Control of n-Butanol Induced Lipidome Adaptations in E. coli
Abstract
1. Introduction
2. Results
2.1. Simplification of the Lipid Extraction Allows for More Detailed Lipidome Analysis
2.2. Random Forest Analysis Identifies Key Lipids and Lipid Genes
2.3. The Lipidome Is Organized in Clusters of Closely Connected Lipid Species
2.4. Validation of Metabolic Pivot Points
2.5. Linking Lipid Related Genes to n-Butanol Tolerance
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Bacterial Strains, Growth Conditions and Plasmids
4.3. Lipid Extraction
4.4. Liquid Chromatography Mass Spectrometry of Lipids
4.5. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, C.; Zhang, Y.; Li, Y. Metabolic Engineering for the Production of Butanol, a Potential Advanced Biofuel, from Renewable Resources. Biochem. Soc. Trans. 2020, 48, 2283–2293. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Yao, M.; Liu, H.; Lee, C.F.; Ji, J. Progress in the Production and Application of N-Butanol as a Biofuel. Renew. Sustain. Energy Rev. 2011, 15, 4080–4106. [Google Scholar] [CrossRef]
- Nair, P.; Meenakshi, H.N. Review on the Synthesis, Performance and Trends of Butanol: A Cleaner Fuel Additive for Gasoline. Int. J. Ambient Energy 2021, 1–17. [Google Scholar] [CrossRef]
- Mehlman, M.A. Dangerous Properties of Petroleum-Refining Products: Carcinogenicity of Motor Fuels (Gasoline). Teratog. Carcinog. Mutagen. 1990, 10, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Caprino, L.; Togna, G.I. Potential Health Effects of Gasoline and Its Constituents: A Review of Current Literature (1990–1997) on Toxicological Data. Environ. Health Perspect. 1998, 106, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Ekpenyong, C.E.; Asuquo, A.E. Recent Advances in Occupational and Environmental Health Hazards of Workers Exposed to Gasoline Compounds. Int. J. Occup. Med. Environ. Health 2017, 30, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kang, S.; Li, J.; Li, Z.; Chang, J.; Xu, Y.; Max Lu, G.; Sun, C. Near Zero-Waste Biofuel Production from Bioderived Polyhydroxybutyrate. Fuel 2021, 286, 119405. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Patel, M.K. Plastics Derived from Biological Sources: Present and Future: A Technical and Environmental Review. Chem. Rev. 2012, 112, 2082–2099. [Google Scholar] [CrossRef]
- Li, S.; Huang, L.; Ke, C.; Pang, Z.; Liu, L. Pathway Dissection, Regulation, Engineering and Application: Lessons Learned from Biobutanol Production by Solventogenic Clostridia. Biotechnol. Biofuels 2020, 13, 39. [Google Scholar] [CrossRef]
- Du, Y.; Zou, W.; Zhang, K.; Ye, G.; Yang, J. Advances and Applications of Clostridium Co-Culture Systems in Biotechnology. Front. Microbiol. 2020, 11, 2824. [Google Scholar] [CrossRef] [PubMed]
- Volesky, B.; Szczesny, T. Bacterial Conversion of Pentose Sugars to Acetone and Butanol. Adv. Biochem. Eng. Biotechnol. 1983, 27, 101–118. [Google Scholar] [CrossRef]
- Azambuja, S.P.H.; Goldbeck, R. Butanol Production by Saccharomyces cerevisiae: Perspectives, Strategies and Challenges. World J. Microbiol. Biotechnol. 2020, 36, 48. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Cann, A.F.; Connor, M.R.; Shen, C.R.; Smith, K.M.; Brynildsen, M.P.; Chou, K.J.Y.; Hanai, T.; Liao, J.C. Metabolic Engineering of Escherichia coli for 1-Butanol Production. Metab. Eng. 2008, 10, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Liao, J.C. Metabolic Engineering of Escherichia coli for 1-Butanol and 1-Propanol Production via the Keto-Acid Pathways. Metab. Eng. 2008, 10, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Sukhozhenko, A.V.; Shakulov, R.S.; Debabov, V.G. Metabolic Engineering of Escherichia coli for 1-Butanol Biosynthesis through the Inverted Aerobic Fatty Acid β-Oxidation Pathway. Biotechnol. Lett. 2012, 34, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Kujawska, A.; Kujawski, J.; Bryjak, M.; Kujawski, W. ABE Fermentation Products Recovery Methods-A Review. Renew. Sustain. Energy Rev. 2015, 48, 648–661. [Google Scholar] [CrossRef]
- Milestone, N.B.; Bibby, D.M. Concentration of Alcohols by Adsorption on Silicate. J. Chem. Technol. Biotechnol. 1981, 31, 732–736. [Google Scholar] [CrossRef]
- Groot, W.J.; Soedjak, H.S.; Donck, P.B.; van der Lans, R.G.J.M.; Luyben, K.C.A.M.; Timmer, J.M.K. Butanol Recovery from Fermentations by Liquid-Liquid Extraction and Membrane Solvent Extraction. Bioprocess Eng. 1990, 5, 203–216. [Google Scholar] [CrossRef]
- Kanno, M.; Katayama, T.; Tamaki, H.; Mitani, Y.; Meng, X.-Y.; Hori, T.; Narihiro, T.; Morita, N.; Hoshino, T.; Yumoto, I.; et al. Isolation of Butanol-and Isobutanol-Tolerant Bacteria and Physiological Characterization of Their Butanol Tolerance. Appl. Environ. Microbiol. 2013, 79, 6998–7005. [Google Scholar] [CrossRef]
- Heipieper, H.J.; Neumann, G.; Cornelissen, S.; Meinhardt, F. Solvent-Tolerant Bacteria for Biotransformations in Two-Phase Fermentation Systems. Appl. Microbiol. Biotechnol. 2007, 74, 961–973. [Google Scholar] [CrossRef]
- Sardessai, Y.; Bhosle, S. Organic Solvent Tolerant Bacteria in Mangrove Ecosystem. Curr. Sci. 2002, 82, 622–623. [Google Scholar]
- Guo, J.; Ho, J.C.S.; Chin, H.; Mark, A.E.; Zhou, C.; Kjelleberg, S.; Liedberg, B.; Parikh, A.N.; Cho, N.-J.; Hinks, J.; et al. Response of Microbial Membranes to Butanol: Interdigitation vs. Disorder. Phys. Chem. Chem. Phys. 2019, 21, 11903–11915. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.A.; Boyarskiy, S.; Yamada, M.R.; Kong, N.; Bauer, S.; Tullman-Ercek, D. Enhancing Tolerance to Short-Chain Alcohols by Engineering the Escherichia coli AcrB Efflux Pump to Secrete the Non-Native Substrate n-Butanol. ACS Synth. Biol. 2013, 3, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Bui, L.M.; Lee, J.Y.; Geraldi, A.; Rahman, Z.; Lee, J.H.; Kim, S.C. Improved n-Butanol Tolerance in Escherichia coli by Controlling Membrane Related Functions. J. Biotechnol. 2015, 204, 33–44. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rau, M.H.; Calero, P.; Lennen, R.M.; Long, K.S.; Nielsen, A.T. Genome-Wide Escherichia coli Stress Response and Improved Tolerance towards Industrially Relevant Chemicals. Microb. Cell Factories 2016, 15, 176. [Google Scholar] [CrossRef]
- Reyes, L.H.; Almario, M.P.; Winkler, J.; Orozco, M.M.; Kao, K.C. Visualizing Evolution in Real Time to Determine the Molecular Mechanisms of N-Butanol Tolerance in Escherichia coli. Metab. Eng. 2012, 14, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Patakova, P.; Kolek, J.; Jureckova, K.; Branska, B.; Sedlar, K.; Vasylkivska, M.; Provaznik, I. Deeper below the Surface-Transcriptional Changes in Selected Genes of Clostridium beijerinckii in Response to Butanol Shock. MicrobiologyOpen 2021, 10, e1146. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006-0008. [Google Scholar] [CrossRef] [PubMed]
- Jeucken, A.; Molenaar, M.R.; van de Lest, C.H.A.; Jansen, J.W.A.; Helms, J.B.; Brouwers, J.F. A Comprehensive Functional Characterization of Escherichia coli Lipid Genes. Cell Rep. 2019, 27, 1597–1606.e2. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.B.; Snyder, A.P.; Harden, C.S. Characterization of Bacterial Phospholipids by Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem 1995, 67, 1824–1830. [Google Scholar] [CrossRef]
- Gidden, J.; Denson, J.; Liyanage, R.; Ivey, D.M.; Lay, J.O. Lipid Compositions in Escherichia coli and Bacillus subtilis During Growth as Determined by MALDI-TOF and TOF/TOF Mass Spectrometry. Int. J. Mass Spectrom. 2009, 283, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Jeucken, A.; Helms, J.B.; Brouwers, J.F. Cardiolipin Synthases of Escherichia coli Have Phospholipid Class Specific Phospholipase D Activity Dependent on Endogenous and Foreign Phospholipids. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2018, 1863, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Yeh, J.I.; Chinte, U.; Du, S. Structure of Glycerol-3-Phosphate Dehydrogenase, an Essential Monotopic Membrane Enzyme Involved in Respiration and Metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 3280–3285. [Google Scholar] [CrossRef] [PubMed]
- Karpievitch, Y.V.; Hill, E.G.; Leclerc, A.P.; Dabney, A.R.; Almeida, J.S. An Introspective Comparison of Random Forest-Based Classifiers for the Analysis of Cluster-Correlated Data by Way of RF++. PLoS ONE 2009, 4, e7087. [Google Scholar] [CrossRef] [PubMed]
- Züllig, T.; Köfeler, H.C. High Resolution Mass Spectrometry in Lipidomics. Mass Spectrom. Rev. 2021, 40, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Poger, D.; Mark, A.E. A Ring to Rule Them All: The Effect of Cyclopropane Fatty Acids on the Fluidity of Lipid Bilayers. J. Phys. Chem. B 2015, 119, 5487–5495. [Google Scholar] [CrossRef] [PubMed]
- Pini, C.-V.; Bernal, P.; Godoy, P.; Ramos, J.-L.; Segura, A. Cyclopropane Fatty Acids Are Involved in Organic Solvent Tolerance but Not in Acid Stress Resistance in Pseudomonas putida DOT-T1E. Microb. Biotechnol. 2009, 2, 253–261. [Google Scholar] [CrossRef]
- Chang, Y.Y.; Cronan, J.E. Membrane Cyclopropane Fatty Acid Content Is a Major Factor in Acid Resistance of Escherichia coli. Mol. Microbiol. 1999, 33, 249–259. [Google Scholar] [CrossRef]
- Kim, B.H.; Kim, S.; Kim, H.G.; Lee, J.; Lee, I.S.; Park, Y.K. The Formation of Cyclopropane Fatty Acids in Salmonella enterica Serovar Typhimurium. Microbiol 2005, 151, 209–218. [Google Scholar] [CrossRef]
- Grogan, D.W.; Cronan, J.E. Cyclopropane Ring Formation in Membrane Lipids of Bacteria. Microbiol. Mol. Biol. Rev. MMBR 1997, 61, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Faber, H.R.; Worthylake, D.; Meadow, N.D.; Roseman, S.; Pettigrew, D.W.; Remington, S.J. Structure of the Regulatory Complex of Escherichia coli IIIGlc with Glycerol Kinase. Science 1993, 259, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Zwaig, N.; Lin, E.C. Feedback Inhibition of Glycerol Kinase, a Catabolic Enzyme in Escherichia coli. Science 1966, 153, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Khil, P.P.; Camerini-Otero, R.D. Over 1000 Genes Are Involved in the DNA Damage Response of Escherichia coli. Mol. Microbiol. 2002, 44, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Thorner, J.W.; Paulus, H. Catalytic and Allosteric Properties of Glycerol Kinase from Escherichia coli. J. Biol. Chem. 1973, 248, 3922–3932. [Google Scholar] [CrossRef]
- Wegener, M.; Vogtmann, K.; Huber, M.; Laass, S.; Soppa, J. The GlpD Gene Is a Novel Reporter Gene for E. coli That Is Superior to Established Reporter Genes like LacZ and GusA. J. Microbiol. Methods 2016, 131, 181–187. [Google Scholar] [CrossRef]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete Set of ORF Clones of Escherichia coli ASKA Library (a Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef]
- Brouwers, J.F.; Aalberts, M.; Jansen, J.W.A.; van Niel, G.; Wauben, M.H.; Stout, T.A.E.; Helms, J.B.; Stoorvogel, W. Distinct Lipid Compositions of Two Types of Human Prostasomes. Proteomics 2013, 13, 10–11. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open Source Software for Rapid Proteomics Tools Development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2011. [Google Scholar]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from Sample Preparation to Data Analysis: A Primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef] [PubMed]
- Kayala, M.A.; Baldi, P. Cyber-T Web Server: Differential Analysis of High-Throughput Data. Nucleic Acids Res. 2012, 40, W553–W559. [Google Scholar] [CrossRef] [PubMed]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining, Fourth Edition: Practical Machine Learning Tools and Techniques, 4th ed.; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2016; ISBN 978-0-12-804291-5. [Google Scholar]

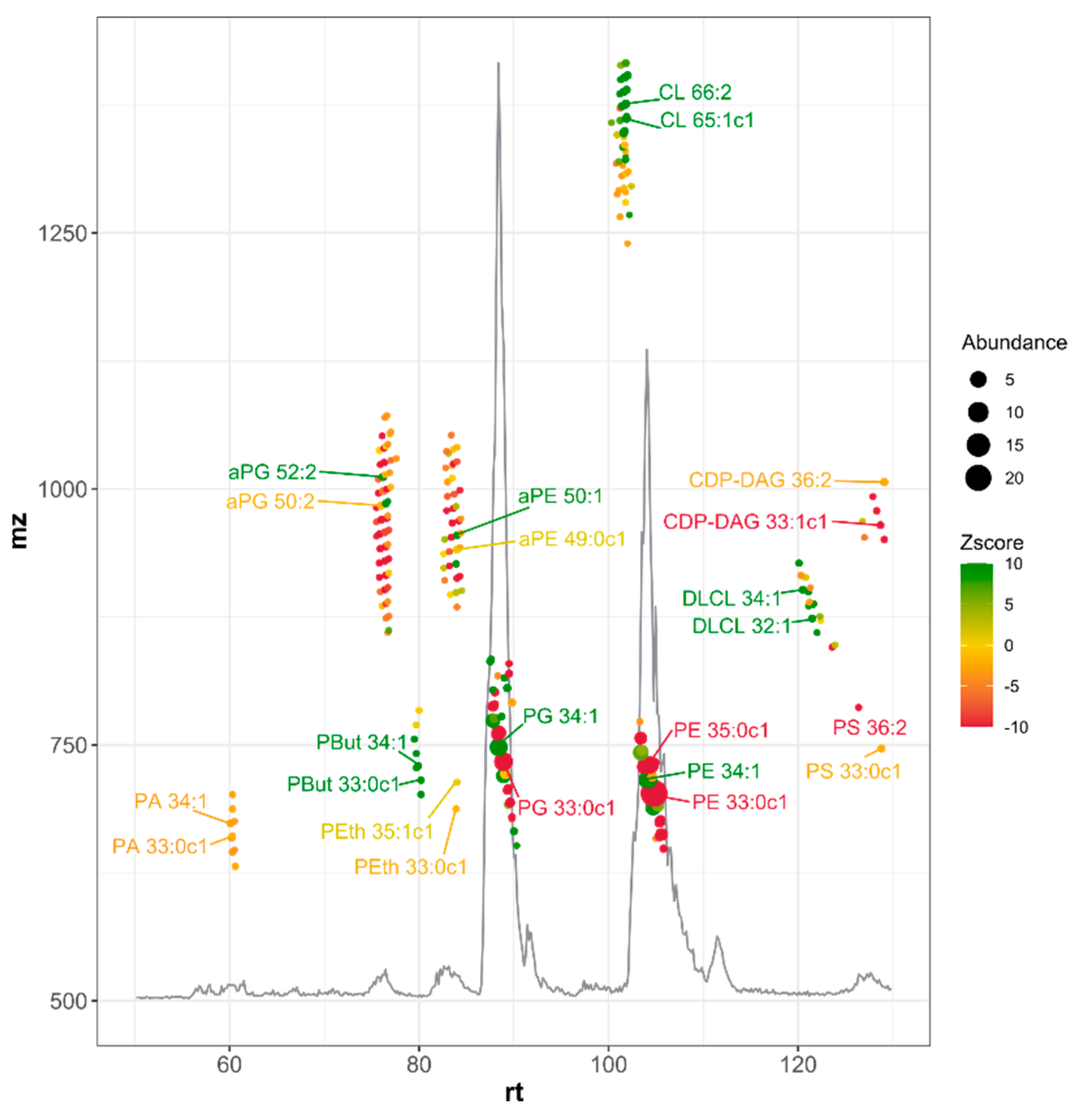

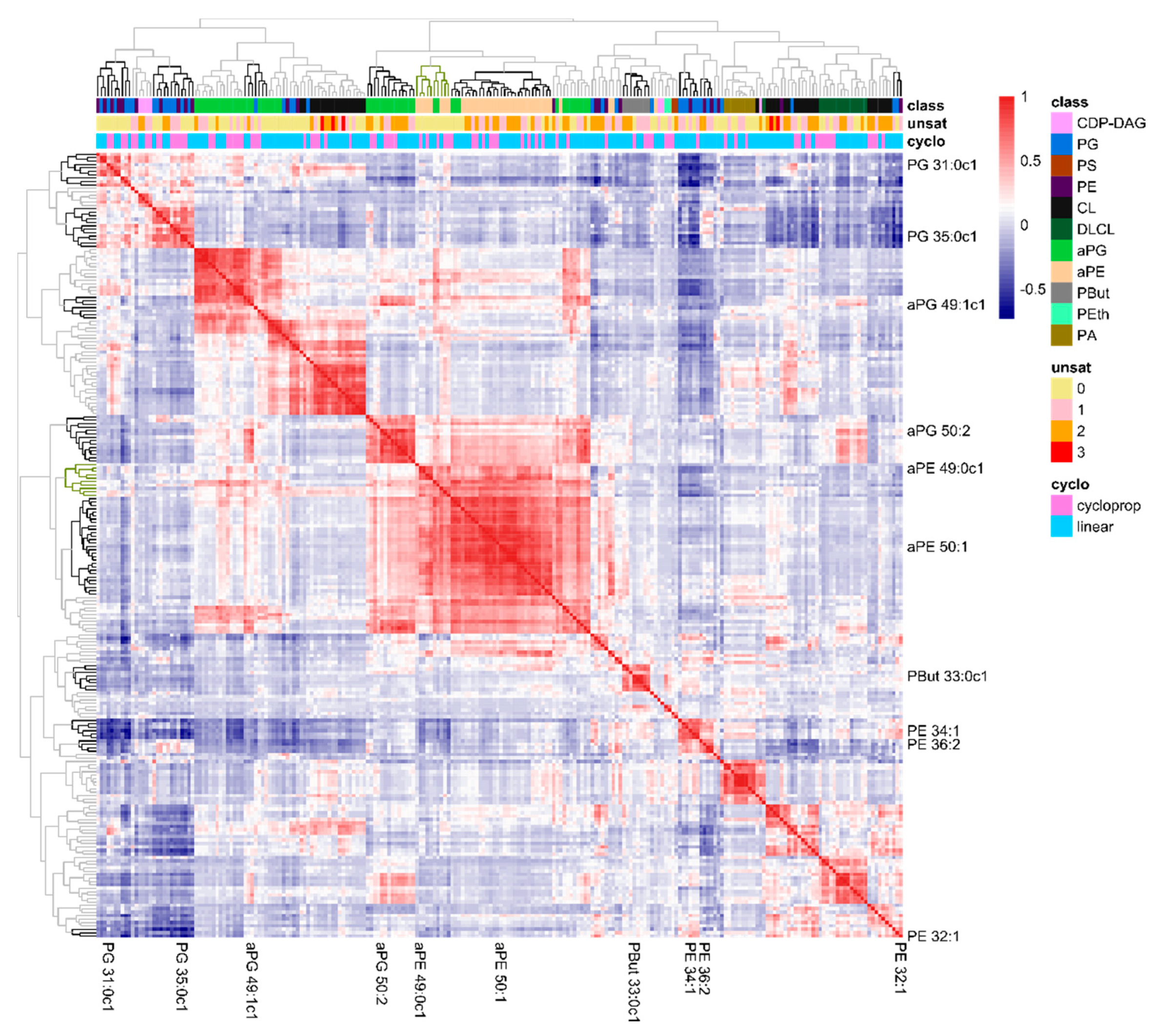
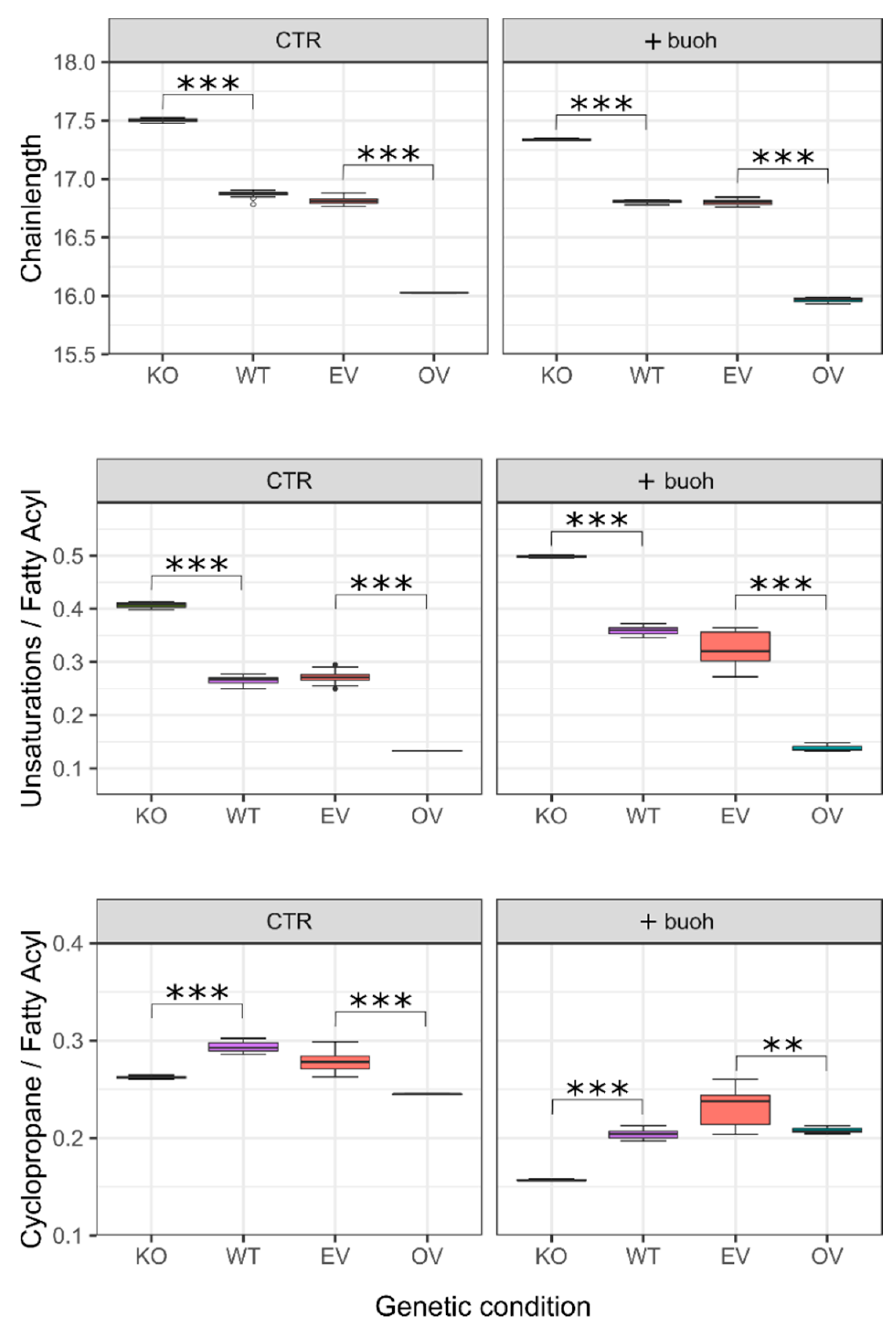

| Lipid Species | Average (%) | Max. (%) | Min. (%) | Genes 1 (Extremes) |
|---|---|---|---|---|
| PE 34:1 | 7.67 | 17.41 | 0.72 | glpD (OV); fabH (OV) |
| PE 32:1 | 5.75 | 24.82 | 0.72 | cfa (KO); cfa (OV) |
| PE 36:2 | 4.20 | 12.09 | 0.84 | fabH (KO); fabH (OV) |
| PG 35:0c1 | 2.60 | 10.47 | 0.20 | glpR (KO); aas (OV) |
| PG 31:0c1 | 0.35 | 1.21 | 0.05 | fabH (OV); fabH (KO) |
| aPG 50:2 | 0.26 | 2.94 | 0.03 | fadL (OV); fabF (OV) |
| aPE 49:0c1 | 0.15 | 0.91 | 0.01 | aas (OV); cfa (KO) |
| aPG 49:1c1 | 0.10 | 1.00 | 0.01 | pldA (OV); fabH (KO) |
| PBut 33:0c1 | 0.07 | 2.99 | 0.00 | clsB (OV); fadD (KO) |
| aPE 50:1 | 0.07 | 1.34 | 0.00 | aas (OV); clsA (OV) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeucken, A.; Zhou, M.; Wösten, M.M.S.M.; Brouwers, J.F. Control of n-Butanol Induced Lipidome Adaptations in E. coli. Metabolites 2021, 11, 286. https://doi.org/10.3390/metabo11050286
Jeucken A, Zhou M, Wösten MMSM, Brouwers JF. Control of n-Butanol Induced Lipidome Adaptations in E. coli. Metabolites. 2021; 11(5):286. https://doi.org/10.3390/metabo11050286
Chicago/Turabian StyleJeucken, Aike, Miaomiao Zhou, Marc M. S. M. Wösten, and Jos F. Brouwers. 2021. "Control of n-Butanol Induced Lipidome Adaptations in E. coli" Metabolites 11, no. 5: 286. https://doi.org/10.3390/metabo11050286
APA StyleJeucken, A., Zhou, M., Wösten, M. M. S. M., & Brouwers, J. F. (2021). Control of n-Butanol Induced Lipidome Adaptations in E. coli. Metabolites, 11(5), 286. https://doi.org/10.3390/metabo11050286






