Analyzing Mass Spectrometry Imaging Data of 13C-Labeled Phospholipids in Camelina sativa and Thlaspi arvense (Pennycress) Embryos
Abstract
1. Introduction
2. Results and Discussion
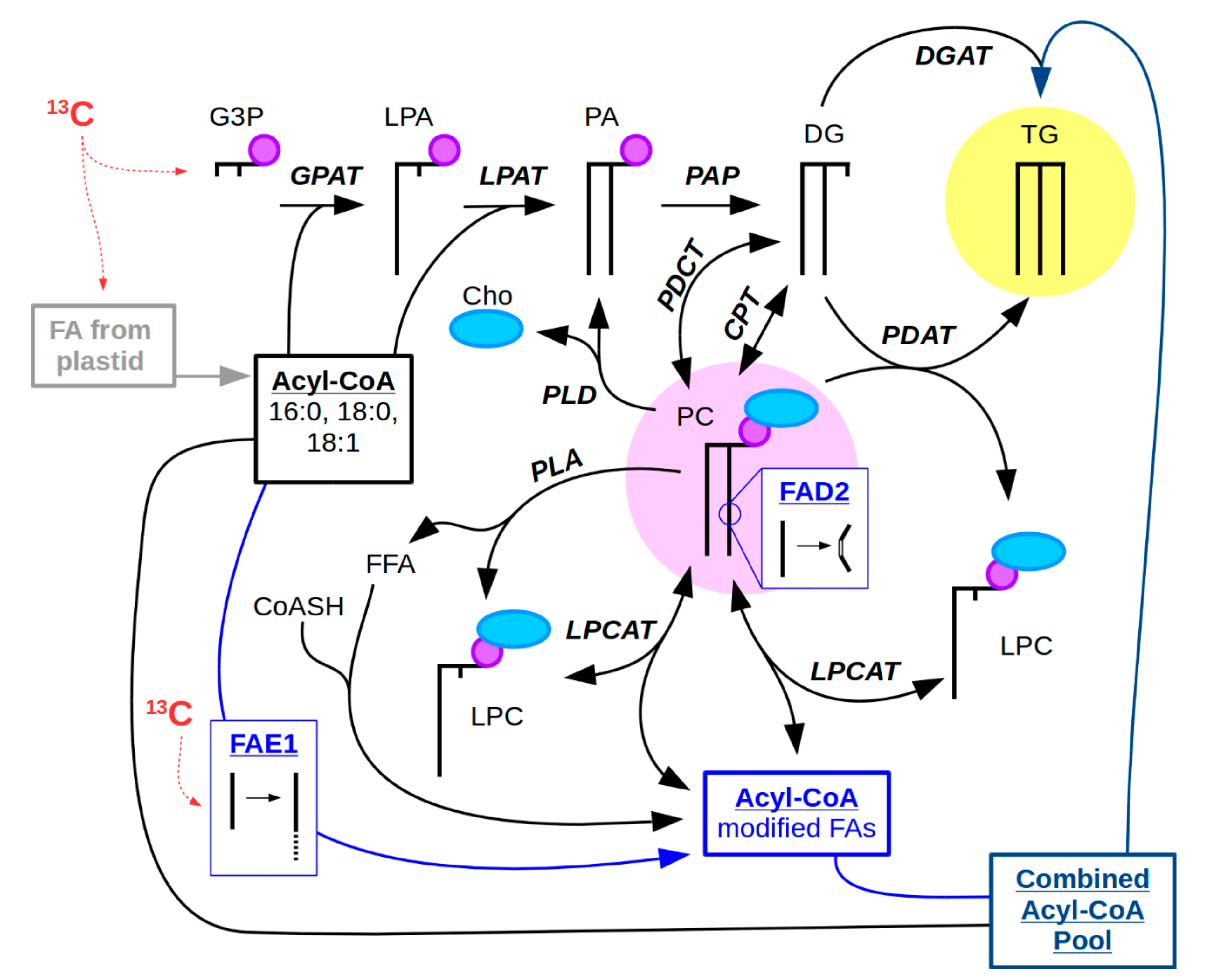
2.1. Research Questions Addressed with 13C-MSI
2.2. Considerations for 13C-Labeling in 13C-MSI Experiments
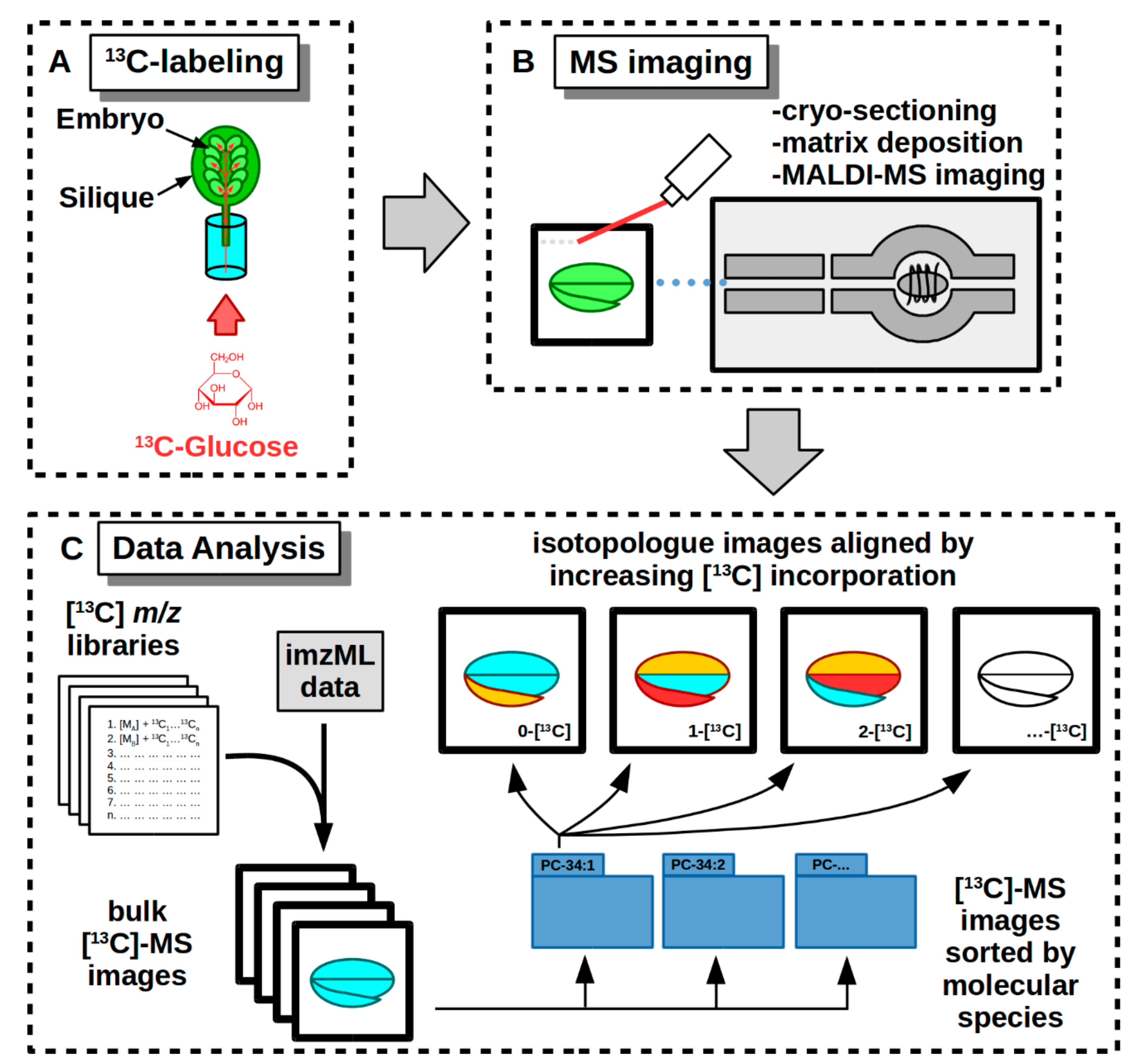
2.3. Labeling and Harvesting Samples for MS Imaging
2.4. Cryo-Sectioning Biological Samples for 13C-MSI
2.5. MS Instrument Parameters
2.6. Data Analysis of 13C-Labeled MS Images
2.7. List of Software Used to Analyze 13C-Labeled MS Images
2.8. Prior to Collecting or Analyzing MS Images
2.8.1. Creating Libraries of Isotopologue m/z Values and Their Respective Adducts for Metabolite Classes of Interest
2.8.2. Custom Scripts for Moving Image Files to Appropriate Directories
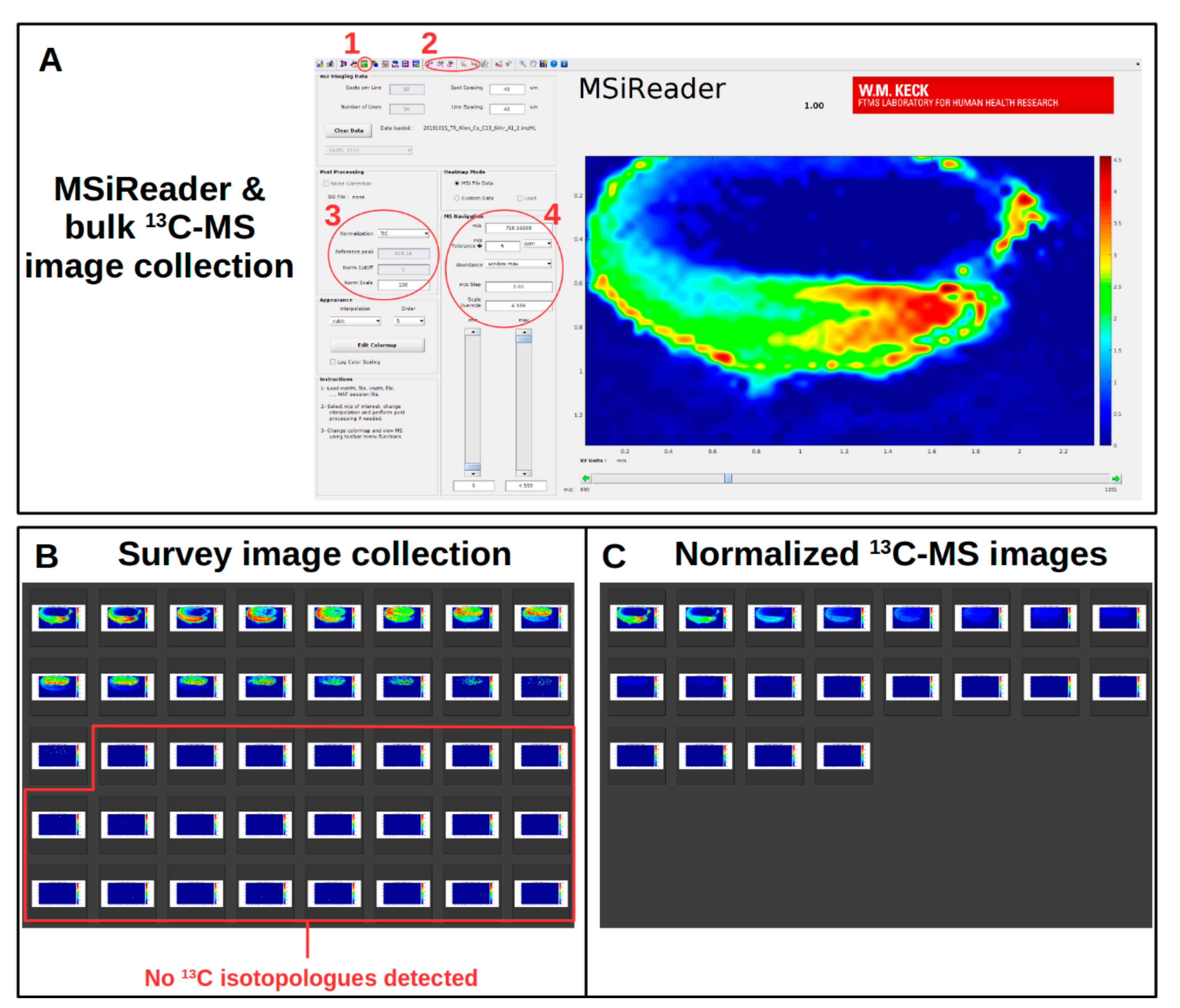
2.9. Initial Survey for Metabolites, Adduct Forms, and Extent of 13C-Labeling
Viewing 13C-Labeled MS Images within MSiReader
2.10. Collection of Normalized 13C-Labeled MS Images
2.10.1. Revising m/z Library Values for Other Replicates
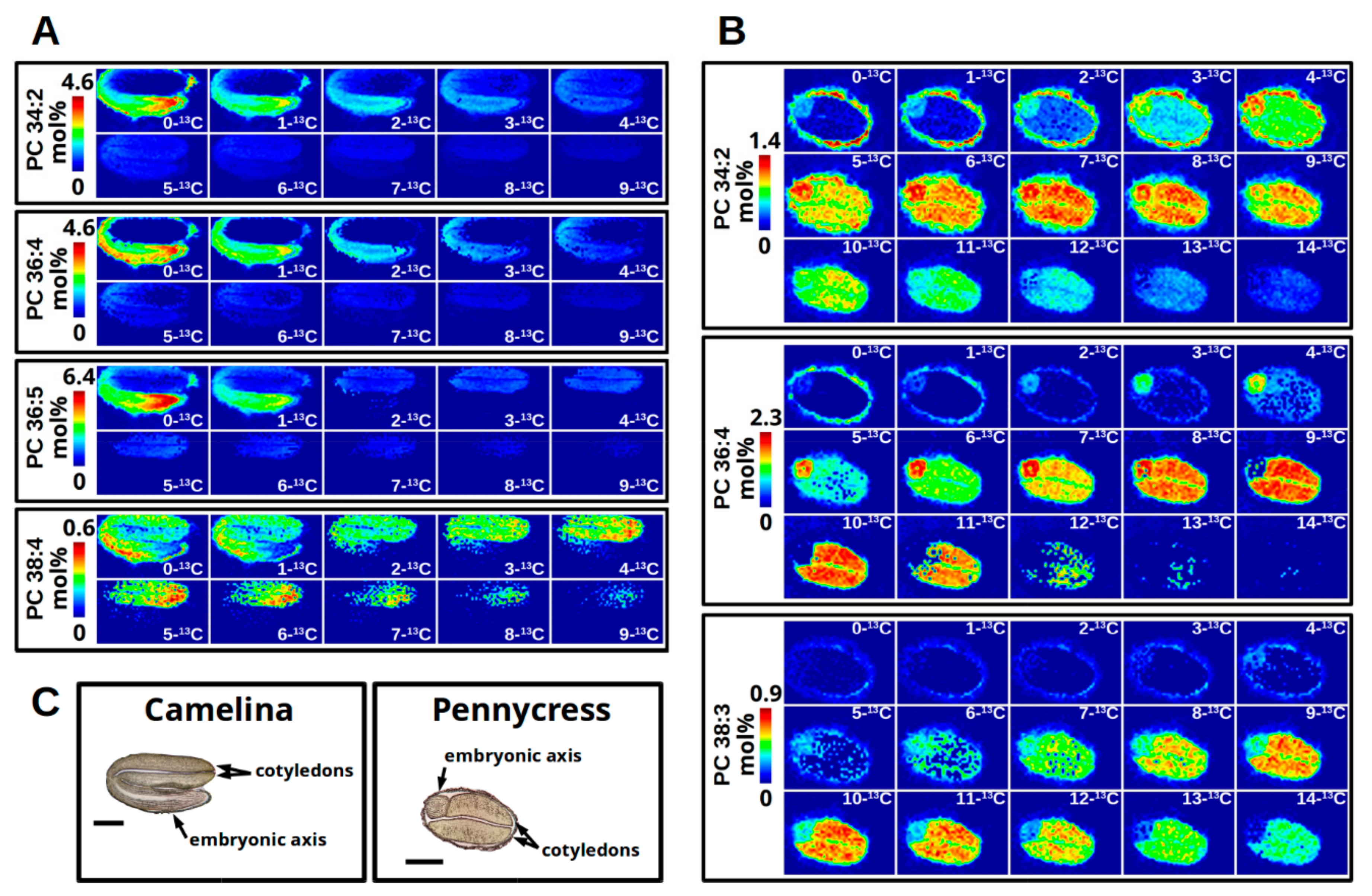
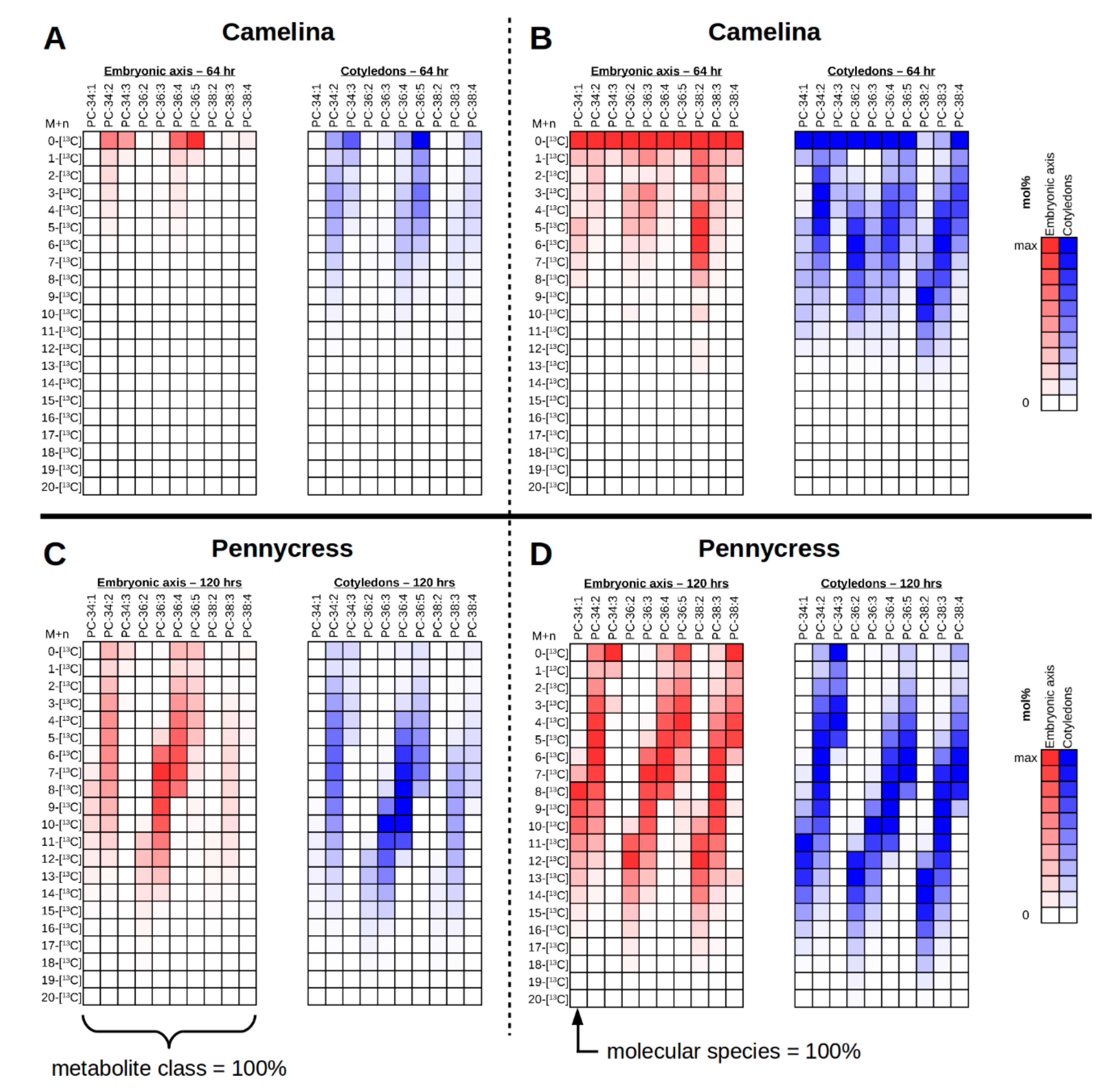
2.10.2. Organizing Collected Images for High-Quality Figures and Interpreting Labeling Patterns
2.11. Interrogating Tissue Specific 13C-Labeling Patterns
3. Materials and Methods
3.1. Chemicals
3.2. Plant Material
3.3. 13C-Labeling and Culturing Conditions
3.4. Fatty Acid Extraction and Derivatization with N-Butylamine from Pennycress Embryos
3.5. MS Imaging
3.5.1. Embedding and Cryo-Sectioning Embryos
3.5.2. Matrix Application and MALDI-MS Imaging Instrument Parameters
3.5.3. Extraction, Chromatographic Separation and Instrumentation Used for LC-MS/MS Analysis
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spengler, B. Mass Spectrometry Imaging of Biomolecular Information. Anal. Chem. 2015, 87, 64–82. [Google Scholar] [CrossRef]
- Sturtevant, D.; Lee, Y.-J.; Chapman, K.D. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr. Opin. Biotechnol. 2016, 37, 53–60. [Google Scholar] [CrossRef]
- Boskamp, M.S.; Soltwisch, J. Charge Distribution between Different Classes of Glycerophospolipids in MALDI-MS Imaging. Anal. Chem. 2020, 92, 5222–5230. [Google Scholar] [CrossRef] [PubMed]
- Boughton, B.A.; Thinagaran, D.; Sarabia, D.; Bacic, A.; Roessner, U. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2016, 15, 445–488. [Google Scholar] [CrossRef]
- Winter, G.; Krömer, J.O. Fluxomics—Connecting ‘omics analysis and phenotypes. Environ. Microbiol. 2013, 15, 1901–1916. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.K. Quantifying plant phenotypes with isotopic labeling & metabolic flux analysis. Curr. Opin. Biotechnol. 2016, 37, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Sauer, U. Metabolic networks in motion: 13 C-based flux analysis. Mol. Syst. Biol. 2006, 2, 62. [Google Scholar] [CrossRef]
- Tsogtbaatar, E.; Cocuron, J.-C.; Alonso, A.P. Non-conventional pathways enable pennycress (Thlaspi arvense L.) embryos to achieve high efficiency of oil biosynthesis. J. Exp. Bot. 2020, 71, 3037–3051. [Google Scholar] [CrossRef]
- Schwender, J.; Shachar-Hill, Y.; Ohlrogge, J.B. Mitochondrial Metabolism in Developing Embryos of Brassica napus. J. Biol. Chem. 2006, 281, 34040–34047. [Google Scholar] [CrossRef] [PubMed]
- Acket, S.; Degournay, A.; Rossez, Y.; Mottelet, S.; Villon, P.; Troncosoponce, A.; Thomasset, B. 13C-metabolic flux analysis in developing flaxseed embryos to understand storage lipid biosynthesis. Metabolites 2019, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.P.; Goffman, F.D.; Ohlrogge, J.B.; Shachar-Hill, Y. Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J. 2007, 52, 296–308. [Google Scholar] [CrossRef]
- Cocuron, J.-C.; Koubaa, M.; Kimmelfield, R.; Ross, Z.; Alonso, A.P. A Combined Metabolomics and Fluxomics Analysis Identifies Steps Limiting Oil Synthesis in Maize Embryos. Plant Physiol. 2019, 181, 961–975. [Google Scholar] [CrossRef]
- Allen, D.K.; Young, J.D. Carbon and Nitrogen Provisions Alter the Metabolic Flux in Developing Soybean Embryos. Plant Physiol. 2013, 161, 1458–1475. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.K.; Ohlrogge, J.B.; Shachar-Hill, Y. The role of light in soybean seed filling metabolism. Plant J. 2009, 58, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.K.; Bates, P.D.; Tjellström, H. Tracking the metabolic pulse of plant lipid production with isotopic labeling and flux analyses: Past, present and future. Prog. Lipid Res. 2015, 58, 97–120. [Google Scholar] [CrossRef]
- Ortiz, R.; Geleta, M.; Gustafsson, C.; Lager, I.; Hofvander, P.; Löfstedt, C.; Cahoon, E.B.; Minina, E.; Bozhkov, P.; Stymne, S. Oil crops for the future. Curr. Opin. Plant Biol. 2020, 56, 181–189. [Google Scholar] [CrossRef]
- Lu, C.; Napier, J.A.; Clemente, T.E.; Cahoon, E.B. New frontiers in oilseed biotechnology: Meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr. Opin. Biotechnol. 2011, 22, 252–259. [Google Scholar] [CrossRef]
- Snapp, A.R.; Lu, C. Engineering industrial fatty acids in oilseeds. Front. Biol. 2012, 8, 323–332. [Google Scholar] [CrossRef]
- Msanne, J.; Kim, H.; Cahoon, E.B. Biotechnology tools and applications for development of oilseed crops with healthy vegetable oils. Biochimie 2020, 178, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Iskandarov, U.; Kim, H.J.; Cahoon, E.B. Camelina: An Emerging Oilseed Platform for Advanced Biofuels and Bio-Based Materials. In Plants and BioEnergy; Springer Science and Business Media LLC: New York, NY, USA, 2013; pp. 131–140. [Google Scholar]
- Chopra, R.; Johnson, E.B.; Emenecker, R.; Cahoon, E.B.; Lyons, J.; Kliebenstein, D.J.; Daniels, E.; Dorn, K.M.; Esfahanian, M.; Folstad, N.; et al. Identification and stacking of crucial traits required for the domestication of pennycress. Nat. Food 2020, 1, 84–91. [Google Scholar] [CrossRef]
- Sturtevant, D.; Romsdahl, T.B.; Yu, X.-H.; Burks, D.J.; Azad, R.K.; Shanklin, J.; Chapman, K.D. Tissue-specific differences in metabolites and transcripts contribute to the heterogeneity of ricinoleic acid accumulation in Ricinus communis L. (castor) seeds. Metabolomics 2019, 15, 6. [Google Scholar] [CrossRef]
- Marmon, S.; Sturtevant, D.; Herrfurth, C.; Chapman, K.; Stymne, S.; Feussner, I. Two Acyltransferases Contribute Differently to Linolenic Acid Levels in Seed Oil. Plant Physiol. 2017, 173, 2081–2095. [Google Scholar] [CrossRef]
- Sturtevant, D.; Dueñas, M.E.; Lee, Y.-J.; Chapman, K.D. Three-dimensional visualization of membrane phospholipid distributions in Arabidopsis thaliana seeds: A spatial perspective of molecular heterogeneity. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 268–281. [Google Scholar] [CrossRef]
- Lu, S.; Aziz, M.; Sturtevant, D.; Chapman, K.D.; Guo, L. Heterogeneous Distribution of Erucic Acid in Brassica napus Seeds. Front. Plant Sci. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and Temporal Mapping of Key Lipid Species in Brassica napus Seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Rolletschek, H.; Schwender, J.; König, C.; Chapman, K.D.; Romsdahl, T.; Lorenz, C.; Braun, H.-P.; Denolf, P.; Van Audenhove, K.; Munz, E.; et al. Cellular Plasticity in Response to Suppression of Storage Proteins in the Brassica napus Embryo. Plant Cell 2020, 32, 2383–2401. [Google Scholar] [CrossRef]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of Triacylglycerol Accumulation in Plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar] [PubMed]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef]
- Bates, P.D.; Fatihi, A.; Snapp, A.R.; Carlsson, A.S.; Browse, J.; Lu, C. Acyl Editing and Headgroup Exchange Are the Major Mechanisms That Direct Polyunsaturated Fatty Acid Flux into Triacylglycerols. Plant Physiol. 2012, 160, 1530–1539. [Google Scholar] [CrossRef]
- Kunst, L.; Taylor, D.; Underhill, E. Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol. Biochem. 1992, 30, 425–434. [Google Scholar]
- Zou, J.; Wei, Y.; Jako, C.; Kumar, A.; Selvaraj, G.; Taylor, D.C. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 1999, 19, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, D.H.; Lu, C.; Hills, M.J. Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett. 1999, 452, 145–149. [Google Scholar] [CrossRef]
- Römpp, A.; Spengler, B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 2013, 139, 759–783. [Google Scholar] [CrossRef]
- Morley-Smith, E.R.; Pike, M.J.; Findlay, K.; Köckenberger, W.; Hill, L.M.; Smith, A.M.; Rawsthorne, S. The Transport of Sugars to Developing Embryos Is Not via the Bulk Endosperm in Oilseed Rape Seeds. Plant Physiol. 2008, 147, 2121–2130. [Google Scholar] [CrossRef]
- Griffith, S.M.; Jones, R.J.; Brenner, M.L. In Vitro Sugar Transport in Zea mays L. Kernels. Plant Physiol. 1987, 84, 472–475. [Google Scholar] [CrossRef]
- Dufresne, M.; Patterson, N.H.; Norris, J.L.; Caprioli, R.M. Combining Salt Doping and Matrix Sublimation for High Spatial Resolution MALDI Imaging Mass Spectrometry of Neutral Lipids. Anal. Chem. 2019, 91, 12928–12934. [Google Scholar] [CrossRef]
- Allen, D.K. Assessing compartmentalized flux in lipid metabolism with isotopes. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 1226–1242. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.M.; Creger, S.; Singla, V.; Kurulugama, R.T.; Fjeldsted, J.C.; Laskin, J. Ion Mobility-Mass Spectrometry Imaging Workflow. J. Am. Soc. Mass Spectrom. 2020, 31, 2437–2442. [Google Scholar] [CrossRef]
- Robichaud, G.; Garrard, K.P.; Barry, J.A.; Muddiman, D.C. MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. J. Am. Soc. Mass Spectrom. 2013, 24, 718–721. [Google Scholar] [CrossRef] [PubMed]
- The ImageMagick Development Team. ImageMagick. Available online: https://imagemagick.org (accessed on 15 October 2018).
- Schramm, T.; Hester, Z.; Klinkert, I.; Both, J.-P.; Heeren, R.M.; Brunelle, A.; Laprévote, O.; Desbenoit, N.; Robbe, M.-F.; Stoeckli, M.; et al. imzML—A common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteom. 2012, 75, 5106–5110. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, P.; Kohler, C.; Ellmann, L.; Kuerner, P.; Spang, R.; Oefner, P.J.; Dettmer, K. Correcting for natural isotope abundance and tracer impurity in MS-, MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci. Rep. 2018, 8, 17910. [Google Scholar] [CrossRef]
- Tsogtbaatar, E.; Cocuron, J.-C.; Sonera, M.C.; Alonso, A.P. Metabolite fingerprinting of pennycress (Thlaspi arvenseL.) embryos to assess active pathways during oil synthesis. J. Exp. Bot. 2015, 66, 4267–4277. [Google Scholar] [CrossRef]
- Allen, D.K.; Shachar-Hill, Y.; Ohlrogge, J.B. Compartment-specific labeling information in 13C metabolic flux analysis of plants. Phytochemisty 2007, 68, 2197–2210. [Google Scholar] [CrossRef]
- Hankin, J.A.; Barkley, R.M.; Murphy, R.C. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007, 18, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Kambhampati, S.; Aznar-Moreno, J.A.; Hostetler, C.; Caso, T.; Bailey, S.R.; Hubbard, A.H.; Durrett, T.P.; Allen, D.K. On the Inverse Correlation of Protein and Oil: Examining the Effects of Altered Central Carbon Metabolism on Seed Composition Using Soybean Fast Neutron Mutants. Metabolites 2019, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Segu, S.; Li, Y.; Irgang, S.; Jueppner, J.; Giavalisco, P. Ultra Performance Liquid Chromatography and High Resolution Mass Spectrometry for the Analysis of Plant Lipids. Front. Plant Sci. 2011, 2, 54. [Google Scholar] [CrossRef] [PubMed]
| 64 h Labeling * in PC 38:4 | ||
|---|---|---|
| Paired Fatty Acids | Cotyledon (%) | Embryo Axis (%) |
| FA 18:2 | 10.37 | 6.54 |
| FA 20:2 | 2.76 | 5.39 |
| FA 18:3 | 7.11 | 1.28 |
| FA 20:1 | 8.08 | 3.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romsdahl, T.B.; Kambhampati, S.; Koley, S.; Yadav, U.P.; Alonso, A.P.; Allen, D.K.; Chapman, K.D. Analyzing Mass Spectrometry Imaging Data of 13C-Labeled Phospholipids in Camelina sativa and Thlaspi arvense (Pennycress) Embryos. Metabolites 2021, 11, 148. https://doi.org/10.3390/metabo11030148
Romsdahl TB, Kambhampati S, Koley S, Yadav UP, Alonso AP, Allen DK, Chapman KD. Analyzing Mass Spectrometry Imaging Data of 13C-Labeled Phospholipids in Camelina sativa and Thlaspi arvense (Pennycress) Embryos. Metabolites. 2021; 11(3):148. https://doi.org/10.3390/metabo11030148
Chicago/Turabian StyleRomsdahl, Trevor B., Shrikaar Kambhampati, Somnath Koley, Umesh P. Yadav, Ana Paula Alonso, Doug K. Allen, and Kent D. Chapman. 2021. "Analyzing Mass Spectrometry Imaging Data of 13C-Labeled Phospholipids in Camelina sativa and Thlaspi arvense (Pennycress) Embryos" Metabolites 11, no. 3: 148. https://doi.org/10.3390/metabo11030148
APA StyleRomsdahl, T. B., Kambhampati, S., Koley, S., Yadav, U. P., Alonso, A. P., Allen, D. K., & Chapman, K. D. (2021). Analyzing Mass Spectrometry Imaging Data of 13C-Labeled Phospholipids in Camelina sativa and Thlaspi arvense (Pennycress) Embryos. Metabolites, 11(3), 148. https://doi.org/10.3390/metabo11030148






