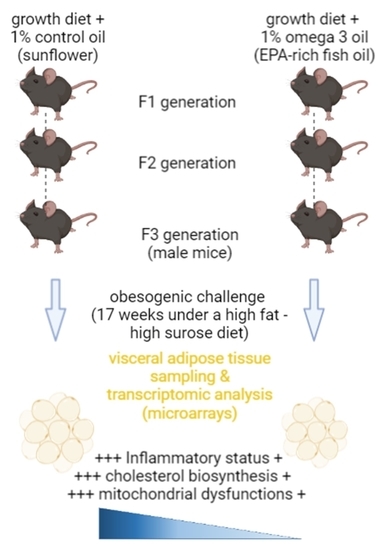
Adipose Tissue Dysfunctions in Response to an Obesogenic Diet Are Reduced in Mice after Transgenerational Supplementation with Omega 3 Fatty Acids
Abstract
:1. Introduction
2. Results
2.1. High-Fat Diet Related Transcriptomic Adaptation in VAT Are Reduced after Transgenerational Intake of EPA
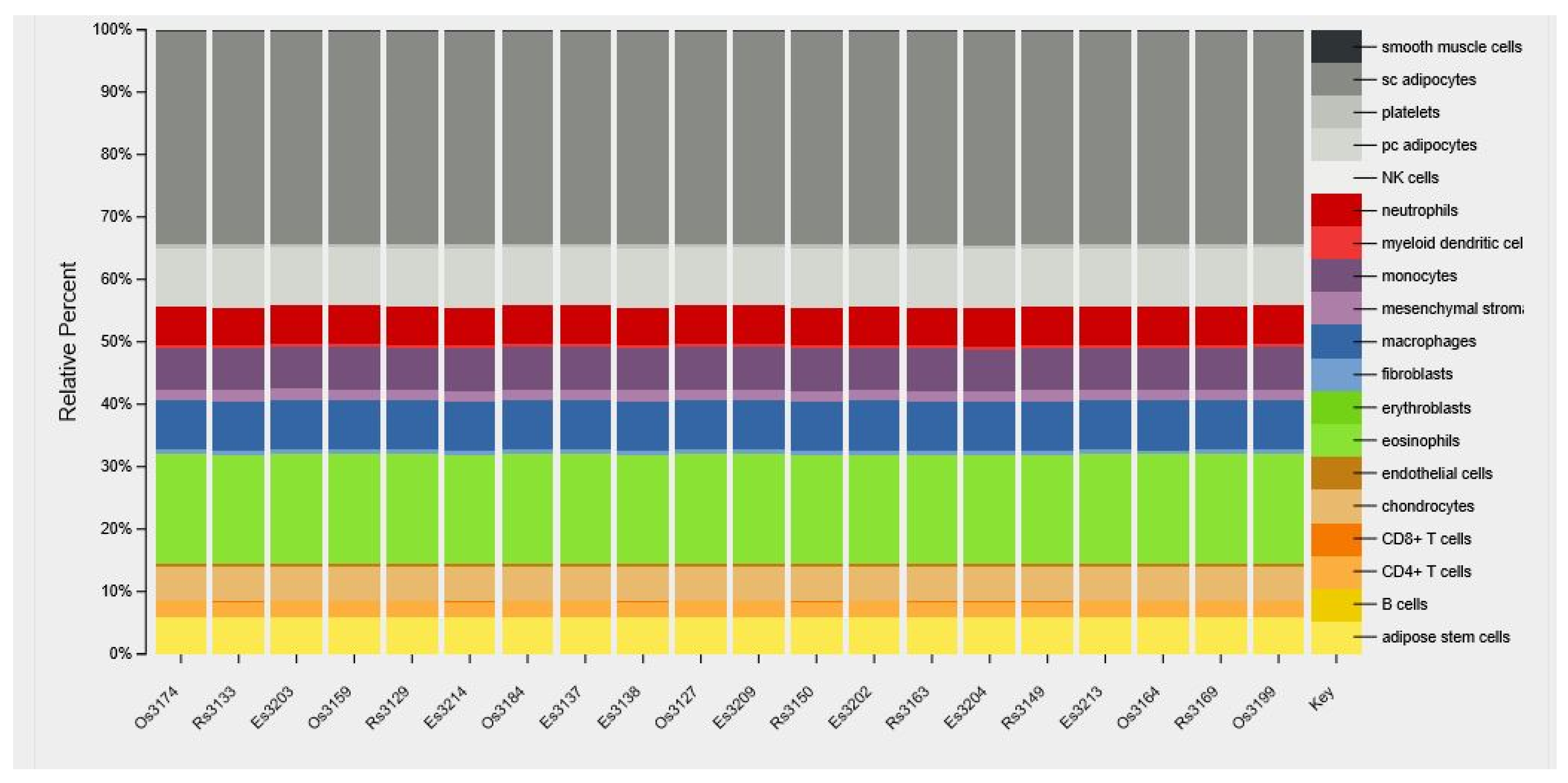
2.2. Comparison of Cell Type Distribution in VAT
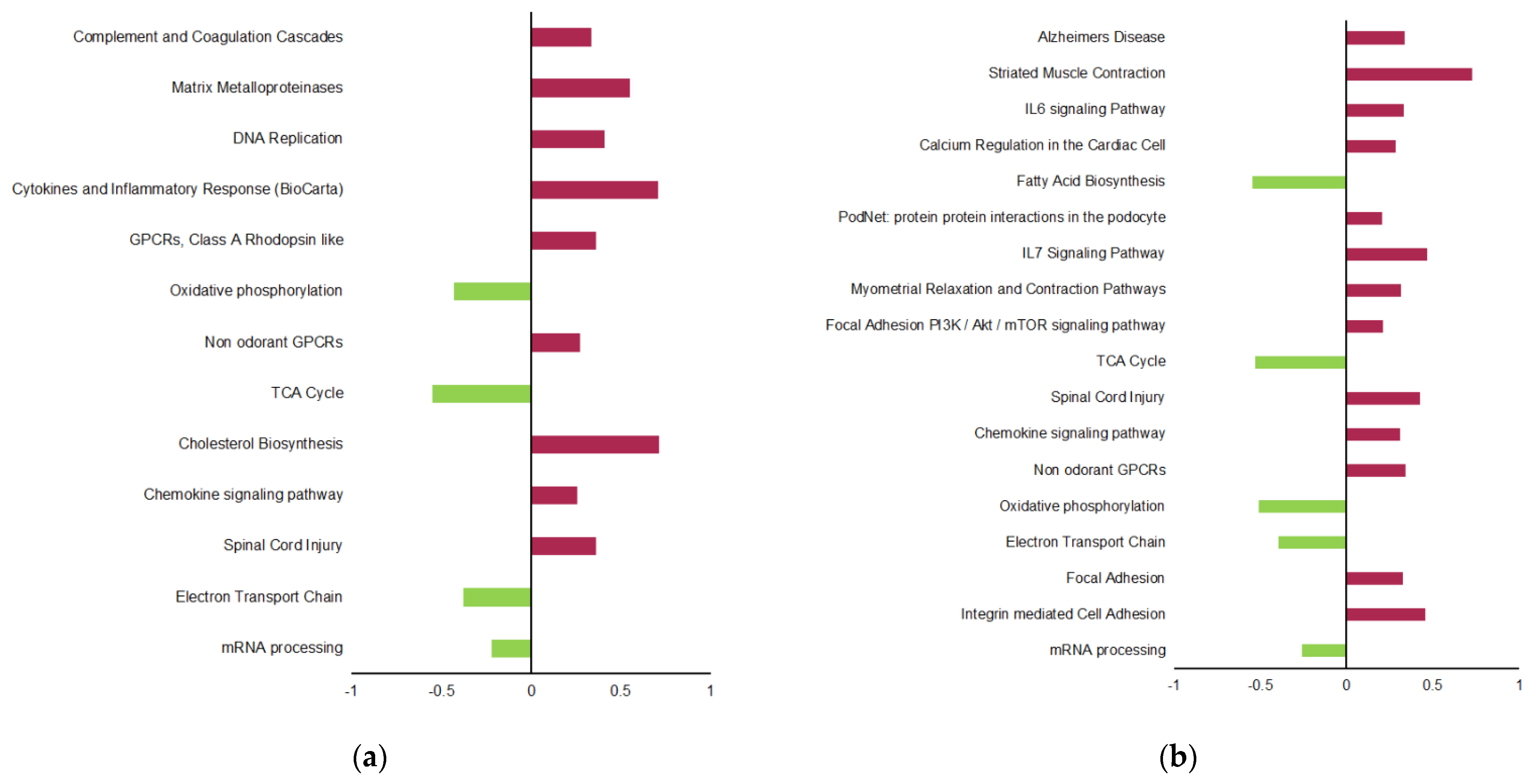
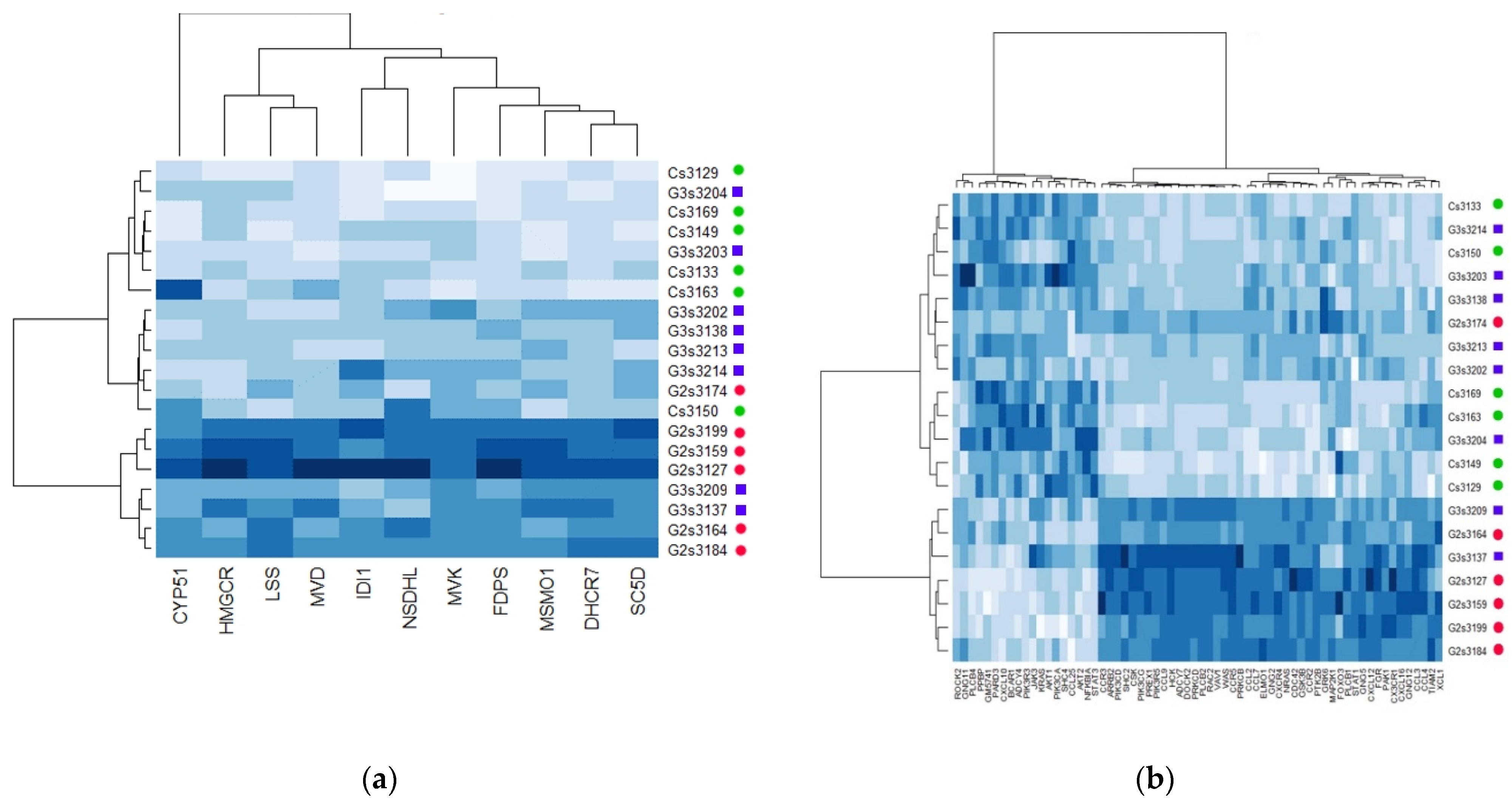
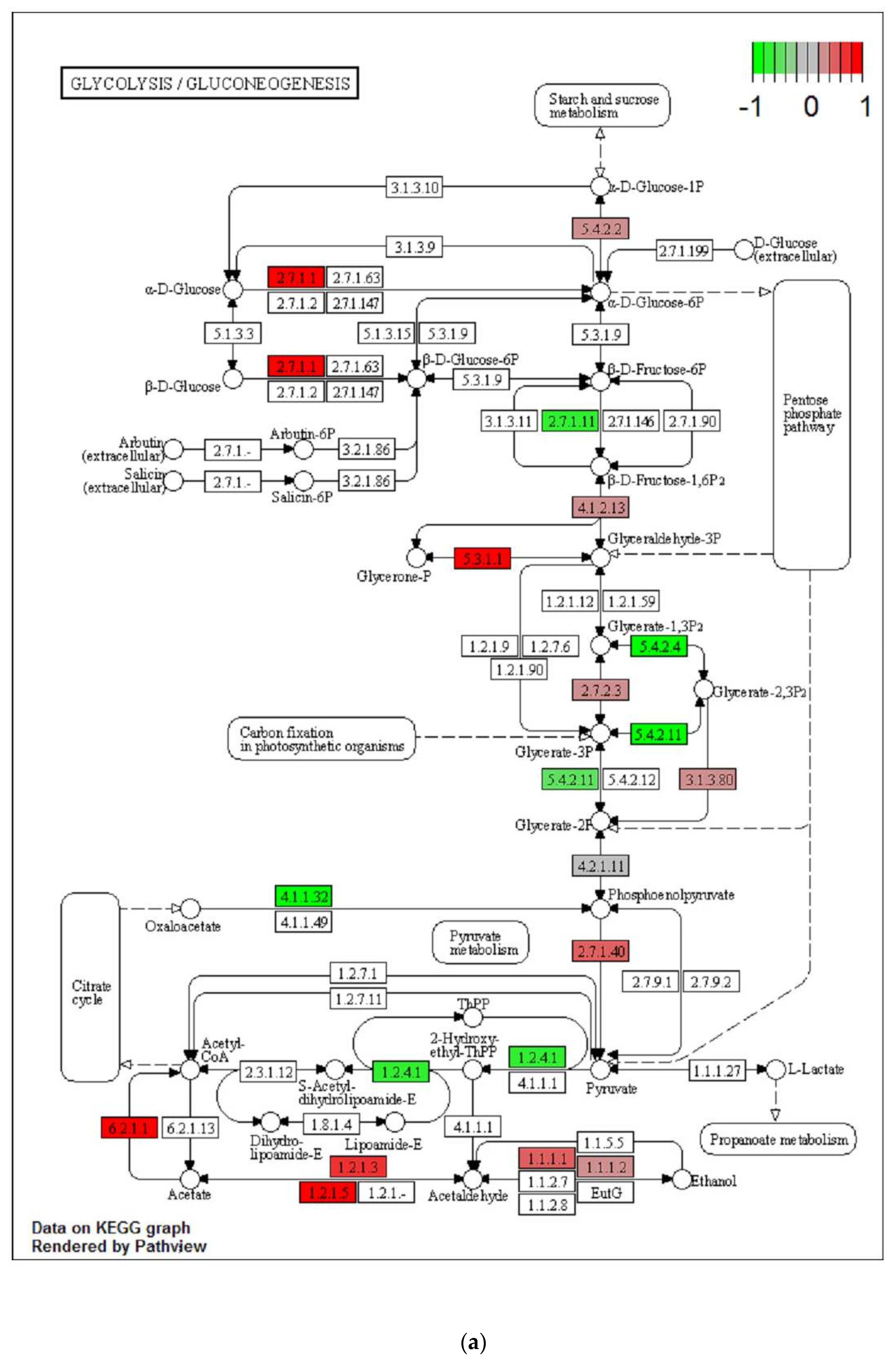
2.3. Transgenerational Supplementation with EPA-Induced Differential Regulation of Biological Pathways and Processes in VAT
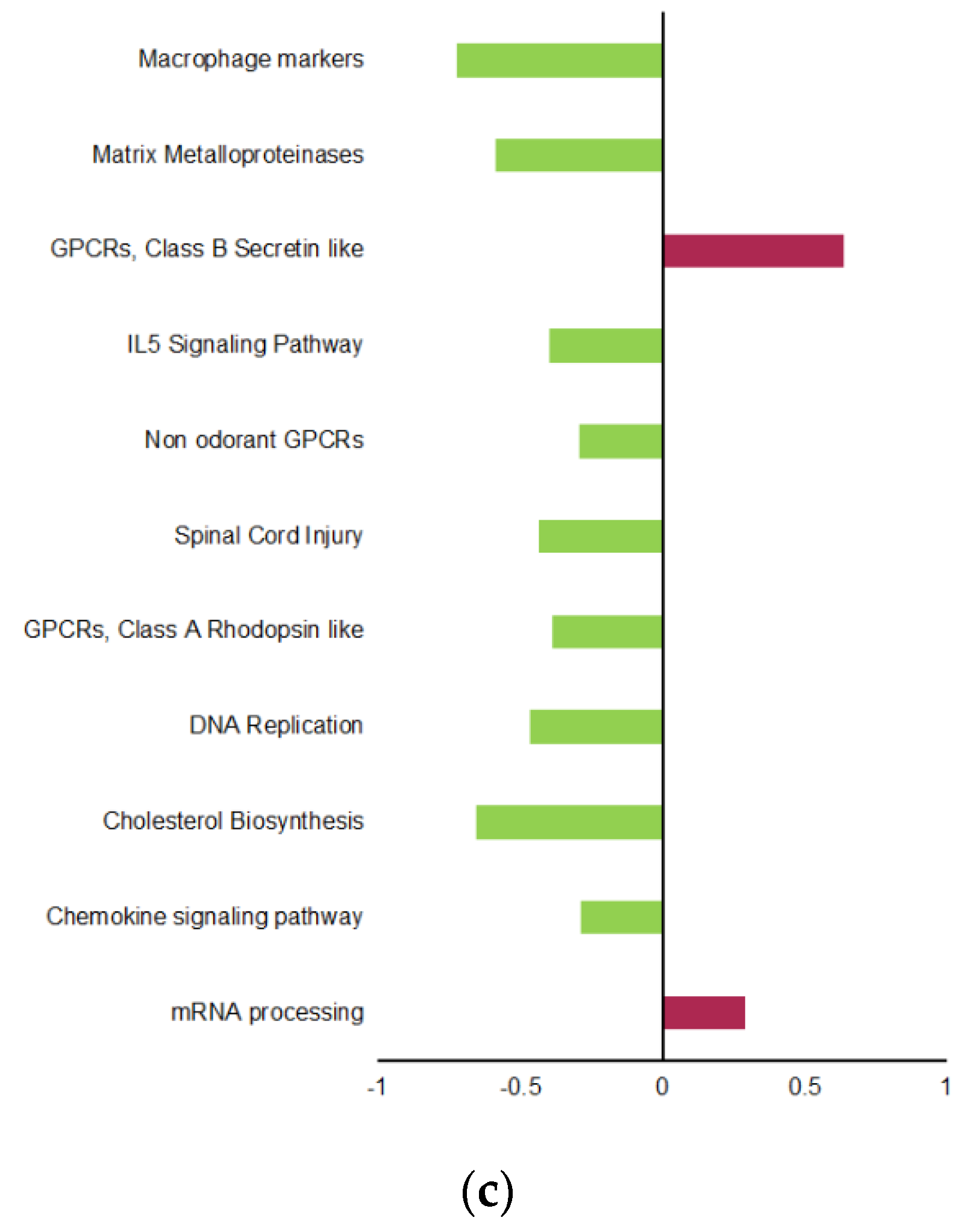
2.4. Differential Regulation of Mitochondrial Processes in VAT between EPA and Control Lineages
2.5. Molecular Mediators of Differential Expression in VAT between Groups
3. Discussion
4. Materials and Methods
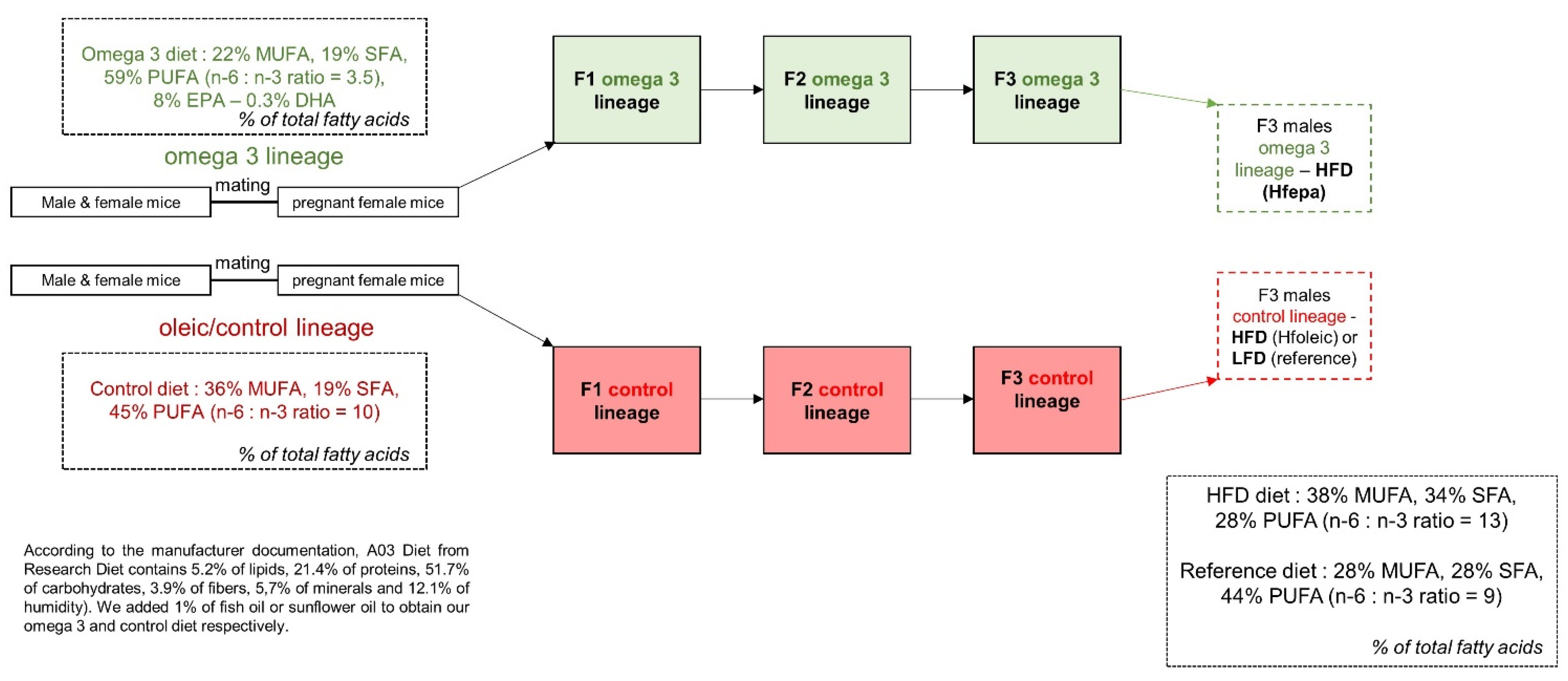
4.1. Dietary Intervention and Sample Collection
4.2. Transcriptomic Analysis
4.3. RT-q PCR Validation of Gene Expression Change
4.4. Statistics
4.5. Characterization of Cell-Type Abundance
4.6. Biological Pathway Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bjerregaard, L.G.; Adelborg, K.; Baker, J.L. Change in body mass index from childhood onwards and risk of adult cardiovascular disease. Trends Cardiovasc. Med. 2020, 30, 39–45. [Google Scholar] [CrossRef]
- Freedman, D.S.; Khan, L.K.; Dietz, W.H.; Srinivasan, S.R.; Berenson, G.S. Relationship of childhood obesity to coronary heart disease risk factors in adulthood: The Bogalusa Heart Study. Pediatrics 2001, 108, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P.; Wood, I.S. Adipokines: Inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr. 2004, 92, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Cusi, K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr. Diab. Rep. 2010, 10, 306–315. [Google Scholar] [CrossRef]
- Kjellberg, J.; Tange Larsen, A.; Ibsen, R.; Hojgaard, B. The socioeconomic burden of obesity. Obes. Facts 2017, 10, 493–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, F.T.; Zulet, M.A.; Marchini, J.S.; Martinez, J.A. Bioactive compounds with effects on inflammation markers in humans. Int. J. Food Sci. Nutr. 2012, 63, 749–765. [Google Scholar] [CrossRef]
- Simopoulos, A.P. The omega-6/omega-3 fatty acid ratio: Health implications. Oilseeds fats Crop. Lipids 2010, 17, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P.; Leaf, A.; Salem, N., Jr. Workshop statement on the essentiality of and recommended dietary intakes for Omega-6 and Omega-3 fatty acids. Prostaglandins Leukot. Essent. Fatty Acids 2000, 63, 119–121. [Google Scholar] [CrossRef]
- ANSES, Rapport d’Expertise Collective. Actualisation des Apports Nutritionnels Conseillés Pour les Acides Gras. Saisine n° 2006-SA-0359. Available online: https://www.anses.fr/fr/system/files/NUT2006sa0359Ra.pdf (accessed on 1 September 2020).
- Poudyal, H.; Panchal, S.K.; Diwan, V.; Brown, L. Omega-3 fatty acids and metabolic syndrome: Effects and emerging mechanisms of action. Prog. Lipid Res. 2011, 50, 372–387. [Google Scholar] [CrossRef]
- Pinel, A.; Morio-Liondore, B.; Capel, F. n-3 Polyunsaturated fatty acids modulate metabolism of insulin-sensitive tissues: Implication for the prevention of type 2 diabetes. J. Physiol. Biochem. 2014, 70, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Massiera, F.; Barbry, P.; Guesnet, P.; Joly, A.; Luquet, S.; Moreilhon-Brest, C.; Mohsen-Kanson, T.; Amri, E.Z.; Ailhaud, G. A western-like fat diet is sufficient to induce a gradual enhancement in fat mass over generations. J. Lipid. Res. 2010, 51, 2352–2361. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 2006, 83, 1505S–1519S. [Google Scholar] [CrossRef] [PubMed]
- Galano, J.M.; Lee, Y.Y.; Oger, C.; Vigor, C.; Vercauteren, J.; Durand, T.; Giera, M.; Lee, J.C. Isoprostanes, neuroprostanes and phytoprostanes: An overview of 25years of research in chemistry and biology. Prog. Lipid Res. 2017, 68, 83–108. [Google Scholar] [CrossRef] [PubMed]
- Kuda, O.; Rossmeisl, M.; Kopecky, J. Omega-3 fatty acids and adipose tissue biology. Mol. Asp. Med. 2018, 64, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Pinel, A.; Rigaudiere, J.P.; Jouve, C.; Capel, F. Modulation of insulin resistance and the adipocyte-skeletal muscle cell cross-talk by LCn-3PUFA. Int. J. Mol. Sci. 2018, 19, 2778. [Google Scholar] [CrossRef] [Green Version]
- Pinel, A.; Pitois, E.; Rigaudiere, J.P.; Jouve, C.; De Saint-Vincent, S.; Laillet, B.; Montaurier, C.; Huertas, A.; Morio, B.; Capel, F. EPA prevents fat mass expansion and metabolic disturbances in mice fed with a Western diet. J. Lipid Res. 2016, 57, 1382–1397. [Google Scholar] [CrossRef] [Green Version]
- Kalupahana, N.S.; Claycombe, K.; Newman, S.J.; Stewart, T.; Siriwardhana, N.; Matthan, N.; Lichtenstein, A.H.; Moustaid-Moussa, N. Eicosapentaenoic acid prevents and reverses insulin resistance in high-fat diet-induced obese mice via modulation of adipose tissue inflammation. J. Nutr. 2010, 140, 1915–1922. [Google Scholar] [CrossRef] [Green Version]
- Madsen, L.; Petersen, R.K.; Kristiansen, K. Regulation of adipocyte differentiation and function by polyunsaturated fatty acids. Biochim. Biophys. Acta 2005, 1740, 266–286. [Google Scholar] [CrossRef] [Green Version]
- Manickam, E.; Sinclair, A.J.; Cameron-Smith, D. Suppressive actions of eicosapentaenoic acid on lipid droplet formation in 3T3-L1 adipocytes. Lipids Health Dis. 2010, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Todoric, J.; Loffler, M.; Huber, J.; Bilban, M.; Reimers, M.; Kadl, A.; Zeyda, M.; Waldhausl, W.; Stulnig, T.M. Adipose tissue inflammation induced by high-fat diet in obese diabetic mice is prevented by n-3 polyunsaturated fatty acids. Diabetologia 2006, 49, 2109–2119. [Google Scholar] [CrossRef] [Green Version]
- White, P.J.; Arita, M.; Taguchi, R.; Kang, J.X.; Marette, A. Transgenic restoration of long-chain n-3 fatty acids in insulin target tissues improves resolution capacity and alleviates obesity-linked inflammation and insulin resistance in high-fat-fed mice. Diabetes 2010, 59, 3066–3073. [Google Scholar] [CrossRef] [Green Version]
- Pinel, A.; Rigaudiere, J.P.; Jouve, C.; Montaurier, C.; Jousse, C.; Lhomme, M.; Morio, B.; Capel, F. Transgenerational supplementation with eicosapentaenoic acid reduced the metabolic consequences on the whole body and skeletal muscle in mice receiving an obesogenic diet. Eur. J. Nutr. 2021, 60, 3143–3157. [Google Scholar] [CrossRef]
- Corral-Jara, K.F.; Cantini, L.; Poupin, N.; Ye, T.; Rigaudiere, J.P.; De Saint Vincent, S.; Pinel, A.; Morio, B.; Capel, F. An integrated analysis of miRNA and gene expression changes in response to an obesogenic diet to explore the impact of transgenerational supplementation with Omega 3 fatty acids. Nutrients 2020, 12, 3864. [Google Scholar] [CrossRef]
- Moreno-Aliaga, M.J.; Lorente-Cebrian, S.; Martinez, J.A. Regulation of adipokine secretion by n-3 fatty acids. Proc. Nutr. Soc. 2010, 69, 324–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Smith, A.C.; Robinson, A.J. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016, 44, D1258–D1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhlhausler, B.S.; Gibson, R.A.; Makrides, M. The effect of maternal omega-3 long-chain polyunsaturated fatty acid (n-3 LCPUFA) supplementation during pregnancy and/or lactation on body fat mass in the offspring: A systematic review of animal studies. Prostaglandins Leukot. Essent. Fatty Acids 2011, 85, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.E.; Magill-Collins, M.J.; Newsom, S.A.; Janssen, R.C.; Friedman, J.E. Maternal fat-1 transgene protects offspring from excess weight gain, oxidative stress, and reduced fatty acid oxidation in response to high-fat Diet. Nutrients 2020, 12, 767. [Google Scholar] [CrossRef] [Green Version]
- Rutkowski, J.M.; Stern, J.H.; Scherer, P.E. The cell biology of fat expansion. J. Cell Biol. 2015, 208, 501–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, S.M.; Saltiel, A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017, 13, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Tordjman, J.; Clement, K.; Scherer, P.E. Fibrosis and adipose tissue dysfunction. Cell Metab. 2013, 18, 470–477. [Google Scholar] [CrossRef] [Green Version]
- Lopategi, A.; Lopez-Vicario, C.; Alcaraz-Quiles, J.; Garcia-Alonso, V.; Rius, B.; Titos, E.; Claria, J. Role of bioactive lipid mediators in obese adipose tissue inflammation and endocrine dysfunction. Mol. Cell. Endocrinol. 2016, 419, 44–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hames, K.C.; Morgan-Bathke, M.; Harteneck, D.A.; Zhou, L.; Port, J.D.; Lanza, I.R.; Jensen, M.D. Very-long-chain omega-3 fatty acid supplements and adipose tissue functions: A randomized controlled trial. Am. J. Clin. Nutr. 2017, 105, 1552–1558. [Google Scholar] [CrossRef] [Green Version]
- Spencer, M.; Finlin, B.S.; Unal, R.; Zhu, B.; Morris, A.J.; Shipp, L.R.; Lee, J.; Walton, R.G.; Adu, A.; Erfani, R.; et al. Omega-3 fatty acids reduce adipose tissue macrophages in human subjects with insulin resistance. Diabetes 2013, 62, 1709–1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardel, M.; Lemas, D.J.; Jackson, K.H.; Friedman, J.E.; Fernandez, J.R. Higher intake of PUFAs is associated with lower total and visceral adiposity and higher lean mass in a racially diverse sample of children. J. Nutr. 2015, 145, 2146–2152. [Google Scholar] [CrossRef]
- Sato, T.; Kameyama, T.; Ohori, T.; Matsuki, A.; Inoue, H. Effects of eicosapentaenoic acid treatment on epicardial and abdominal visceral adipose tissue volumes in patients with coronary artery disease. J. Atheroscler. Thromb. 2014, 21, 1031–1043. [Google Scholar] [CrossRef] [Green Version]
- Gray, B.; Steyn, F.; Davies, P.S.; Vitetta, L. Omega-3 fatty acids: A review of the effects on adiponectin and leptin and potential implications for obesity management. Eur. J. Clin. Nutr. 2013, 67, 1234–1242. [Google Scholar] [CrossRef] [Green Version]
- Prasun, P. Mitochondrial dysfunction in metabolic syndrome. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165838. [Google Scholar] [CrossRef]
- Worsch, S.; Heikenwalder, M.; Hauner, H.; Bader, B.L. Dietary n-3 long-chain polyunsaturated fatty acids upregulate energy dissipating metabolic pathways conveying anti-obesogenic effects in mice. Nutr. Metab. 2018, 15, 65. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, T.E.; Castro, E.; Belchior, T.; Andrade, M.L.; Chaves-Filho, A.B.; Peixoto, A.S.; Moreno, M.F.; Ortiz-Silva, M.; Moreira, R.J.; Inague, A.; et al. Fish oil protects wild type and uncoupling protein 1-deficient mice from obesity and glucose intolerance by increasing energy expenditure. Mol. Nutr. Food Res. 2019, 63, e1800813. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.A.; She, P.; Peroni, O.D.; Lynch, C.J.; Kahn, B.B. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. J. Biol. Chem. 2010, 285, 11348–11356. [Google Scholar] [CrossRef] [Green Version]
- Itariu, B.K.; Zeyda, M.; Hochbrugger, E.E.; Neuhofer, A.; Prager, G.; Schindler, K.; Bohdjalian, A.; Mascher, D.; Vangala, S.; Schranz, M.; et al. Long-chain n-3 PUFAs reduce adipose tissue and systemic inflammation in severely obese nondiabetic patients: A randomized controlled trial. Am. J. Clin. Nutr. 2012, 96, 1137–1149. [Google Scholar] [CrossRef] [Green Version]
- Eldar-Finkelman, H.; Schreyer, S.A.; Shinohara, M.M.; LeBoeuf, R.C.; Krebs, E.G. Increased glycogen synthase kinase-3 activity in diabetes- and obesity-prone C57BL/6J mice. Diabetes 1999, 48, 1662–1666. [Google Scholar] [CrossRef] [PubMed]
- Rayasam, G.V.; Tulasi, V.K.; Sodhi, R.; Davis, J.A.; Ray, A. Glycogen synthase kinase 3: More than a namesake. Br. J. Pharmacol. 2009, 156, 885–898. [Google Scholar] [CrossRef] [Green Version]
- Mraz, M.; Haluzik, M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J. Endocrinol. 2014, 222, R113–R127. [Google Scholar] [CrossRef] [Green Version]
- Corvera, S. Cellular heterogeneity in adipose tissues. Annu. Rev. Physiol. 2021, 83, 257–278. [Google Scholar] [CrossRef]
- Bordoni, A.; Di Nunzio, M.; Danesi, F.; Biagi, P.L. Polyunsaturated fatty acids: From diet to binding to ppars and other nuclear receptors. Genes Nutr. 2006, 1, 95–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, R.C.; Kaliannan, K.; Strain, C.R.; Ross, R.P.; Stanton, C.; Kang, J.X. Maternal omega-3 fatty acids regulate offspring obesity through persistent modulation of gut microbiota. Microbiome 2018, 6, 95. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2009; ISBN 3-900051-07-0. Available online: http://www.R-project.org (accessed on 1 September 2018).
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Society. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Lenz, M.; Arts, I.C.W.; Peeters, R.L.M.; de Kok, T.M.; Ertaylan, G. Adipose tissue in health and disease through the lens of its building blocks. Sci. Rep. 2020, 10, 10433. [Google Scholar] [CrossRef]
- Stockel, D.; Kehl, T.; Trampert, P.; Schneider, L.; Backes, C.; Ludwig, N.; Gerasch, A.; Kaufmann, M.; Gessler, M.; Graf, N.; et al. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics 2016, 32, 1502–1508. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Microarray | RT-qPCR | |||||
|---|---|---|---|---|---|---|
| Genes | HFoleic vs. Ref | HFepa vs. HFoleic | HFepa vs. Ref | HFoleic vs. Ref | HFepa vs. HFoleic | HFepa vs. Ref |
| Dnajb1 | 1.00 | 1.20 a | 1.20a | 0.93 | 1.12 b | 1.05 |
| Dock8 | 4.38 * | 0.81 | 3.53 * | 3.56 b | 0.82 c | 2.92 b |
| Foxo3 | 1.10 | 0.86 a | 0.95 | 0.91 | 0.85 c | 0.77 b |
| Irs2 | 0.51 * | 1.54 a | 0.79 | 0.49 b | 1.38 b | 0.68 b |
| Leptin | 2.18 * | 0.74 a | 1.62 | 4.97 a | 0.46 a | 2.30 |
| Mmp13 | 1.80 * | 0.63 a | 1.13 | 4.40 b | 0.38 b | 1.66 |
| Nupl2 | 0.96 | 0.83 a | 0.79 * | 0.86 c | 0.82 b | 0.71 b |
| Plin2 | 1.94 * | 0.76 | 1.47 a | 1.65 b | 0.66 b | 1.09 |
| Serpine1 | 0.38 * | 2.02 a | 0.77 | 0.37 b | 1.94 c | 0.72 |
| Stat3 | 0.78 * | 1.13 | 0.88 | 0.63 b | 0.96 | 0.60 b |
| Timp3 | 0.61 * | 1.45 a | 0.88 | 0.59 b | 1.18 | 0.70 b |
| Name | Hits (n) | Adjusted p-Value | |
|---|---|---|---|
| Transcription factors | SP1 | 104 | <0.001 |
| SP3 | 45 | <0.001 | |
| SPI1 | 27 | <0.001 | |
| CEBPB | 31 | <0.001 | |
| IRF1 | 16 | <0.01 | |
| IRF8 | 12 | <0.01 | |
| HIF1A | 16 | <0.01 | |
| STAT1 | 15 | <0.01 | |
| YY1 | 18 | <0.01 | |
| USF1 | 15 | <0.01 | |
| CEBPA | 17 | <0.01 | |
| EGR1 | 15 | <0.01 | |
| RELA | 15 | <0.05 | |
| ETS1 | 12 | <0.05 | |
| POU2F1 | 14 | <0.05 | |
| STAT3 | 13 | <0.05 | |
| NFYA | 12 | <0.05 | |
| Transcription factor complexes | AP-1 | 24 | <0.001 |
| NF-kappaB | 14 | <0.05 | |
| PPARgamma:RXR-alpha | 8 | <0.05 | |
| RelA-p65:p50 | 9 | <0.05 | |
| USF2:usf1 | 8 | <0.05 | |
| Transcription factor families | NF-1 | 14 | <0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinel, A.; Rigaudière, J.-P.; Morio, B.; Capel, F. Adipose Tissue Dysfunctions in Response to an Obesogenic Diet Are Reduced in Mice after Transgenerational Supplementation with Omega 3 Fatty Acids. Metabolites 2021, 11, 838. https://doi.org/10.3390/metabo11120838
Pinel A, Rigaudière J-P, Morio B, Capel F. Adipose Tissue Dysfunctions in Response to an Obesogenic Diet Are Reduced in Mice after Transgenerational Supplementation with Omega 3 Fatty Acids. Metabolites. 2021; 11(12):838. https://doi.org/10.3390/metabo11120838
Chicago/Turabian StylePinel, Alexandre, Jean-Paul Rigaudière, Béatrice Morio, and Frédéric Capel. 2021. "Adipose Tissue Dysfunctions in Response to an Obesogenic Diet Are Reduced in Mice after Transgenerational Supplementation with Omega 3 Fatty Acids" Metabolites 11, no. 12: 838. https://doi.org/10.3390/metabo11120838
APA StylePinel, A., Rigaudière, J.-P., Morio, B., & Capel, F. (2021). Adipose Tissue Dysfunctions in Response to an Obesogenic Diet Are Reduced in Mice after Transgenerational Supplementation with Omega 3 Fatty Acids. Metabolites, 11(12), 838. https://doi.org/10.3390/metabo11120838