornnell, H.H.; Pratley, R.E.; Smith, S.R.; Wang, M.; Han, X.; et al. Endurance training remodels skeletal muscle phospholipid composition and increases intrinsic mitochondrial respiration in men with Type 2 diabetes. Physiol. Genom. 2019, 51, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Fikenzer, K.; Fikenzer, S.; Laufs, U.; Werner, C. Effects of endurance training on serum lipids. Vasc. Pharm. 2018, 101, 9–20. [Google Scholar] [CrossRef]
- Chua, E.C.; Shui, G.; Lee, I.T.; Lau, P.; Tan, L.C.; Yeo, S.C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473. [Google Scholar] [CrossRef] [PubMed]
- Gooley, J.J. Circadian regulation of lipid metabolism. Proc. Nutr. Soc. 2016, 75, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Hyötyläinen, T.; Orešič, M. Optimizing the lipidomics workflow for clinical studies—Practical considerations. Anal. Bioanal. Chem. 2015, 407, 4973–4993. [Google Scholar] [CrossRef]
- Schielzeth, H. Simple means to improve the interpretability of regression coefficients. Methods Ecol. Evol. 2010, 1, 103–113. [Google Scholar] [CrossRef]
- Introduction to SAS. UCLA: Statistical Consulting Group. Available online: https://stats.idre.ucla.edu/other/mult-pkg/faq/general/faqhow-do-i-interpret-a-regression-model-when-some-variables-are-log-transformed/ (accessed on 16 September 2020).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 April 2020).
- Henglin, M.; Niiranen, T.; Watrous, J.D.; Lagerborg, K.A.; Antonelli, J.; Claggett, B.L.; Demosthenes, E.J.; von Jeinsen, B.; Demler, O.; Vasan, R.S.; et al. A Single Visualization Technique for Displaying Multiple Metabolite-Phenotype Associations. Metabolites 2019, 9, 128. [Google Scholar] [CrossRef] [PubMed]
| Young | Aged | |||
|---|---|---|---|---|
| Female | Male | Female | Male | |
| Participants, n (%) | 32 (21.3) | 41 (27.3) | 37 (24.7) | 40 (26.7) |
| Anthropometry, mean ± SD | ||||
| Age (years) | 25.1 ± 2.3 | 25.1 ± 2.8 | 74.0 ± 2.4 | 73.9 ± 2.5 |
| Body mass (kg) | 60.5 ± 9.0 | 76.7 ± 9.5 | 61.8 ± 7.5 | 74.9 ± 8.3 |
| Body fat mass (%) | 23.1 ± 6.9 | 14.7 ± 5.3 | 30.4 ± 7.5 | 24.3 ± 6.1 |
| Body mass index (kg/m2) | 21.5 ± 2.9 | 23.7 ± 2.3 | 23.5 ± 3.0 | 24.9 ± 2.6 |
| Systolic blood pressure (mmHg) | 111 ± 8 | 126 ± 10 | 137 ± 12 | 133 ± 13 |
| Diastolic blood pressure (mmHg) | 71 ± 8 | 71 ± 7 | 80 ± 8 | 82 ± 8 |
| Smoking status, n (%) | ||||
| Never smoked | 31 (97) | 40 (98) | 26 (70) | 21 (52) |
| Ex-smokers (quit > 10 years ago) | 1 (3) | 1 (2) | 11 (30) | 19 (48) |
| Physical activity (PA) levels, mean ± SD | ||||
| Daily total PA (min) | 282.3 ± 56.1 | 274.7 ± 69.3 | 257.2 ± 87.3 | 237.7 ± 75.2 |
| Daily moderate-to-vigorous PA (min) | 190.7 ± 45.1 | 186.7 ± 53.1 | 142.4 ± 63.0 | 140.8 ± 55.3 |
| Biochemical parameters, mean ± SD | ||||
| Fasting time prior to blood sampling (h) | 6.0 ± 1.6 | 5.6 ± 2.0 | 6.6 ± 3.7 | 7.4 ± 4.5 |
| Total cholesterol (mmol/L) | 4.96 ± 0.78 | 4.68 ± 0.95 | 6.49 ± 0.80 | 5.96 ± 1.11 |
| LDL-C (mmol/L) | 2.62 ± 0.48 | 2.52 ± 0.56 | 3.56 ± 0.59 | 3.40 ± 0.73 |
| HDL-C (mmol/L) | 1.81 ± 0.42 | 1.43 ± 0.24 | 1.91 ± 0.35 | 1.58 ± 0.33 |
| Triglycerides (mmol/L) | 1.08 ± 0.53 | 1.33 ± 0.80 | 1.41 ± 1.06 | 1.35 ± 0.45 |
| HbA1c (%) | 5.0 ± 0.2 | 5.0 ± 0.2 | 5.4 ± 0.3 | 5.3 ± 0.3 |
| Cardiovascular medications, n (%) | ||||
| Antihypertensives | 0 (0) | 0(0) | 9 (16) | 19 (35) |
| Low-dose aspirin | 0 (0) | 0 (0) | 2 (5) | 4 (10) |
| Statins | 0 (0) | 0 (0) | 3 (8) | 6 (15) |
| Hormonal medications, n (%) | ||||
| Oestrogen/HRT | 4 (13) | 0 (0) | 5 (14) | 0 (0) |
| 5α-reductase inhibitors | 0 (0) | 0 (0) | 0 (0) | 4 (10) |
| Thyroid hormones | 0 (0) | 0 (0) | 3 (8) | 3 (8) |
| Psychiatric medications, n (%) | ||||
| Antidepressants | 1 (3) | 1 (2) | 1 (3) | 0 (0) |
| Z-drugs | 0 (0) | 0 (0) | 3 (8) | 0 (0) |
| Other medications, n (%) | 3 (9) | 6 (12) | 31 (49) | 24 (42) |
| Lipid Subclass, Full Name | Lipid Subclass, Abbreviation | Identified Lipid Species, n |
|---|---|---|
| Diglycerides | DG | 2 |
| Triglycerides | TG | 58 |
| Cholesterol esters | CE | 5 |
| Glycerophosphocholines | PC | 42 |
| Alkyl-glycerophosphocholines | PC-O | 16 |
| Lyso-glycerophosphocholines | LPC | 15 |
| Glycerophosphoinositols | PI | 7 |
| Lyso-glycerophosphoinositols | LPI | 2 |
| Glycerophospoethanolamines | PE | 11 |
| Alkyl-glycerophosphoethanolamines | PE-O | 15 |
| Alkenyl-glycerophosphoethanolamines | PE-P | 1 |
| Lyso-glycerophosphoethanolamines | LPE | 7 |
| Lyso-alkyl-glycerophosphoethanolamines | LPE-O | 2 |
| Ceramides | Cer | 6 |
| Sphingomyelins | SM | 24 |
| Glycosphingolipids | GSL | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrard, J.; Gallart-Ayala, H.; Infanger, D.; Teav, T.; Wagner, J.; Knaier, R.; Colledge, F.; Streese, L.; Königstein, K.; Hinrichs, T.; et al. Metabolic View on Human Healthspan: A Lipidome-Wide Association Study. Metabolites 2021, 11, 287. https://doi.org/10.3390/metabo11050287
Carrard J, Gallart-Ayala H, Infanger D, Teav T, Wagner J, Knaier R, Colledge F, Streese L, Königstein K, Hinrichs T, et al. Metabolic View on Human Healthspan: A Lipidome-Wide Association Study. Metabolites. 2021; 11(5):287. https://doi.org/10.3390/metabo11050287
Chicago/Turabian StyleCarrard, Justin, Hector Gallart-Ayala, Denis Infanger, Tony Teav, Jonathan Wagner, Raphael Knaier, Flora Colledge, Lukas Streese, Karsten Königstein, Timo Hinrichs, and et al. 2021. "Metabolic View on Human Healthspan: A Lipidome-Wide Association Study" Metabolites 11, no. 5: 287. https://doi.org/10.3390/metabo11050287
APA StyleCarrard, J., Gallart-Ayala, H., Infanger, D., Teav, T., Wagner, J., Knaier, R., Colledge, F., Streese, L., Königstein, K., Hinrichs, T., Hanssen, H., Ivanisevic, J., & Schmidt-Trucksäss, A. (2021). Metabolic View on Human Healthspan: A Lipidome-Wide Association Study. Metabolites, 11(5), 287. https://doi.org/10.3390/metabo11050287













