Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of Enrolled Patients
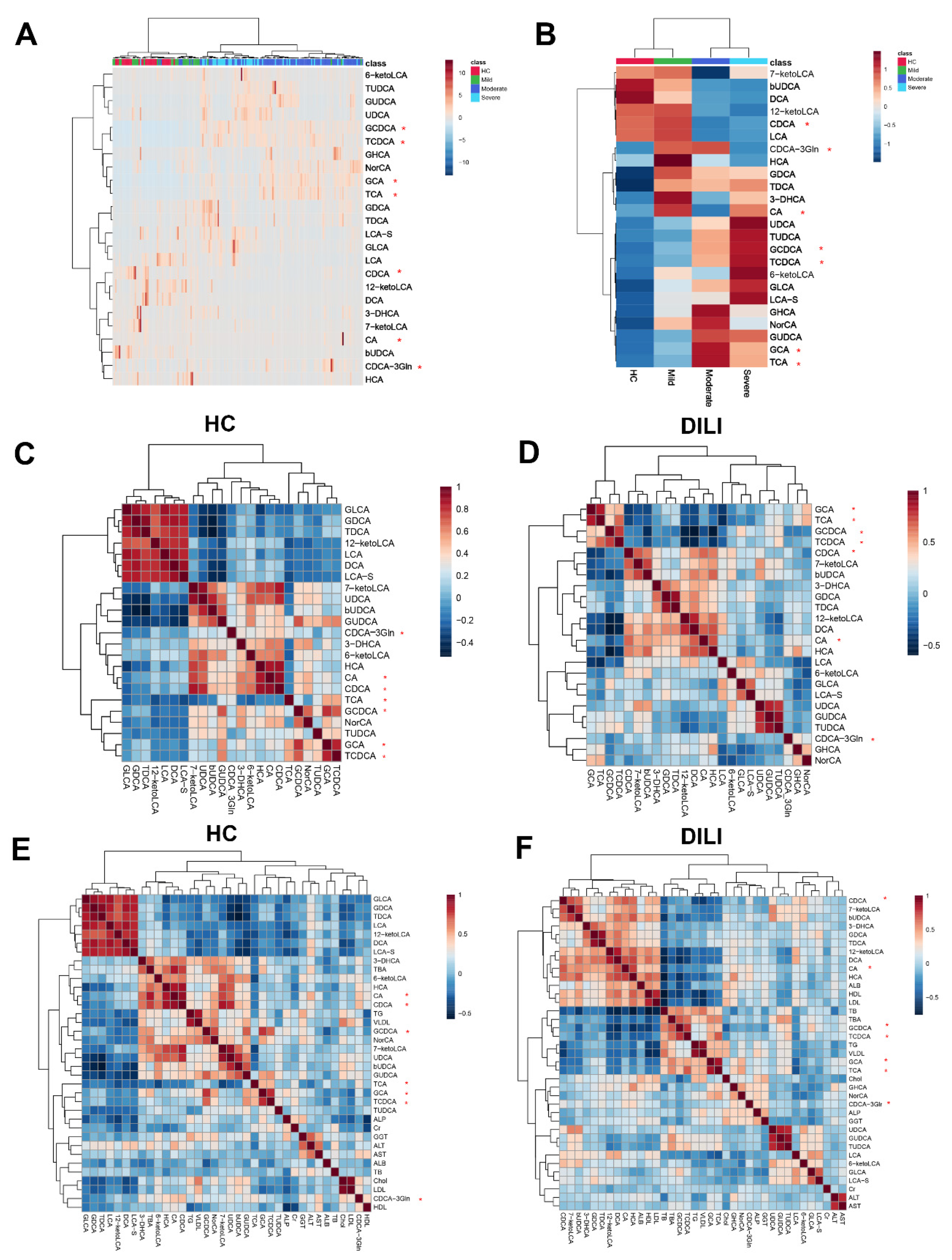
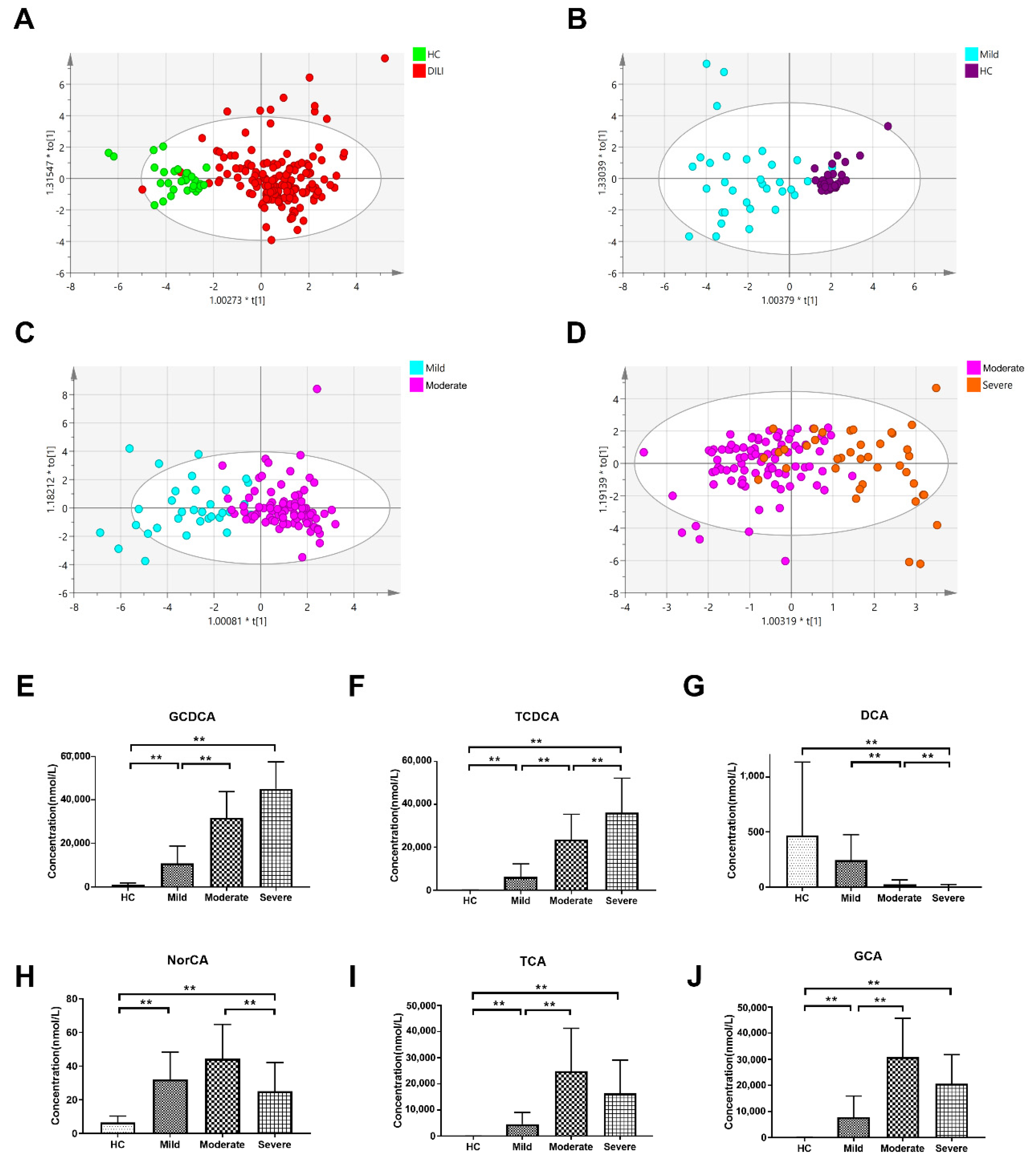
2.2. Multivariate Analysis of Targeted Metabolomics Data of Bile Acids
2.3. Differential Bile Acid Analysis of DILI Patients with Different Severity
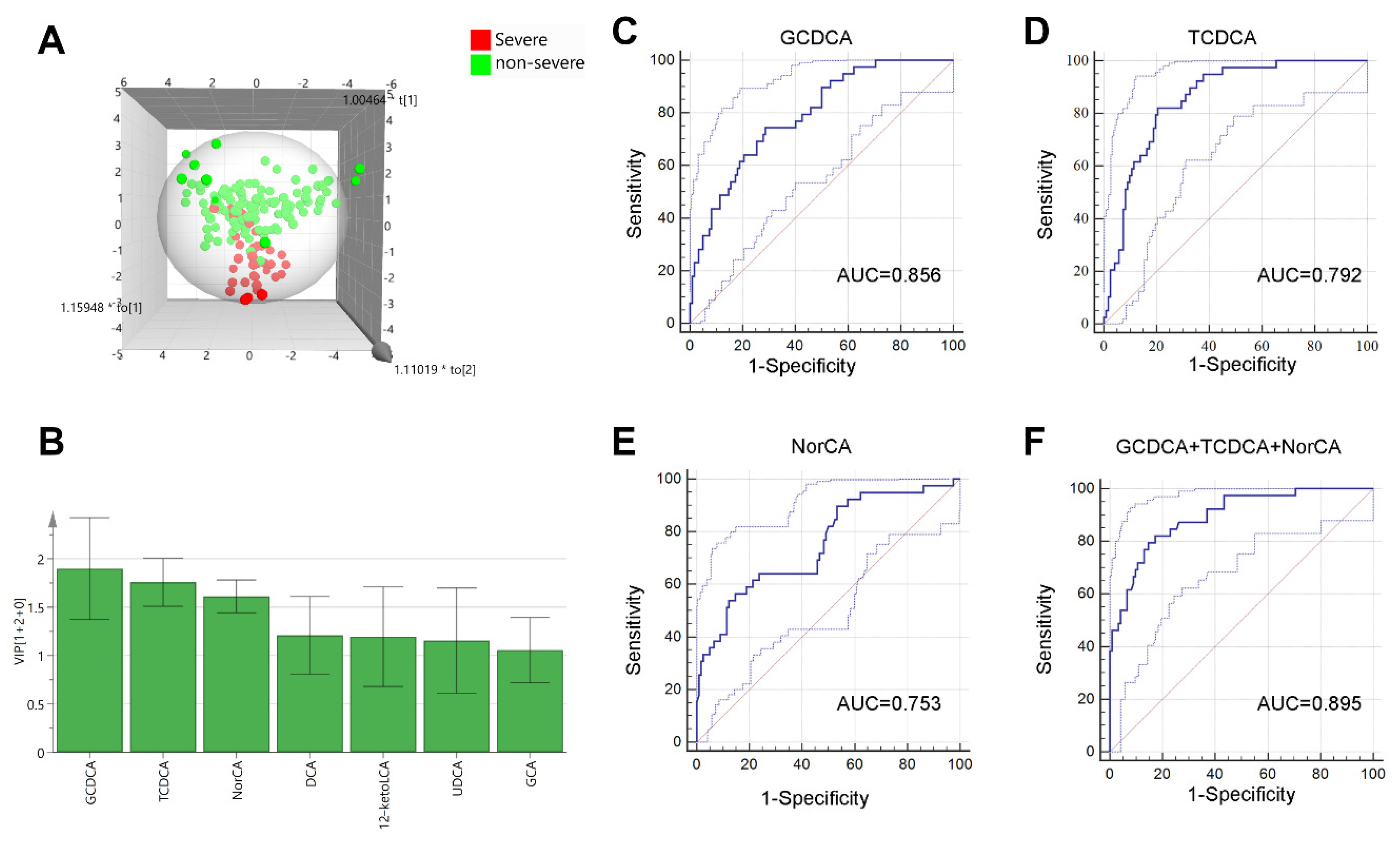
2.4. Bile Acids for Predicting Severe DILI Patients
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Criteria
4.3. Targeted Metabolomic Analysis
4.3.1. Sample Preparation
4.3.2. On-Board Testing
4.4. Statistic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, Y.C.; Mao, Y.M.; Chen, C.W.; Chen, J.J.; Chen, J.; Cong, W.M.; Ding, Y.; Duan, Z.P.; Fu, Q.C.; Guo, X.Y.; et al. CSH guidelines for the diagnosis and treatment of drug-induced liver injury. Hepatol. Int. 2017, 11, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cortes, M.; Robles-Diaz, M.; Stephens, C.; Ortega-Alonso, A.; Lucena, M.I.; Andrade, R.J. Drug induced liver injury: An update. Arch. Toxicol. 2020, 94, 3381–3407. [Google Scholar] [CrossRef]
- Shen, T.; Liu, Y.; Shang, J.; Xie, Q.; Li, J.; Yan, M.; Xu, J.; Niu, J.; Liu, J.; Watkins, P.B.; et al. Incidence and Etiology of Drug-Induced Liver Injury in Mainland China. Gastroenterology 2019, 156, 2230–2241. [Google Scholar] [CrossRef] [Green Version]
- Hillman, L.; Gottfried, M.; Whitsett, M.; Rakela, J.; Schilsky, M.; Lee, W.M.; Ganger, D. Clinical Features and Outcomes of Complementary and Alternative Medicine Induced Acute Liver Failure and Injury. Am. J. Gastroenterol. 2016, 111, 958–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalasani, N.; Bonkovsky, H.L.; Fontana, R.; Lee, W.; Stolz, A.; Talwalkar, J.; Reddy, K.R.; Watkins, P.B.; Navarro, V.; Barnhart, H.; et al. Features and Outcomes of 899 Patients with Drug-Induced Liver Injury: The DILIN Prospective Study. Gastroenterology 2015, 148, 1340–1352.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Huang, A.; Wang, J.B.; Zou, Z. Chronic Drug-Induced Liver Injury: Updates and Future Challenges. Front. Pharmacol. 2021, 12, 627133. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver; Andrade, R.J.; Aithal, G.P.; Björnsson, E.S.; Kaplowitz, N.; Kullak-Ublick, G.A.; Larrey, D.; Karlsen, T.H. EASL Clinical Practice Guidelines: Drug-induced liver injury. J. Hepatol. 2019, 70, 1222–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devarbhavi, H.; Aithal, G.; Treeprasertsuk, S.; Takikawa, H.; Mao, Y.; Shasthry, S.M.; Hamid, S.; Tan, S.S.; Philips, C.A.; George, J.; et al. Drug-induced liver injury: Asia Pacific Association of Study of Liver consensus guidelines. Hepatol. Int. 2021, 15, 258–282. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, E.; Ouyang, X.; Xu, X.; Ma, S.; Ji, F.; Wu, D.; Zhang, S.; Zhao, Y.; Li, L. Metabolomics and Cytokine Analysis for Identification of Severe Drug-Induced Liver Injury. J. Proteome Res. 2019, 18, 2514–2524. [Google Scholar] [CrossRef]
- Cuykx, M.; Rodrigues, R.M.; Laukens, K.; Vanhaecke, T.; Covaci, A. In vitro assessment of hepatotoxicity by metabolomics: A review. Arch. Toxicol. 2018, 92, 3007–3029. [Google Scholar] [CrossRef] [PubMed]
- Watkins, P.B. Biomarkers for the diagnosis and management of drug-induced liver injury. Semin. Liver Dis. 2009, 29, 393–399. [Google Scholar] [CrossRef]
- Soga, T.; Sugimoto, M.; Honma, M.; Mori, M.; Igarashi, K.; Kashikura, K.; Ikeda, S.; Hirayama, A.; Yamamoto, T.; Yoshida, H.; et al. Serum metabolomics reveals gamma-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J. Hepatol. 2011, 55, 896–905. [Google Scholar] [CrossRef]
- Quintas, G.; Martinez-Sena, T.; Conde, I.; Pareja Ibars, E.; Kleinjans, J.; Castell, J.V. Metabolomic analysis to discriminate drug-induced liver injury (DILI) phenotypes. Arch. Toxicol. 2021, 95, 3049–3062. [Google Scholar] [CrossRef]
- Zhang, L.; Niu, M.; Wei, A.-W.; Tang, J.-F.; Tu, C.; Bai, Z.-F.; Zou, Z.-S.; Xiao, X.-H.; Liu, Y.-P.; Wang, J.-B. Risk profiling using metabolomic characteristics for susceptible individuals of drug-induced liver injury caused by Polygonum multiflorum. Arch. Toxicol. 2019, 94, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef]
- Klepacki, J.; Klawitter, J.; Klawitter, J.; Karimpour-Fard, A.; Thurman, J.; Ingle, G.; Patel, D.; Christians, U. Amino acids in a targeted versus a non-targeted metabolomics LC-MS/MS assay. Are the results consistent? Clin. Biochem. 2016, 49, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Fickert, P.; Wagner, M. Biliary bile acids in hepatobiliary injury—What is the link? J. Hepatol. 2017, 67, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, S.; Li, Q.; Cai, L.; Wang, C.; Lei, X. Chemoproteomic Profiling of Bile Acid Interacting Proteins. ACS Cent. Sci. 2017, 3, 501–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, S.; Tsuji, M.; Morio, Y.; Oguchi, K. Involvement of endoplasmic reticulum in glycochenodeoxycholic acid-induced apoptosis in rat hepatocytes. Toxicol. Lett. 2006, 166, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Utanohara, S.; Tsuji, M.; Momma, S.; Morio, Y.; Oguchi, K. The effect of ursodeoxycholic acid on glycochenodeoxycholic acid-induced apoptosis in rat hepatocytes. Toxicology 2005, 214, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, J.; Hu, L.; Liao, B.; Qiao, L.; Shen, W.; Xie, F.; Zhu, G. Glycochenodeoxycholic acid induces stemness and chemoresistance via the STAT3 signaling pathway in hepatocellular carcinoma cells. Aging 2020, 12, 15546–15555. [Google Scholar] [CrossRef] [PubMed]
- Rust, C.; Bauchmuller, K.; Fickert, P.; Fuchsbichler, A.; Beuers, U. Phosphatidylinositol 3-kinase-dependent signaling modulates taurochenodeoxycholic acid-induced liver injury and cholestasis in perfused rat livers. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 289, G88–G94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hata, S.; Wang, P.; Eftychiou, N.; Ananthanarayanan, M.; Batta, A.; Salen, G.; Pang, K.S.; Wolkoff, A.W. Substrate specificities of rat oatp1 and ntcp: Implications for hepatic organic anion uptake. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G829–G839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kihira, K.; Shimazu, K.; Kuwabara, M.; Yoshii, M.; Takeuchi, H.; Nakano, I.; Ozawa, S.; Onuki, M.; Hatta, Y.; Hoshita, T. Bile acid profiles in bile, urine, and feces of a patient with cerebrotendinous xanthomatosis. Steroids 1986, 48, 109–119. [Google Scholar] [CrossRef]
- Kuramoto, T.; Furukawa, Y.; Nishina, T.; Sugimoto, T.; Mahara, R.; Tohma, M.; Kihira, K.; Hoshita, T. Identification of short side chain bile acids in urine of patients with cerebrotendinous xanthomatosis. J. Lipid Res. 1990, 31, 1895–1902. [Google Scholar] [CrossRef]
- Almé, B.; Bremmelgaard, A.; Sjövall, J.; Thomassen, P. Analysis of metabolic profiles of bile acids in urine using a lipophilic anion exchanger and computerized gas-liquid chromatorgaphy-mass spectrometry. J. Lipid Res. 1977, 18, 339–362. [Google Scholar] [CrossRef]
- Amuro, Y.; Hayashi, E.; Endo, T.; Higashino, K.; Kishimoto, S. Unusual trihydroxylated bile acids in urine of patients with liver cirrhosis. Clin. Chim. Acta 1983, 127, 61–67. [Google Scholar] [CrossRef]
- Tian, Q.; Yang, R.; Wang, Y.; Liu, J.; Wee, A.; Saxena, R.; Wang, L.; Li, M.; Liu, L.; Shan, S.; et al. A High Serum Level of Taurocholic Acid is Correlated with the Severity and Resolution of Drug-induced Liver Injury. Clin. Gastroenterol. Hepatol. 2020, 19, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.R.; Soroka, C.J.; Hagey, L.R.; Boyer, J.L. Sirtuin 1 activation alleviates cholestatic liver injury in a cholic acid-fed mouse model of cholestasis. Hepatology 2016, 64, 2151–2164. [Google Scholar] [CrossRef] [Green Version]
- Baghdasaryan, A.; Fuchs, C.D.; Osterreicher, C.H.; Lemberger, U.J.; Halilbasic, E.; Pahlman, I.; Graffner, H.; Krones, E.; Fickert, P.; Wahlstrom, A.; et al. Inhibition of intestinal bile acid absorption improves cholestatic liver and bile duct injury in a mouse model of sclerosing cholangitis. J. Hepatol. 2016, 64, 674–681. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.; Wang, X.; Jiang, R.; Zhao, A.; Yan, J.; Zheng, X.; Huang, F.; Liu, X.; Panee, J.; Rajani, C.; et al. Dysregulated bile acid signaling contributes to the neurological impairment in murine models of acute and chronic liver failure. EBioMedicine 2018, 37, 294–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biagioli, M.; Carino, A.; Fiorucci, C.; Marchiano, S.; Di Giorgio, C.; Bordoni, M.; Roselli, R.; Baldoni, M.; Distrutti, E.; Zampella, A.; et al. The Bile Acid Receptor GPBAR1 Modulates CCL2/CCR2 Signaling at the Liver Sinusoidal/Macrophage Interface and Reverses Acetaminophen-Induced Liver Toxicity. J. Immunol. 2020, 204, 2535–2551. [Google Scholar] [CrossRef] [PubMed]
- Watkins, P.B.; Seeff, L.B. Drug-induced liver injury: Summary of a single topic clinical research conference. Hepatology 2006, 43, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Benichou, C.; Danan, G.; Flahault, A. Causality assessment of adverse reactions to drug—II. An original model for validation of drug causality assessment methods: Case reports with positive rechallenge. J. Clin. Epidemiol. 1993, 46, 1331–1336. [Google Scholar] [CrossRef]
- Aithal, G.P.; Watkins, P.B.; Andrade, R.J.; Larrey, D.; Molokhia, M.; Takikawa, H.; Hunt, C.M.; Wilke, R.A.; Avigan, M.; Kaplowitz, N.; et al. Case definition and phenotype standardization in drug-induced liver injury. Clin. Pharmacol. Ther. 2011, 89, 806–815. [Google Scholar] [CrossRef]
- Lan, K.; Su, M.; Xie, G.; Ferslew, B.C.; Brouwer, K.L.; Rajani, C.; Liu, C.; Jia, W. Key Role for the 12-Hydroxy Group in the Negative Ion Fragmentation of Unconjugated C24 Bile Acids. Anal. Chem. 2016, 88, 7041–7048. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ni, Y.; Qian, L.; Fang, Q.; Zheng, T.; Zhang, M.; Gao, Q.; Zhang, Y.; Ni, J.; Hou, X.; et al. Decreased Abundance of Akkermansia muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv. Sci. 2021, 8, e2100536. [Google Scholar] [CrossRef]

| Health Control (n = 31) | Grade 1 (Mild) n = 32 | Grade 2 (Moderate) n = 90 | Grade 3 (Severe) n = 39 | p Value | |
|---|---|---|---|---|---|
| Age, years | 50.1 ± 14.8 | 51.4 ± 14.9 | 51.1 ± 13.8 | 52.1 ± 15.5 | 0.937 |
| Female% | 16(51.6) | 26(81.3) | 55(61.1) | 29(74.4) | 0.071 |
| Alcohol use | 7(22.6) | 5(15.6) | 17(18.9) | 6(15.4) | 0.852 |
| Hypertension | 7(22.6) | 7(21.9) | 20(22.2) | 6(15.4) | 0.661 |
| Latency | |||||
| <5 days | / | 6(18.8) | 7(7.8) | 1(2.6) | 0.179 |
| 5 days–90 days | 23(71.9) | 76(84.4) | 34(87.2) | ||
| >90 days | 3(9.4) | 7(7.8) | 4(10.3) | ||
| Liver biochemistries | |||||
| WBC (109/L) | 6.6(5.4, 7.6) | 4.6(3.8, 5.3) | 5(4.3, 6.8) | 7.2(4.7, 9.3) ## | <0.001 |
| ALB (g/L) | 47.5(45.1, 48.8) | 40.6(38, 42.6) | 36.4(32.5, 39.1) *** | 31.4(28.2, 33.5) ### | <0.001 |
| ALT (U/L) | 14(12, 22) | 401(246, 655.8) | 360.5(136.8, 715.8) | 337(131, 617) | 0.448 |
| AST (U/L) | 19(17, 21.5) | 200.5(96.3, 318.3) | 248(88.8, 474.3) | 211(117, 336) | 0.65 |
| TBA (μmol/L) | 5(3, 6) | 13.6(7, 20.6) | 151.4(117.9, 194.5) *** | 177(137, 210) | <0.001 |
| ALP (U/L) | 66(55, 82) | 120.5(98.3, 147.3) | 147.5(113.5, 217.8) | 116(99, 134) ### | <0.001 |
| GGT (U/L) | 24(14, 35) | 124(70, 194.8) | 128(73.5, 305) ** | 72(41, 122) ### | <0.001 |
| TB (μmol/L) | 12(8, 16) | 13.3(9.7, 18.6) | 241.3(111.6, 358.6) *** | 291(240.2, 443) | <0.001 |
| TG (mmol/L) | 1.7(0.97, 2.31) | 1.15(0.74, 1.49) | 2.54(1.78, 3.52) *** | 1.63(1.06, 2.4) ### | <0.001 |
| Cholesterol (mmol/L) | 4.54(4.06, 5.08) | 3.41(3.18, 4.05) | 3.52(2.81, 4.61) | 2.71(2.25, 3.19) ### | <0.001 |
| HDL (mmol/L) | 1.13(0.97, 1.31) | 1.07(0.79, 1.24) | 0.19(0.13, 0.43) *** | 0.15(0.12, 0.33) | <0.001 |
| LDL (mmol/L) | 2.34(2.08, 3.01) | 1.78(1.37, 2.22) | 0.67(0.23, 1.42) *** | 0.68(0.18, 1.37) | <0.001 |
| VLDL (mmol/L) | 0.88(0.65, 1.13) | 0.69(0.55, 0.91) | 2.26(1.44, 3.45) *** | 1.45(0.6, 2.26) ### | <0.001 |
| INR | / | 0.99(0.96, 1.05) | 1.08(0.97, 1.23) * | 1.62(1.53, 2.16) ### | <0.001 |
| Pattern of liver injury (%) | |||||
| Hepatocellular | / | 26(81.3) | 57(63.3) | 25(64.1) | 0.025 |
| Cholestatic | 6(18.8) | 12(13.3) | 9(23.1) | ||
| Mixed | 0(0) | 21(23.3) | 5(12.8) | ||
| RUCAM score | |||||
| Highly probable (>8) | / | 3(9.4) | 1(1.1) | 1(2.6) | 0.094 |
| Probable (6–8) | 21(65.6) | 58(64.4) | 30(76.9) | ||
| Possible (3–5) | 8(25) | 31(34.4) | 8(20.5) |
| Bile Acid | Type | VIP | FC | log2(FC) | Differentially Expression | p Value |
|---|---|---|---|---|---|---|
| DILI vs. HC | ||||||
| 12-ketoLCA | Secondary | 1.109 | 0.359 | −1.476 | ↓ | 7.28 × 10−5 |
| bUDCA | Secondary | 1.043 | 0.185 | −2.436 | ↓ | 4.61 × 10−8 |
| DCA | Secondary | 1.301 | 0.139 | −2.846 | ↓ | 3.28 × 10−11 |
| GCA | Primary | 1.501 | 147.95 | 7.209 | ↑ | 1.05 × 10−14 |
| GCDCA | Primary | 1.551 | 34.106 | 5.092 | ↑ | 4.94 × 10−20 |
| NorCA | Secondary | 1.4 | 5.645 | 2.497 | ↑ | 2.47 × 10−14 |
| TCA | Primary | 1.353 | 926.55 | 9.856 | ↑ | 5.05 × 10−10 |
| TCDCA | Primary | 1.446 | 618.02 | 9.272 | ↑ | 3.21 × 10−14 |
| Mild vs. HC | ||||||
| 6-ketoLCA | Secondary | 1.026 | 3.526 | 1.818 | ↑ | 7.68 × 10−5 |
| GCA | Primary | 1.218 | 48.529 | 5.601 | ↑ | 2.36 × 10−6 |
| GCDCA | Primary | 1.408 | 11.881 | 3.571 | ↑ | 4.05 × 10−9 |
| GDCA | Secondary | 1.215 | 4.964 | 2.312 | ↑ | 8.28 × 10−7 |
| GHCA | Secondary | 1.294 | 4544 | 12.15 | ↑ | 1.66 × 10−7 |
| GLCA | Secondary | 1.015 | 2.865 | 1.518 | ↑ | 5.03 × 10−3 |
| GUDCA | Secondary | 1.135 | 45.726 | 5.515 | ↑ | 3.20 × 10−4 |
| HCA | Secondary | 1.017 | 5.565 | 2.476 | ↑ | 5.91 × 10−5 |
| NorCA | Secondary | 1.402 | 4.88 | 2.287 | ↑ | 5.27 × 10−12 |
| TCA | Primary | 1.284 | 226.42 | 7.823 | ↑ | 4.68 × 10−7 |
| TCDCA | Primary | 1.33 | 163.05 | 7.349 | ↑ | 6.90 × 10−7 |
| TDCA | Secondary | 1.325 | 18.522 | 4.211 | ↑ | 7.58 × 10−8 |
| TUDCA | Secondary | 1.162 | 363.28 | 8.505 | ↑ | 4.18 × 10−4 |
| Moderate vs. Mild | ||||||
| 12-ketoLCA | Secondary | 1.217 | 0.223 | −2.164 | ↓ | 6.47 × 10−8 |
| 7-ketoLCA | Secondary | 1.239 | 0.221 | −2.18 | ↓ | 4.48 × 10−9 |
| CDCA | Secondary | 1.081 | 0.186 | −2.423 | ↓ | 1.90 × 10−6 |
| DCA | Secondary | 1.471 | 0.108 | −3.212 | ↓ | 2.79 × 10−14 |
| GCA | Primary | 1.395 | 3.955 | 1.984 | ↑ | 1.40 × 10−13 |
| GCDCA | Primary | 1.49 | 2.947 | 1.559 | ↑ | 2.37 × 10−15 |
| LCA | Secondary | 1.084 | 0.355 | −1.492 | ↓ | 7.76 × 10−4 |
| TCA | Primary | 1.351 | 5.406 | 2.435 | ↑ | 3.24 × 10−10 |
| TCDCA | Primary | 1.428 | 3.842 | 1.942 | ↑ | 1.00 × 10−12 |
| Severe vs. Moderate | ||||||
| CDCA-3Gln | Primary | 1.136 | 0.433 | −1.207 | ↓ | 1.16 × 10−4 |
| DCA | Secondary | 1.177 | 0.368 | −1.442 | ↓ | 0.010523 |
| NorCA | Secondary | 1.692 | 0.565 | −0.824 | ↓ | 6.78 × 10−7 |
| TCDCA | Primary | 1.398 | 1.537 | 0.62 | ↑ | 1.59 × 10−6 |
| UDCA | Secondary | 1.037 | 1.808 | 0.854 | ↑ | 0.014075 |
| Variable | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | |
| 12-ketoLCA (nmol/L) | 0.665(0.486, 0.908) | 0.01 | ||
| HCA (nmol/L) | 0.847(0.775, 0.925) | <0.001 | ||
| NorCA (nmol/L) | 0.945(0.919, 0.972) | <0.001 | 0.941(0.912, 0.972) | <0.001 |
| DCA (nmol/L) | 0.965(0.943, 0.987) | 0.002 | ||
| bUDCA (nmol/L) | 0.981(0.967, 0.995) | 0.008 | ||
| CDCA-3Gln (nmol/L) | 0.995(0.992, 0.998) | <0.001 | ||
| 6-ketoLCA (nmol/L) | 1.042(1.003, 1.082) | 0.034 | ||
| TCDCA (umol/L) | 1.084(1.051, 1.117) | <0.001 | 1.061(1.016, 1.107) | 0.007 |
| GCDCA (umol/L) | 1.111(1.067, 1.156) | <0.001 | 1.064(1.020, 1.110) | <0.001 |
| UDCA (umol/L) | 1.365(1.121, 1.663) | 0.002 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Zhang, L.; Chen, E.; Lu, J.; Xiao, L.; Liu, Q.; Zhu, D.; Zhang, F.; Xu, X.; Li, L. Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury. Metabolites 2021, 11, 852. https://doi.org/10.3390/metabo11120852
Xie Z, Zhang L, Chen E, Lu J, Xiao L, Liu Q, Zhu D, Zhang F, Xu X, Li L. Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury. Metabolites. 2021; 11(12):852. https://doi.org/10.3390/metabo11120852
Chicago/Turabian StyleXie, Zhongyang, Lingjian Zhang, Ermei Chen, Juan Lu, Lanlan Xiao, Qiuhong Liu, Danhua Zhu, Fen Zhang, Xiaowei Xu, and Lanjuan Li. 2021. "Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury" Metabolites 11, no. 12: 852. https://doi.org/10.3390/metabo11120852
APA StyleXie, Z., Zhang, L., Chen, E., Lu, J., Xiao, L., Liu, Q., Zhu, D., Zhang, F., Xu, X., & Li, L. (2021). Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury. Metabolites, 11(12), 852. https://doi.org/10.3390/metabo11120852






