Metabolic Classification and Intervention Opportunities for Tumor Energy Dysfunction
Abstract
1. Introduction
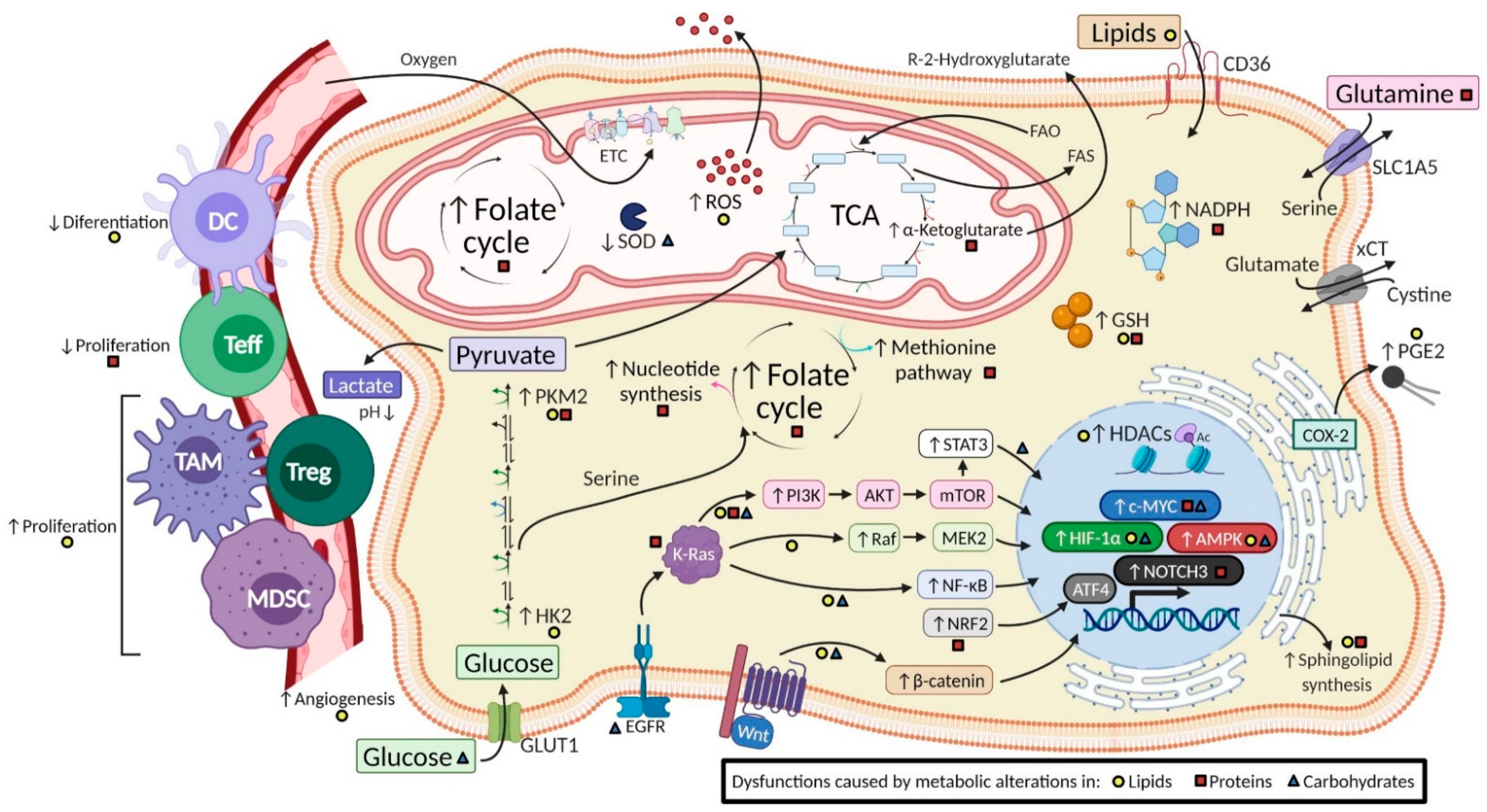
2. Evidence of Changes in the Metabolic Pathways of Tumor Energy Dysfunction
2.1. Tumors with Transformed Lipid Metabolism Pathways
2.2. Tumors with Altered Amino Acid Metabolism
2.3. Tumors with Carbohydrate Pathway Modifications
2.4. Dysregulated pH as a Hallmark of Cancer
2.5. Tumors with Hypoxic Adaptation
3. Intervention Opportunities from a Metabolic View of Cancer
3.1. Intervention on the Fatty Acid Pathway
3.2. Intervention in the Protein Pathway
3.3. Intervention in the Carbohydrate Pathway
3.4. Chemotherapy and the Fundamental Role of Immunotherapy
3.5. Critical Role of the Tumor Microenvironment
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, X.; Chen, S.; Yu, D. Metabolic reprogramming of chemoresistant cancer cells and the potential significance of metabolic regulation in the reversal of cancer chemoresistance. Metabolites 2020, 10, 289. [Google Scholar] [CrossRef]
- Elia, I.; Haigis, M.C. Metabolites and the tumour microenvironment: From cellular mechanisms to systemic metabolism. Nat. Metab. 2021, 3. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Shelton, L.M. Cancer as a metabolic disease. Nutr. Metab. 2010, 7, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Lehúede, C.; Dupuy, F.; Rabinovitch, R.; Jones, R.G.; Siegel, P.M. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 2016, 76, 5201–5208. [Google Scholar] [CrossRef] [PubMed]
- De Berardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef]
- Borriello, A.; Della Ragione, F. The new anticancer era: Tumor metabolism targeting. Cell Cycle 2017, 16, 310–311. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften 1924, 12, 1131–1137. [Google Scholar] [CrossRef]
- Ben-Haim, S.; Ell, P. 18F-FDG PET and PET/CT in the evaluation of cancer treatment response. J. Nucl. Med. 2009, 50, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, S.; Li, Y.; Tang, Z.; Kong, W. Hexokinase 2 overexpression promotes the proliferation and survival of laryngeal squamous cell carcinoma. Tumor Biol. 2014, 35, 3743–3753. [Google Scholar] [CrossRef]
- Roda, N.; Gambino, V.; Giorgio, M. Metabolic Constrains Rule Metastasis Progression. Cells 2020, 9, 2081. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Rose, C.J.; Waterton, J.C.; Carano, R.A.D.; Parker, G.J.M.; Jackson, A. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin. Cancer Res. 2015, 21, 249–257. [Google Scholar] [CrossRef]
- Yang, C.; Huang, X.; Liu, Z.; Qin, W.; Wang, C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 2020, 14, 896–913. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, R.; Liang, F.; Zhang, L.; Liang, X. Identification of Metabolism-Associated Prostate Cancer Subtypes and Construction of a Prognostic Risk Model. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, H.Z.; Peng, R.Y.; Xu, F.; Wang, F.; Zhao, Q. Metabolism-Associated Molecular Classification of Colorectal Cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Vander Linden, C.; Corbet, C. Reconciling environment-mediated metabolic heterogeneity with the oncogene-driven cancer paradigm in precision oncology. Semin. Cell Dev. Biol. 2020, 98, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Camelo, F.; Le, A. The intricate metabolism of pancreatic cancers. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2018; Volume 1063, pp. 73–81. [Google Scholar]
- Martinez, C.A.; Scafoglio, C. Heterogeneity of glucose transport in lung cancer. Biomolecules 2020, 10, 868. [Google Scholar] [CrossRef]
- Cappelletti, V.; Iorio, E.; Miodini, P.; Silvestri, M.; Dugo, M.; Daidone, M.G. Metabolic Footprints and Molecular Subtypes in Breast Cancer. Dis. Markers 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Kidd, E.A.; Grigsby, P.W. Intratumoral metabolic heterogeneity of cervical cancer. Clin. Cancer Res. 2008, 14, 5236–5241. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef] [PubMed]
- Diedrich, J.D.; Herroon, M.K.; Rajagurubandara, E.; Podgorski, I. The Lipid Side of Bone Marrow Adipocytes: How Tumor Cells Adapt and Survive in Bone. Curr. Osteoporos. Rep. 2018, 16, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Bai, L.; Li, W.; Cui, J. The Lipid Metabolic Landscape of Cancers and New Therapeutic Perspectives. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Corn, K.C.; Windham, M.A.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80, 101055. [Google Scholar] [CrossRef] [PubMed]
- Attané, C.; Milhas, D.; Hoy, A.J.; Muller, C. Metabolic Remodeling Induced by Adipocytes: A New Achilles’ Heel in Invasive Breast Cancer? Curr. Med. Chem. 2018, 27, 3984–4001. [Google Scholar] [CrossRef] [PubMed]
- Menendez, J.A.; Lupu, R. Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin. Ther. Targets 2017, 21, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Fako, V.E.; Wu, X.; Pflug, B.; Liu, J.Y.; Zhang, J.T. Repositioning proton pump inhibitors as anticancer drugs by targeting the thioesterase domain of human fatty acid synthase. J. Med. Chem. 2015, 58, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Zadra, G.; Ribeiro, C.F.; Chetta, P.; Ho, Y.; Cacciatore, S.; Gao, X.; Syamala, S.; Bango, C.; Photopoulos, C.; Huang, Y.; et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 631–640. [Google Scholar] [CrossRef]
- Flavin, R.; Peluso, S.; Nguyen, P.L.; Loda, M. Fatty acid synthase as a potential therapeutic target in cancer. Futur. Oncol. 2010, 6, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Gil-De-gómez, L.; Balgoma, D.; Montero, O. Lipidomic-based advances in diagnosis and modulation of immune response to cancer. Metabolites 2020, 10, 332. [Google Scholar] [CrossRef]
- Long, Q.Q.; Yi, Y.X.; Qiu, J.; Xu, C.J.; Huang, P.L. Fatty acid synthase (FASN) levels in serum of colorectal cancer patients: Correlation with clinical outcomes. Tumor Biol. 2014, 35, 3855–3859. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.C.; Lin, C.L.; Hsu, W.Y.; Lu, I.T.; Yeh, H.Z.; Chang, C.-S.; Kao, C.H. Proton Pump Inhibitor Use is Associated with Risk of Pancreatic Cancer: A Nested Case–Control Study. Dose-Response 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Schcolnik-Cabrera, A.; Chavez-Blanco, A.; Dominguez-Gomez, G.; Juarez, M.; Lai, D.; Hua, S.; Tovar, A.R.; Diaz-Chavez, J.; Duenas-Gonzalez, A. The combination of orlistat, lonidamine and 6-diazo-5-oxo-L-norleucine induces a quiescent energetic phenotype and limits substrate flexibility in colon cancer cells. Oncol. Lett. 2020, 20, 3053–3060. [Google Scholar] [CrossRef] [PubMed]
- Schcolnik-Cabrera, A.; Chávez-Blanco, A.; Domínguez-Gómez, G.; Taja-Chayeb, L.; Morales-Barcenas, R.; Trejo-Becerril, C.; Perez-Cardenas, E.; Gonzalez-Fierro, A.; Dueñas-González, A. Orlistat as a FASN inhibitor and multitargeted agent for cancer therapy. Expert Opin. Investig. Drugs 2018, 27, 475–489. [Google Scholar] [CrossRef]
- Wang, B.Y.; Zhang, J.; Wang, J.L.; Sun, S.; Wang, Z.H.; Wang, L.P.; Zhang, Q.L.; Lv, F.F.; Cao, E.Y.; Shao, Z.M.; et al. Intermittent high dose proton pump inhibitor enhances the antitumor effects of chemotherapy in metastatic breast cancer. J. Exp. Clin. Cancer Res. 2015, 34. [Google Scholar] [CrossRef]
- Estañ, M.C.; Calviño, E.; Calvo, S.; Guillén-Guío, B.; Boyano-Adánez, M.D.C.; De Blas, E.; Rial, E.; Aller, P. Apoptotic efficacy of etomoxir in human acute myeloid leukemia cells. Cooperation with arsenic trioxide and glycolytic inhibitors, and regulation by oxidative stress and protein kinase activities. PLoS ONE 2014, 9, e0115250. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.S.; Smift, A.L.; Croteau, N.J.; Ferrick, D.A.; Wu, M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 726–734. [Google Scholar] [CrossRef]
- Loew, A.; Köhnke, T.; Rehbeil, E.; Pietzner, A.; Weylandt, K.H. A Role for Lipid Mediators in Acute Myeloid Leukemia. Int. J. Mol. Sci. 2019, 20, 2425. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.J.; Thomas, R.; Kingsley, P.J.; Shimizu, F.; Montrose, D.C.; Marnett, L.J.; Tabar, V.S.; Dannenberg, A.J.; Benezra, R. Cox-2-derived PGE2 induces Id1-dependent radiation resistance and self-renewal in experimental glioblastoma. Neuro. Oncol. 2016, 18, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fu, L.; Sun, H.; Guo, L.; Dubois, R.N. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology 2015, 149, 1884–1895. [Google Scholar] [CrossRef]
- Finetti, F.; Travelli, C.; Ercoli, J.; Colombo, G.; Buoso, E.; Trabalzini, L. Prostaglandin E2 and cancer: Insight into tumor progression and immunity. Biology 2020, 9, 434. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Svahn, S.L.; Johansson, M.E. Effects of omega-3 fatty acids on immune cells. Int. J. Mol. Sci. 2019, 20, 5028. [Google Scholar] [CrossRef] [PubMed]
- Laviano, A.; Rianda, S.; Molfino, A.; Fanelli, F.R. Omega-3 fatty acids in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 156–161. [Google Scholar] [CrossRef] [PubMed]
- García-Barros, M.; Coant, N.; Kawamori, T.; Wada, M.; Snider, A.J.; Truman, J.P.; Wu, B.X.; Furuya, H.; Clarke, C.J.; Bialkowska, A.B.; et al. Role of neutral ceramidase in colon cancer. FASEB J. 2016, 30, 4159–4171. [Google Scholar] [CrossRef] [PubMed]
- Furuya, H.; Shimizu, Y.; Kawamori, T. Sphingolipids in cancer. Cancer Metastasis Rev. 2011, 30, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, Y.; Lankadasari, M.B.; Harikumar, K.B. Acid Ceramidase: A Novel Therapeutic Target in Cancer. Curr. Top. Med. Chem. 2019, 19, 1512–1520. [Google Scholar] [CrossRef]
- Shimizu, Y.; Furuya, H.; Tamashiro, P.M.; Iino, K.; Chan, O.T.M.; Goodison, S.; Pagano, I.; Hokutan, K.; Peres, R.; Loo, L.W.M.; et al. Genetic deletion of sphingosine kinase 1 suppresses mouse breast tumor development in an HER2 transgenic model. Carcinogenesis 2018, 39, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Furuya, H.; Shimizu, Y.; Tamashiro, P.M.; Iino, K.; Bielawski, J.; Chan, O.T.M.; Pagano, I.; Kawamori, T. Sphingosine kinase 1 expression enhances colon tumor growth. J. Transl. Med. 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Furuya, H.; Tamashiro, P.M.; Shimizu, Y.; Iino, K.; Peres, R.; Chen, R.; Sun, Y.; Hannun, Y.A.; Obeid, L.M.; Kawamori, T. Sphingosine Kinase 1 expression in peritoneal macrophages is required for colon carcinogenesis. Carcinogenesis 2017, 38, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.M.; Kwon, H.J. Acid ceramidase, an emerging target for anti-cancer and anti-angiogenesis. Arch. Pharm. Res. 2019, 42, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Klobučar, M.; Grbčić, P.; Pavelić, S.K.; Jonjić, N.; Visentin, S.; Sedić, M. Acid ceramidase inhibition sensitizes human colon cancer cells to oxaliplatin through downregulation of transglutaminase 2 and β1 integrin/FAK−mediated signalling. Biochem. Biophys. Res. Commun. 2018, 503, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Pchejetski, D.; Böhler, T.; Stebbing, J.; Waxman, J. Therapeutic potential of targeting sphingosine kinase 1 in prostate cancer. Nat. Rev. Urol. 2011, 8, 569–578. [Google Scholar] [CrossRef]
- Tan, S.F.; Liu, X.; Fox, T.E.; Barth, B.M.; Sharma, A.; Turner, S.D.; Awwad, A.; Dewey, A.; Doi, K.; Spitzer, B.; et al. Acid ceramidase is upregulated in AML and represents a novel therapeutic target. Oncotarget 2016, 7, 83208–83222. [Google Scholar] [CrossRef]
- Kapitonov, D.; Allegood, J.C.; Mitchell, C.; Hait, N.C.; Almenara, J.A.; Adams, J.K.; Zipkin, R.E.; Dent, P.; Kordula, T.; Milstien, S.; et al. Targeting sphingosine kinase 1 inhibits Akt signaling, induces apoptosis, and suppresses growth of human glioblastoma cells and xenografts. Cancer Res. 2009, 69, 6915–6923. [Google Scholar] [CrossRef] [PubMed]
- Dillehay, D.L.; Webb, S.K.; Schmelz, E.M.; Merrill, A.H. Dietary sphingomyelin inhibits 1,2-dimethylhydrazine-induced colon cancer in CF1 mice. J. Nutr. 1994, 124, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.M.; Sullards, M.C.; Dillehay, D.L.; Merrill, A.H. Colonic cell proliferation and aberrant crypt foci formation are inhibited by dairy glycosphingolipids in 1,2-dimethylhydrazine-treated CF1 mice. J. Nutr. 2000, 130, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.F.; Infante, J.R. Molecular pathways: Fatty acid synthase. Clin. Cancer Res. 2015, 21, 5434–5438. [Google Scholar] [CrossRef] [PubMed]
- Kuhajda, F.P. Fatty acid synthase and cancer: New application of an old pathway. Cancer Res. 2006, 66, 5977–5980. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, M.P.; Liesa, M. The Role of Mitochondrial Fat Oxidation in Cancer Cell Proliferation and Survival. Cells 2020, 9, 2600. [Google Scholar] [CrossRef] [PubMed]
- Samudio, I.; Harmancey, R.; Fiegl, M.; Kantarjian, H.; Konopleva, M.; Korchin, B.; Kaluarachchi, K.; Bornmann, W.; Duvvuri, S.; Taegtmeyer, H.; et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Investig. 2010, 120, 142–156. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef]
- Lee, W.K.; Kolesnick, R.N. Sphingolipid abnormalities in cancer multidrug resistance: Chicken or egg? Cell. Signal. 2017, 38, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Fung, M.K.L.; Chan, G.C.F. Drug-induced amino acid deprivation as strategy for cancer therapy. J. Hematol. Oncol. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef]
- DeNicola, G.M.; Chen, P.H.; Mullarky, E.; Sudderth, J.A.; Hu, Z.; Wu, D.; Tang, H.; Xie, Y.; Asara, J.M.; Huffman, K.E.; et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 2015, 47, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Vié, N.; Copois, V.; Bascoul-Mollevi, C.; Denis, V.; Bec, N.; Robert, B.; Fraslon, C.; Conseiller, E.; Molina, F.; Larroque, C.; et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol. Cancer 2008, 7. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Chen, X.; Li, J.; Sun, S. Serine and metabolism regulation: A novel mechanism in antitumor immunity and senescence. Aging Dis. 2020, 11, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.H.; Coloff, J.L. The diverse functions of non-essential amino acids in cancer. Cancers 2019, 11, 675. [Google Scholar] [CrossRef] [PubMed]
- Ndaru, E.; Garibsingh, R.A.A.; Shi, Y.Y.; Wallace, E.; Zakrepine, P.; Wang, J.; Schlessinger, A.; Grewer, C. Novel alanine serine cysteine transporter 2 (ASCT2) inhibitors based on sulfonamide and sulfonic acid ester scaffolds. J. Gen. Physiol. 2019, 151, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Anzawa, H.; Liu, Z.; Ota, N.; Kitamura, H.; Onodera, Y.; Alam, M.M.; Matsumaru, D.; Suzuki, T.; Katsuoka, F.; et al. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.; Dinkova-Kostova, A.T.; Hayes, J.D. Nrf2 and the ambiguous consequences of its activation during initiation and the subsequent stages of tumourigenesis. Cancers 2020, 12, 3609. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.H.; Bantug, G.; Griss, T.; Condotta, S.; Johnson, R.M.; Samborska, B.; Mainolfi, N.; Suri, V.; Guak, H.; Balmer, M.L.; et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017, 25, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Possemato, R.; Marks, K.M.; Shaul, Y.D.; Pacold, M.E.; Kim, D.; Birsoy, K.; Sethumadhavan, S.; Woo, H.K.; Jang, H.G.; Jha, A.K.; et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011, 476, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Tabe, Y.; Lorenzi, P.L.; Konopleva, M. Amino acid metabolism in hematologic malignancies and the era of targeted therapy. Blood 2019, 134, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Pacold, M.E.; Brimacombe, K.R.; Chan, S.H.; Rohde, J.M.; Lewis, C.A.; Swier, L.J.Y.M.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef]
- Gao, X.; Lee, K.; Reid, M.A.; Sanderson, S.M.; Qiu, C.; Li, S.; Liu, J.; Locasale, J.W. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep. 2018, 22, 3507–3520. [Google Scholar] [CrossRef]
- Bernfeld, E.; Foster, D.A. Glutamine as an Essential Amino Acid for KRas-Driven Cancer Cells. Trends Endocrinol. Metab. 2019, 30, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Meng, Y.; Li, Z.; Dai, W.; Xu, X.; Bi, X.; Bian, J. Discovery and development of small molecule modulators targeting glutamine metabolism. Eur. J. Med. Chem. 2019, 163, 215–242. [Google Scholar] [CrossRef]
- Wu, S.; Fukumoto, T.; Lin, J.; Nacarelli, T.; Wang, Y.; Ong, D.; Liu, H.; Fatkhutdinov, N.; Zundell, J.A.; Karakashev, S.; et al. Targeting glutamine dependence through GLS1 inhibition suppresses ARID1A-inactivated clear cell ovarian carcinoma. Nat. Cancer 2021, 2, 189–200. [Google Scholar] [CrossRef]
- Shen, Y.A.; Hong, J.; Asaka, R.; Asaka, S.; Hsu, F.C.; Rahmanto, Y.S.; Jung, J.G.; Chen, Y.W.; Yen, T.T.; Tomaszewski, A.; et al. Inhibition of the MYC-regulated glutaminase metabolic axis is an effective synthetic lethal approach for treating chemoresistant ovarian cancers. Cancer Res. 2021, 80, 4415–4526. [Google Scholar] [CrossRef]
- Wu, C.R.; Chen, L.X.; Jin, S.; Li, H. Glutaminase inhibitors: A patent review. Expert Opin. Ther. Pat. 2018, 28, 823–835. [Google Scholar] [CrossRef]
- Gutierrez, J.A.; Pan, Y.X.; Koroniak, L.; Hiratake, J.; Kilberg, M.S.; Richards, N.G.J. An Inhibitor of Human Asparagine Synthetase Suppresses Proliferation of an L-Asparaginase-Resistant Leukemia Cell Line. Chem. Biol. 2006, 13, 1339–1347. [Google Scholar] [CrossRef]
- Toda, K.; Kawada, K.; Iwamoto, M.; Inamoto, S.; Sasazuki, T.; Shirasawa, S.; Hasegawa, S.; Sakai, Y. Metabolic Alterations Caused by KRAS Mutations in Colorectal Cancer Contribute to Cell Adaptation to Glutamine Depletion by Upregulation of Asparagine Synthetase. Neoplasia 2016, 18, 654–665. [Google Scholar] [CrossRef]
- Schulte, M.L.; Fu, A.; Zhao, P.; Li, J.; Geng, L.; Smith, S.T.; Kondo, J.; Coffey, R.J.; Johnson, M.O.; Rathmell, J.C.; et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 2018, 24, 194–202. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Diabetes and Digestive and Kidney Diseases. Asparaginase; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012. [Google Scholar]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef]
- Li, C.X.; Sun, J.L.; Gong, Z.C.; Lin, Z.Q.; Liu, H. Prognostic value of GLUT-1 expression in oral squamous cell carcinoma A prisma-compliant meta-analysis. Medicine 2016, 95. [Google Scholar] [CrossRef]
- Koh, Y.W.; Lee, S.J.; Park, S.Y. Differential expression and prognostic significance of GLUT1 according to histologic type of non-small-cell lung cancer and its association with volume-dependent parameters. Lung Cancer 2017, 104, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.S.; Wahl, R.L. Overexpression of glut-1 glucose transporter in human breast cancer an immunohistochemical study. Cancer 1993, 72, 2979–2985. [Google Scholar] [CrossRef]
- Kurahara, H.; Maemura, K.; Mataki, Y.; Sakoda, M.; Iino, S.; Kawasaki, Y.; Arigami, T.; Mori, S.; Kijima, Y.; Ueno, S.; et al. Significance of Glucose Transporter Type 1 (GLUT-1) Expression in the Therapeutic Strategy for Pancreatic Ductal Adenocarcinoma. Ann. Surg. Oncol. 2018, 25, 1432–1439. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jin, R.; Wang, W.; Zhang, T.; Sang, J.; Li, N.; Han, Q.; Zhao, W.; Li, C.; Liu, Z. STAT3 regulates glycolysis via targeting hexokinase 2 in hepatocellular carcinoma cells. Oncotarget 2017, 8, 24777–24784. [Google Scholar] [CrossRef]
- Crabtree, H.G. The carbohydrate metabolism of certain pathological overgrowths. Biochem. J. 1928, 22, 1289–1298. [Google Scholar] [CrossRef]
- He, X.; Lin, X.; Cai, M.; Zheng, X.; Lian, L.; Fan, D.; Wu, X.; Lan, P.; Wang, J. Overexpression of Hexokinase 1 as a poor prognosticator in human colorectal cancer. Tumor Biol. 2016, 37, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hu, L.; Wu, F.; Zou, L.; He, T. Poor prognosis of hexokinase 2 overexpression in solid tumors of digestive system: A meta-analysis. Oncotarget 2017, 8, 32332–32344. [Google Scholar] [CrossRef]
- Palmieri, D.; Fitzgerald, D.; Shreeve, S.M.; Hua, E.; Bronder, J.L.; Weil, R.J.; Davis, S.; Stark, A.M.; Merino, M.J.; Kurek, R.; et al. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Mol. Cancer Res. 2009, 7, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, M.; Sun, H.; Wang, F.; Xie, X.; Chen, X.; Su, J.; He, Y.; Dai, Y.; Wu, H.; et al. HK2 is a radiation resistant and independent negative prognostic factor for patients with locally advanced cervical squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 4054–4063. [Google Scholar] [PubMed]
- Zhang, X.Y.; Zhang, M.; Cong, Q.; Zhang, M.X.; Zhang, M.Y.; Lu, Y.Y.; Xu, C.J. Hexokinase 2 confers resistance to cisplatin in ovarian cancer cells by enhancing cisplatin-induced autophagy. Int. J. Biochem. Cell Biol. 2018, 95, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, H.; Liu, Z.; Ding, Y.; LeDoux, S.P.; Wilson, G.L.; Voellmy, R.; Lin, Y.; Lin, W.; Nahta, R.; et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res. 2011, 71, 4585–4597. [Google Scholar] [CrossRef] [PubMed]
- Deligiorgi, M.V.; Liapi, C.; Trafalis, D.T. How far are we from prescribing fasting as anticancer medicine? Int. J. Mol. Sci. 2020, 21, 9175. [Google Scholar] [CrossRef]
- Goncalves, M.D.; Lu, C.; Tutnauer, J.; Hartman, T.E.; Hwang, S.K.; Murphy, C.J.; Pauli, C.; Morris, R.; Taylor, S.; Bosch, K.; et al. High-fructose corn syrup enhances intestinal tumor growth in mice. Science 2019, 363, 1345–1349. [Google Scholar] [CrossRef]
- Goncalves, M.D.; Maddocks, O.D. Engineered diets to improve cancer outcomes. Curr. Opin. Biotechnol. 2021, 70, 29–35. [Google Scholar] [CrossRef]
- Li, W.; Ma, X.; Li, N.; Liu, H.; Dong, Q.; Zhang, J.; Yang, C.; Liu, Y.; Liang, Q.; Zhang, S.; et al. Resveratrol inhibits Hexokinases II mediated glycolysis in non-small cell lung cancer via targeting Akt signaling pathway. Exp. Cell Res. 2016, 349, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.H.; Smith, B.L.; Wang, Y.; Pomper, M.G.; Rini, D.A.; Torbenson, M.S.; Hullihen, J.; Pedersen, P.L. Advanced cancers: Eradication in all cases using 3-bromopyruvate therapy to deplete ATP. Biochem. Biophys. Res. Commun. 2004, 324, 269–275. [Google Scholar] [CrossRef]
- Liu, W.; Yu, X.; Zhou, L.; Li, J.; Li, M.; Li, W.; Gao, F. Sinomenine inhibits non-small cell lung cancer via downregulation of hexokinases II-mediated aerobic glycolysis. Onco. Targets. Ther. 2020, 13, 3209–3221. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xie, G.; Tong, J.; Peng, Y.; Huang, H.; Li, J.; Wang, N.; Liang, H. Overexpression of MicroRNA-122 Re-sensitizes 5-FU-Resistant Colon Cancer Cells to 5-FU Through the Inhibition of PKM2 In Vitro and In Vivo. Cell Biochem. Biophys. 2014, 70, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; He, B.; Lai, L.; Chen, Q.; Liu, Y.; Guo, Q.; Wang, Q. Cyclosporine A inhibits breast cancer cell growth by downregulating the expression of pyruvate kinase subtype M2. Int. J. Mol. Med. 2012, 30, 302–308. [Google Scholar] [CrossRef]
- Deng, Y.; Li, X.; Feng, J.; Zhang, X. Overexpression of miR-202 resensitizes imatinib resistant chronic myeloid leukemia cells through targetting Hexokinase 2. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Yang, L.; Liao, H.; Liang, X.; Xie, B.; Xiong, J.; Tao, X.; Chen, X.; Cheng, Y.; Chen, X.; et al. Metformin sensitizes endometrial cancer cells to chemotherapy through IDH1-induced Nrf2 expression via an epigenetic mechanism. Oncogene 2018, 37, 5666–5681. [Google Scholar] [CrossRef] [PubMed]
- Saed, G.M.; Fletcher, N.M.; Jiang, Z.L.; Abu-Soud, H.M.; Diamond, M.P. Dichloroacetate induces apoptosis of epithelial ovarian cancer cells through a mechanism involving modulation of oxidative stress. Reprod. Sci. 2011, 18, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Kumar, S. Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: Pivotal role of superoxide dismutase (SOD). Cell. Oncol. 2018, 41, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Kitson, S.J.; Rosser, M.; Fischer, D.P.; Marshall, K.M.; Clarke, R.B.; Crosbie, E.J. Targeting endometrial cancer stem cell activity with metformin is inhibited by patient-derived adipocyte-secreted factors. Cancers 2019, 11, 653. [Google Scholar] [CrossRef]
- Song, H.J.; Rhew, K.; Lee, Y.J.; Ha, I.H. Acid-suppressive agents and survival outcomes in patients with cancer: A systematic review and meta-analysis. Int. J. Clin. Oncol. 2021, 26, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Koltai, T. Targeting the ph paradigm at the bedside: A practical approach. Int. J. Mol. Sci. 2020, 21, 9221. [Google Scholar] [CrossRef]
- Fais, S.; Marunaka, Y. The acidic microenvironment: Is it a phenotype of all cancers? a focus on multiple myeloma and some analogies with diabetes mellitus. Cancers 2020, 12, 3226. [Google Scholar] [CrossRef] [PubMed]
- Belisario, D.C.; Kopecka, J.; Pasino, M.; Akman, M.; De Smaele, E.; Donadelli, M.; Riganti, C. Hypoxia Dictates Metabolic Rewiring of Tumors: Implications for Chemoresistance. Cells 2020, 9, 2598. [Google Scholar] [CrossRef]
- Battello, N.; Zimmer, A.D.; Goebel, C.; Dong, X.; Behrmann, I.; Haan, C.; Hiller, K.; Wegner, A. The role of HIF-1 in oncostatin M-dependent metabolic reprogramming of hepatic cells. Cancer Metab. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Audrito, V.; Managò, A.; Gaudino, F.; Sorci, L.; Messana, V.G.; Raffaelli, N.; Deaglio, S. NAD-Biosynthetic and Consuming Enzymes as Central Players of Metabolic Regulation of Innate and Adaptive Immune Responses in Cancer. Front. Immunol. 2019, 10, 1720. [Google Scholar] [CrossRef] [PubMed]
- Audrito, V.; Managò, A.; Gaudino, F.; Deaglio, S. Targeting metabolic reprogramming in metastatic melanoma: The key role of nicotinamide phosphoribosyltransferase (NAMPT). Semin. Cell Dev. Biol. 2020, 98, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-inducible factors: Coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 2017, 36, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Jiang, L.; Zhang, H.; Shimoda, L.A.; Deberardinis, R.J.; Semenza, G.L. Analysis of hypoxia-induced metabolic reprogramming. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 542, pp. 425–455. ISBN 9780124166189. [Google Scholar]
- Cha, J.-Y.; Lee, H.-J. Targeting Lipid Metabolic Reprogramming as Anticancer Therapeutics. J. Cancer Prev. 2016, 21, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Aragaki, A.K.; Tang, J.Y.; Kurian, A.W.; Manson, J.A.E.; Chlebowski, R.T.; Simon, M.; Desai, P.; Wassertheil-Smoller, S.; Liu, S.; et al. Statin use and all-cancer survival: Prospective results from the Women’s Health Initiative. Br. J. Cancer 2016, 115, 129–135. [Google Scholar] [CrossRef]
- Chen, Y.A.; Shih, H.W.; Lin, Y.C.; Hsu, H.Y.; Wu, T.F.; Tsai, C.H.; Wu, C.L.; Wu, H.Y.; Hsieh, J.T.; Tang, C.H.; et al. Simvastatin sensitizes radioresistant prostate cancer cells by compromising DNA double-strand break repair. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Babcook, M.A.; Sramkoski, R.M.; Fujioka, H.; Daneshgari, F.; Almasan, A.; Shukla, S.; Nanavaty, R.R.; Gupta, S. Combination simvastatin and metformin induces G1-phase cell cycle arrest and Ripk1- and Ripk3-dependent necrosis in C4-2B osseous metastatic castration-resistant prostate cancer cells. Cell Death Dis. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- de van der Schueren, M.A.E.; Laviano, A.; Blanchard, H.; Jourdan, M.; Arends, J.; Baracos, V.E. Systematic review and meta-analysis of the evidence for oral nutritional intervention on nutritional and clinical outcomes during chemo(radio)therapy: Current evidence and guidance for design of future trials. Ann. Oncol. 2018, 29, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Muhamad, R.; Yan, G.; Yu, J.; Fan, Q.; Wang, Z.; Li, X.; Purnomoadi, A.; Achmadi, J.; Yan, X. Quantitative proteomics analysis reveals glutamine deprivation activates fatty acid β-oxidation pathway in HepG2 cells. Amino Acids 2016, 48, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- De Cedrón, M.G.; De Molina, A.R. Microtargeting cancer metabolism:Opening new therapeutic windows based on lipid metabolism. J. Lipid Res. 2016, 57, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ren, W.; Huang, X.; Li, T.; Yin, Y. Protein restriction and cancer. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 256–262. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Athineos, D.; Cheung, E.C.; Lee, P.; Zhang, T.; Van Den Broek, N.J.F.; Mackay, G.M.; Labuschagne, C.F.; Gay, D.; Kruiswijk, F.; et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 2017, 544, 372–376. [Google Scholar] [CrossRef]
- Bröer, S. Amino acid transporters as targets for cancer therapy: Why, where, when, and how. Int. J. Mol. Sci. 2020, 21, 6156. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. The harmonious interplay of amino acid andmonocarboxylate transporters induces the robustness of cancer cells. Metabolites 2021, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Ollivier, L.; Forgez, P.; Otz, J.; Alifano, M.; Fournel, L.; Loi, M.; Thariat, J. Perspective: Do Fasting, Caloric Restriction, and Diets Increase Sensitivity to Radiotherapy? A Literature Review. Adv. Nutr. 2020, 11, 1089–1101. [Google Scholar] [CrossRef]
- Evans, J.M.M.; Donnelly, L.A.; Emslie-Smith, A.M.; Alessi, D.R.; Morris, A.D. Metformin and reduced risk of cancer in diabetic patients. Br. Med. J. 2005, 330, 1304–1305. [Google Scholar] [CrossRef]
- Poljsak, B.; Kovac, V.; Dahmane, R.; Levec, T.; Starc, A. Cancer Etiology: A Metabolic Disease Originating from Life’s Major Evolutionary Transition? Oxid. Med. Cell. Longev. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Men, Y.; Lu, L.; Geng, T.; Zhou, J.; Mitsuhashi, A.; Shozu, M.; Maihle, N.J.; Carmichael, G.G.; Taylor, H.S.; et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene 2017, 36, 2345–2354. [Google Scholar] [CrossRef]
- Pernicova, I.; Korbonits, M. Metformin-Mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014, 10, 143–156. [Google Scholar] [CrossRef]
- Hu, L.; Zeng, Z.; Xia, Q.; Liu, Z.; Feng, X.; Chen, J.; Huang, M.; Chen, L.; Fang, Z.; Liu, Q.; et al. Metformin attenuates hepatoma cell proliferation by decreasing glycolytic flux through the HIF-1α/PFKFB3/PFK1 pathway. Life Sci. 2019, 239. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Li, G.Y.; Wang, B.; Han, S.X.; Sun, X.; Jiang, Y.N.; Shen, Y.W.; Zhou, C.; Feng, J.; Lu, S.Y.; et al. Metformin inhibits metastatic breast cancer progression and improves chemosensitivity by inducing vessel normalization via PDGF-B downregulation. J. Exp. Clin. Cancer Res. 2019, 38, 235. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.J.; Thierbach, R.; Voigt, A.; Drewes, G.; Mietzner, B.; Steinberg, P.; Pfeiffer, A.F.H.; Ristow, M. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J. Biol. Chem. 2006, 281, 977–981. [Google Scholar] [CrossRef]
- Yoo, J.J.; Yu, S.J.; Na, J.; Kim, K.; Cho, Y.Y.; Lee, Y.B.; Cho, E.J.; Lee, J.H.; Kim, Y.J.; Youn, H.; et al. Hexokinase-II inhibition synergistically augments the anti-tumor efficacy of sorafenib in hepatocellular carcinoma. Int. J. Mol. Sci. 2019, 20, 1292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, Y.; Zhang, P.; Chao, Z.; Xia, F.; Jiang, C.; Zhang, X.; Jiang, Z.; Liu, H. Hexokinase II inhibitor, 3-BrPA induced autophagy by stimulating ROS formation in human breast cancer cells. Genes Cancer 2014, 5, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Thamrongwaranggoon, U.; Seubwai, W.; Phoomak, C.; Sangkhamanon, S.; Cha’on, U.; Boonmars, T.; Wongkham, S. Targeting hexokinase II as a possible therapy for cholangiocarcinoma. Biochem. Biophys. Res. Commun. 2017, 484, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ding, B.Z.; Lin, Y.P.; Wang, H.B. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene 2018, 644, 56–65. [Google Scholar] [CrossRef]
- Kroemer, G.; López-Otín, C.; Madeo, F.; de Cabo, R. Carbotoxicity—Noxious Effects of Carbohydrates. Cell 2018, 175, 605–614. [Google Scholar] [CrossRef]
- Ludwig, D.S.; Hu, F.B.; Tappy, L.; Brand-Miller, J. Dietary carbohydrates: Role of quality and quantity in chronic disease. BMJ 2018, 361. [Google Scholar] [CrossRef] [PubMed]
- Obrist, F.; Michels, J.; Durand, S.; Chery, A.; Pol, J.; Levesque, S.; Joseph, A.; Astesana, V.; Pietrocola, F.; Wu, G.S.; et al. Metabolic vulnerability of cisplatin-resistant cancers. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Kouidhi, S.; Ayed, F.B.; Elgaaied, A.B. Targeting tumor metabolism: A new challenge to improve immunotherapy. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; Sowell, R.T.; Kaech, S.M.; Pearce, E.L. Metabolic Instruction of Immunity. Cell 2017, 169, 570–586. [Google Scholar] [CrossRef]
- Benavente, S.; Sánchez-García, A.; Naches, S.; LLeonart, M.E.; Lorente, J. Therapy-Induced Modulation of the Tumor Microenvironment: New Opportunities for Cancer Therapies. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Moehler, M.; Delic, M.; Goepfert, K.; Aust, D.; Grabsch, H.I.; Halama, N.; Heinrich, B.; Julie, C.; Lordick, F.; Lutz, M.P.; et al. Immunotherapy in gastrointestinal cancer: Recent results, current studies and future perspectives. Eur. J. Cancer 2016, 59, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jang, J.Y.; Koh, J.; Kwon, D.; Kim, Y.A.; Paeng, J.C.; Ock, C.Y.; Keam, B.; Kim, M.; Kim, T.M.; et al. Programmed cell death ligand-1-mediated enhancement of hexokinase 2 expression is inversely related to T-cell effector gene expression in non-small-cell lung cancer. J. Exp. Clin. Cancer Res. 2019, 38. [Google Scholar] [CrossRef]
- Turbitt, W.J.; Demark-Wahnefried, W.; Peterson, C.M.; Norian, L.A. Targeting glucose metabolism to enhance immunotherapy: Emerging evidence on intermittent fasting and calorie restriction mimetics. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Daillère, R.; Derosa, L.; Bonvalet, M.; Segata, N.; Routy, B.; Gariboldi, M.; Budinská, E.; De Vries, I.J.M.; Naccarati, A.G.; Zitvogel, V.; et al. Trial watch: The gut microbiota as a tool to boost the clinical efficacy of anticancer immunotherapy. Oncoimmunology 2020, 9. [Google Scholar] [CrossRef]
- Drijvers, J.M.; Sharpe, A.H.; Haigis, M.C. The effects of age and systemic metabolism on anti-tumor t cell responses. Elife 2020, 9, 1–29. [Google Scholar] [CrossRef]
- Sosa, V.; Moliné, T.; Somoza, R.; Paciucci, R.; Kondoh, H.; LLeonart, M.E. Oxidative stress and cancer: An overview. Ageing Res. Rev. 2013, 12, 376–390. [Google Scholar] [CrossRef]
- Ghoneum, A.; Abdulfattah, A.Y.; Warren, B.O.; Shu, J.; Said, N. Redox homeostasis and metabolism in cancer: A complex mechanism and potential targeted therapeutics. Int. J. Mol. Sci. 2020, 21, 3100. [Google Scholar] [CrossRef]
- Carmona-Fontaine, C.; Deforet, M.; Akkari, L.; Thompson, C.B.; Joyce, J.A.; Xavier, J.B. Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2017, 114, 2934–2939. [Google Scholar] [CrossRef]
- Xiang, W.; Shi, R.; Zhang, D.; Kang, X.; Zhang, L.; Yuan, J.; Zhang, X.; Miao, H. Dietary fats suppress the peritoneal seeding of colorectal cancer cells through the TLR4/Cxcl10 axis in adipose tissue macrophages. Signal Transduct. Target. Ther. 2020, 5. [Google Scholar] [CrossRef]
- Kerr, J.; Anderson, C.; Lippman, S.M. Physical activity, sedentary behaviour, diet, and cancer: An update and emerging new evidence. Lancet Oncol. 2017, 18, e457–e471. [Google Scholar] [CrossRef]
- Lévesque, S.; Pol, J.G.; Ferrere, G.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Trial watch: Dietary interventions for cancer therapy. Oncoimmunology 2019, 8. [Google Scholar] [CrossRef]
- Mehra, K.; Berkowitz, A.; Sanft, T. Diet, Physical Activity, and Body Weight in Cancer Survivorship. Med. Clin. N. Am. 2017, 101, 1151–1165. [Google Scholar] [CrossRef]
- Mosher, C.E.; Sloane, R.; Morey, M.C.; Snyder, D.C.; Cohen, H.J.; Miller, P.E.; Demark-Wahnefried, W. Associations between lifestyle factors and quality of life among older long-term breast, prostate, and colorectal cancer survivors. Cancer 2009, 115, 4001–4009. [Google Scholar] [CrossRef]
- Ligibel, J. Lifestyle factors in cancer survivorship. J. Clin. Oncol. 2012, 30, 3697–3704. [Google Scholar] [CrossRef]
- Kenzik, K.M.; Morey, M.C.; Cohen, H.J.; Sloane, R.; Demark-Wahnefried, W. Symptoms, weight loss, and physical function in a lifestyle intervention study of older cancer survivors. J. Geriatr. Oncol. 2015, 6, 424–432. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altea-Manzano, P.; Cuadros, A.M.; Broadfield, L.A.; Fendt, S. Nutrient metabolism and cancer in the in vivo context: A metabolic game of give and take. EMBO Rep. 2020, 21. [Google Scholar] [CrossRef]
- Ibrahim, E.M.; Al-Foheidi, M.H.; Al-Mansour, M.M. Energy and caloric restriction, and fasting and cancer: A narrative review. Support. Care Cancer 2020. [Google Scholar] [CrossRef]
- Ngo, B.; Van Riper, J.M.; Cantley, L.C.; Yun, J. Targeting cancer vulnerabilities with high-dose vitamin C. Nat. Rev. Cancer 2019, 19, 271–282. [Google Scholar] [CrossRef]
- Bhoora, S.; Punchoo, R. Policing cancer: Vitamin d arrests the cell cycle. Int. J. Mol. Sci. 2020, 21, 9296. [Google Scholar] [CrossRef]
- Walia, S.; Kamal, R.; Dhawan, D.K.; Kanwar, S.S. Chemoprevention by Probiotics During 1,2-Dimethylhydrazine-Induced Colon Carcinogenesis in Rats. Dig. Dis. Sci. 2018, 63, 900–909. [Google Scholar] [CrossRef]
- Gillberg, L.; Ørskov, A.D.; Liu, M.; Harsløf, L.B.S.; Jones, P.A.; Grønbæk, K. Vitamin C—A new player in regulation of the cancer epigenome. Semin. Cancer Biol. 2018, 51, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Blelloch, R.H.; Hochedlinger, K.; Yamada, Y.; Brennan, C.; Kim, M.; Mintz, B.; Chin, L.; Jaenisch, R. Nuclear cloning of embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 2004, 101, 13985–13990. [Google Scholar] [CrossRef]
- Burgos-Panadero, R.; Lucantoni, F.; Gamero-Sandemetrio, E.; de la Cruz-Merino, L.; Álvaro, T.; Noguera, R. The tumour microenvironment as an integrated framework to understand cancer biology. Cancer Lett. 2019, 461, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sanegre, S.; Lucantoni, F.; Burgos-Panadero, R.; de La Cruz-Merino, L.; Noguera, R.; Naranjo, T.Á. Integrating the tumor microenvironment into cancer therapy. Cancers 2020, 12, 1677. [Google Scholar] [CrossRef] [PubMed]
- Crippa, S.; Ancey, P.; Vazquez, J.; Angelino, P.; Rougemont, A.; Guettier, C.; Zoete, V.; Delorenzi, M.; Michielin, O.; Meylan, E. Mutant CTNNB 1 and histological heterogeneity define metabolic subtypes of hepatoblastoma. EMBO Mol. Med. 2017, 9, 1589–1604. [Google Scholar] [CrossRef] [PubMed]
- Nimmakayala, R.K.; Leon, F.; Rachagani, S.; Rauth, S.; Nallasamy, P.; Marimuthu, S.; Shailendra, G.K.; Chhonker, Y.S.; Chugh, S.; Chirravuri, R.; et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene 2021, 40, 215–231. [Google Scholar] [CrossRef] [PubMed]
| Targeted Pathway | Biomarkers | Tumor Types Affected | Reported Therapeutic Agents |
|---|---|---|---|
| Cancer-associated adipose tissue Extracellular lipid uptake | Tumor [21,22,23]: CD36 increase FABP4 increase LPL increase Adipocyte [21,24]: ATGL increase HSL increase | Breast adenocarcinoma [21] Leukemia [21] Multiple myeloma [21] Prostate adenocarcinoma [21] Ovarian adenocarcinoma [21] Gastric adenocarcinoma [22] Pancreatic adenocarcinoma [22] Small cell lung cancer [22] Squamous cell carcinoma [22] | 3-Bromopyruvate [21] CD36 inhibitors [22] |
| Fatty acid synthesis | FASN [25,26] SREBP1 [27,28] LXR [29] SCD-1 [29] | Breast adenocarcinoma [25] Colon adenocarcinoma [28,30] Pancreatic adenocarcinoma [26,31] Ovarian carcinoma [28] Prostate adenocarcinoma [27] | TVB-3136 [25] TVB-2640 [25] IPI-9119 [27] Cerulenin [28] Orlistat [28,32,33] C93 [28] Proton pump inhibitors [26,34] |
| Fatty acid oxidation | CPT1 [35] IDH2 [36] | Glioblastoma [36] Acute myeloid leukemia [35] | Etomoxir [35,36] |
| Prostaglandin E2 | COX-2 [37,38] mPGES-1 [37] ID1 [38] ARC [37] EP4 [39] | Glioblastoma [38] Acute myeloid leukemia [37] Colorectal adenocarcinoma [39] | Prostaglandine receptors inhibitors [37,39,40] Omega-3 PUFA [37,41,42] Nonsteroidal anti-inflammatory drugs [39] |
| Bioactive sphingolipids | S1P increase [43] Ceramide decrease [43] Neutral ceramidase [43] Acid ceramidase [44,45] Sphk1 [44,46,47,48] S1PR1 [49] S1PR3 [49] | Colorectal adenocarcinoma [43,50] Prostate adenocarcinoma [44,51] Breast adenocarcinoma [44,46] Head and neck squamous carcinoma [44] Ovarian adenocarcinoma [44] Uterus adenocarcinoma [44] Acute myeloid leukemia [52] Glioblastoma [53] | C6 urea-ceramide [43] Dietary sphingomyelin [44,54,55] LCL385 [44] Fingolimod (FTY720) [44] L-t-C6-Pyr-Cer [44] LCL204 [52] Ceranib-b2 [49] |
| Targeted Pathway | Biomarkers | Tumor Type Affected | Reported Therapeutic Agents |
|---|---|---|---|
| Serine Glycine | PHGDH [64,65] PSAT1 [65,66] PSPH [67,68] SLC1A4(ASCT-1) [67] SLC1A5(ASCT-2) [67,69] SHMT1 [67] SHMT2 [64,67] NFR2 [70,71,72,73,74,75] | Melanoma [68] Breast adenocarcinoma [67,74,76] Acute myeloid leukemia [75,76] Mesothelioma [64] Lung adenocarcinoma [67] Non-small cell lung cancer [65] Lymphomas [76] Colorectal adenocarcinoma [66] | Methotrexate [76,77] Pemetrexed [76] NCT-503 [77] Serine/glycine deprivation [68,78] Sulfonamide sulfonic ester scaffolds [69] |
| Glutamine Glutamate | xCT [79] SLC7A5/SLC3A2 [79] GLS1/2 [80,81,82] SLC1A5(ASCT-2) [69,80] | KRAS-driven cancer cells [79,81,82] | Glutamine deprivation [79] Aminooxyacetate (AOA) [79] CB-839 [80] BPTES [80] Sulfasalazine [80] V-9302 [80] Compound-968 [83] Sulfonamide sulfonic ester scaffolds [69] |
| Asparagine | ASNS [68,84,85] SLC1A5(ASCT-2) [69,86,87] | Acute lymphoblastic leukemia [68,84] | Adenylated sulfoximine 1 [84] Asparaginase [68] Sulfonamide sulfonic ester scaffolds [69] |
| Targeted Pathway | Biomarkers | Tumors Affected | Reported Therapeutic Agents |
|---|---|---|---|
| Glucose uptake | GLUT1 [88,89,90,91,92] | Hepatocellular carcinoma [93] Renal cell carcinoma [88] Oral squamous cell carcinoma [89] Non-small cell lung cancer [90] Breast adenocarcinoma [91] Pancreatic adenocarcinoma [92] | Neoadjuvant chemoradiotherapy [92] Trastuzumab [100] Fasting [101] Fasting mimicking diet [101,102] Calorie restriction [103] |
| Glycolysis and TCA | HK1 [95] HK2 [9,93,96,97,98,99,104,105,106] PFKFB3 [88] PFK1 [88] PKM2 [107,108,109] IDH1 [110] PDK [111] | Laryngeal squamous cell carcinoma [9] Cervical squamous cell carcinoma [98] Hepatocellular carcinoma [96,105] Endometrial cancer [110] Breast adenocarcinoma [97,112] Epithelial ovarian cancer [99,111] Non-small cell lung cancer [106] Colon adenocarcinoma [107] Chronic myeloid leukemia [109] | Metformin [110] 2-Deoxyglucose [109] 3-Bromopyruvate [105] Increased frataxin [112] Dichloroacetate [111] Resveratrol [104] Sinomenine [106] Cyclosporine A [108] Overexpression of miR-122 [107] Overexpression of miR-202 [109] |
| Lactate production/extraction | LDHA [88,100] MCT1 [88] MCT4 [88] | Breast adenocarcinoma [100] | Metformin [113] Trastuzumab [100] Oxamate [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monferrer, E.; Vieco-Martí, I.; López-Carrasco, A.; Fariñas, F.; Abanades, S.; de la Cruz-Merino, L.; Noguera, R.; Álvaro Naranjo, T. Metabolic Classification and Intervention Opportunities for Tumor Energy Dysfunction. Metabolites 2021, 11, 264. https://doi.org/10.3390/metabo11050264
Monferrer E, Vieco-Martí I, López-Carrasco A, Fariñas F, Abanades S, de la Cruz-Merino L, Noguera R, Álvaro Naranjo T. Metabolic Classification and Intervention Opportunities for Tumor Energy Dysfunction. Metabolites. 2021; 11(5):264. https://doi.org/10.3390/metabo11050264
Chicago/Turabian StyleMonferrer, Ezequiel, Isaac Vieco-Martí, Amparo López-Carrasco, Fernando Fariñas, Sergio Abanades, Luis de la Cruz-Merino, Rosa Noguera, and Tomás Álvaro Naranjo. 2021. "Metabolic Classification and Intervention Opportunities for Tumor Energy Dysfunction" Metabolites 11, no. 5: 264. https://doi.org/10.3390/metabo11050264
APA StyleMonferrer, E., Vieco-Martí, I., López-Carrasco, A., Fariñas, F., Abanades, S., de la Cruz-Merino, L., Noguera, R., & Álvaro Naranjo, T. (2021). Metabolic Classification and Intervention Opportunities for Tumor Energy Dysfunction. Metabolites, 11(5), 264. https://doi.org/10.3390/metabo11050264






