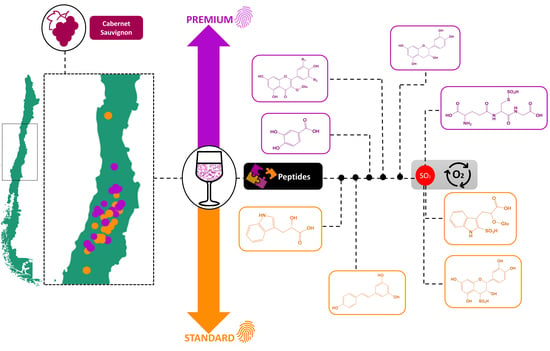
LC–MS-Based Metabolomics Discriminates Premium from Standard Chilean cv. Cabernet Sauvignon Wines from Different Valleys
Abstract
:1. Introduction
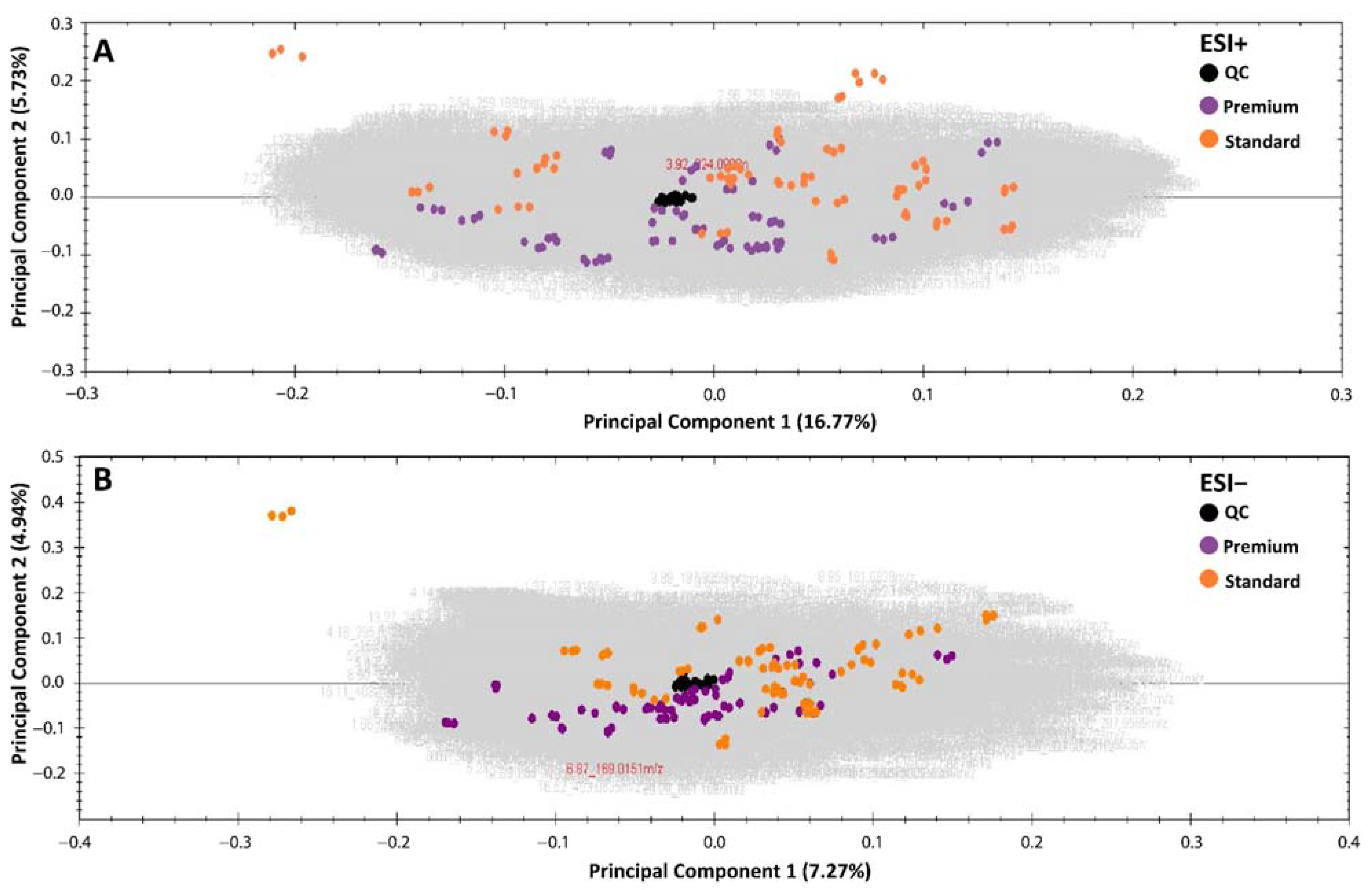
2. Results
2.1. Standard versus Premium Wines
2.2. Super Premium versus Premium versus Standard plus versus Standard Quality Wine Groups
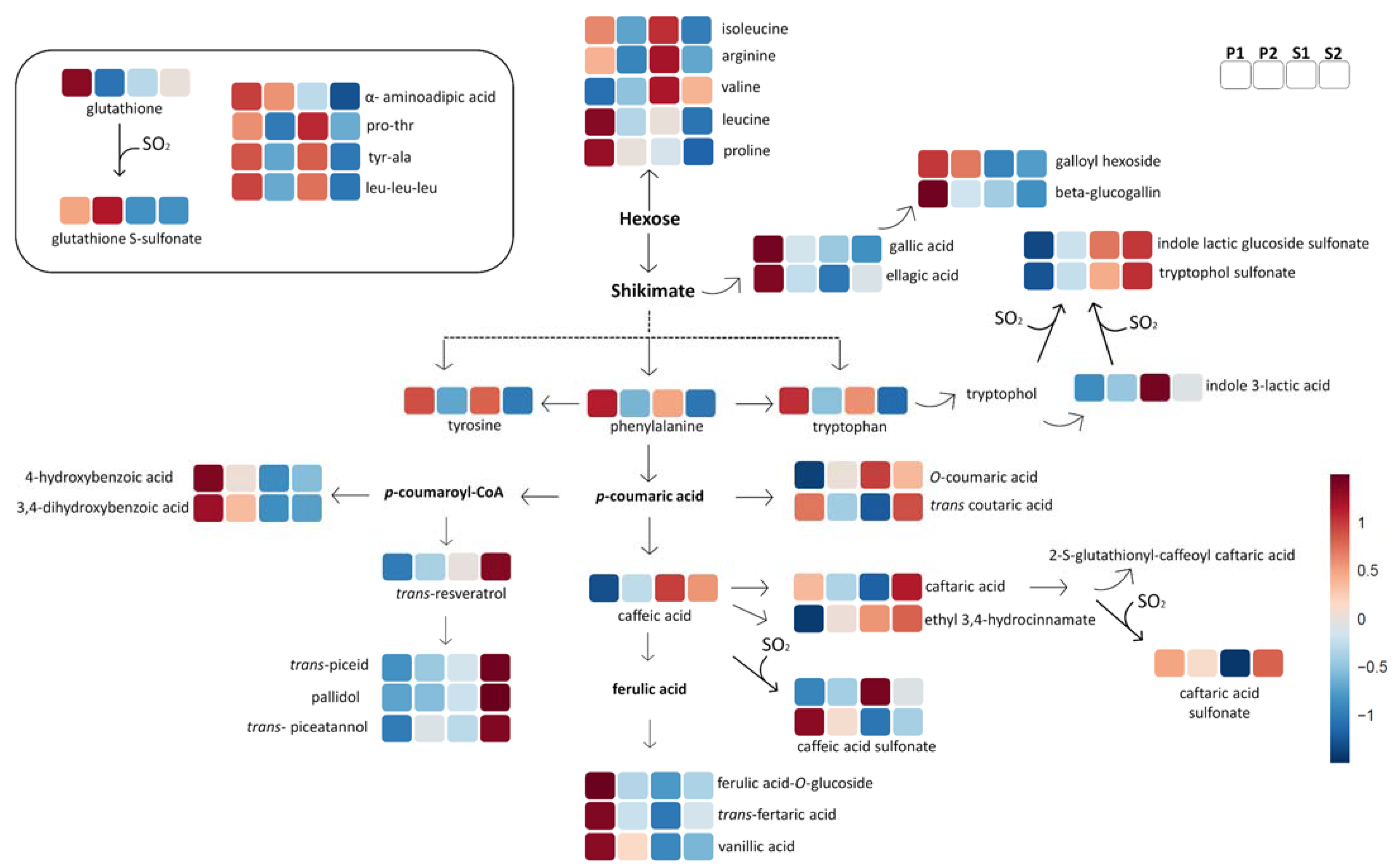
2.2.1. Amino Acids and Peptides
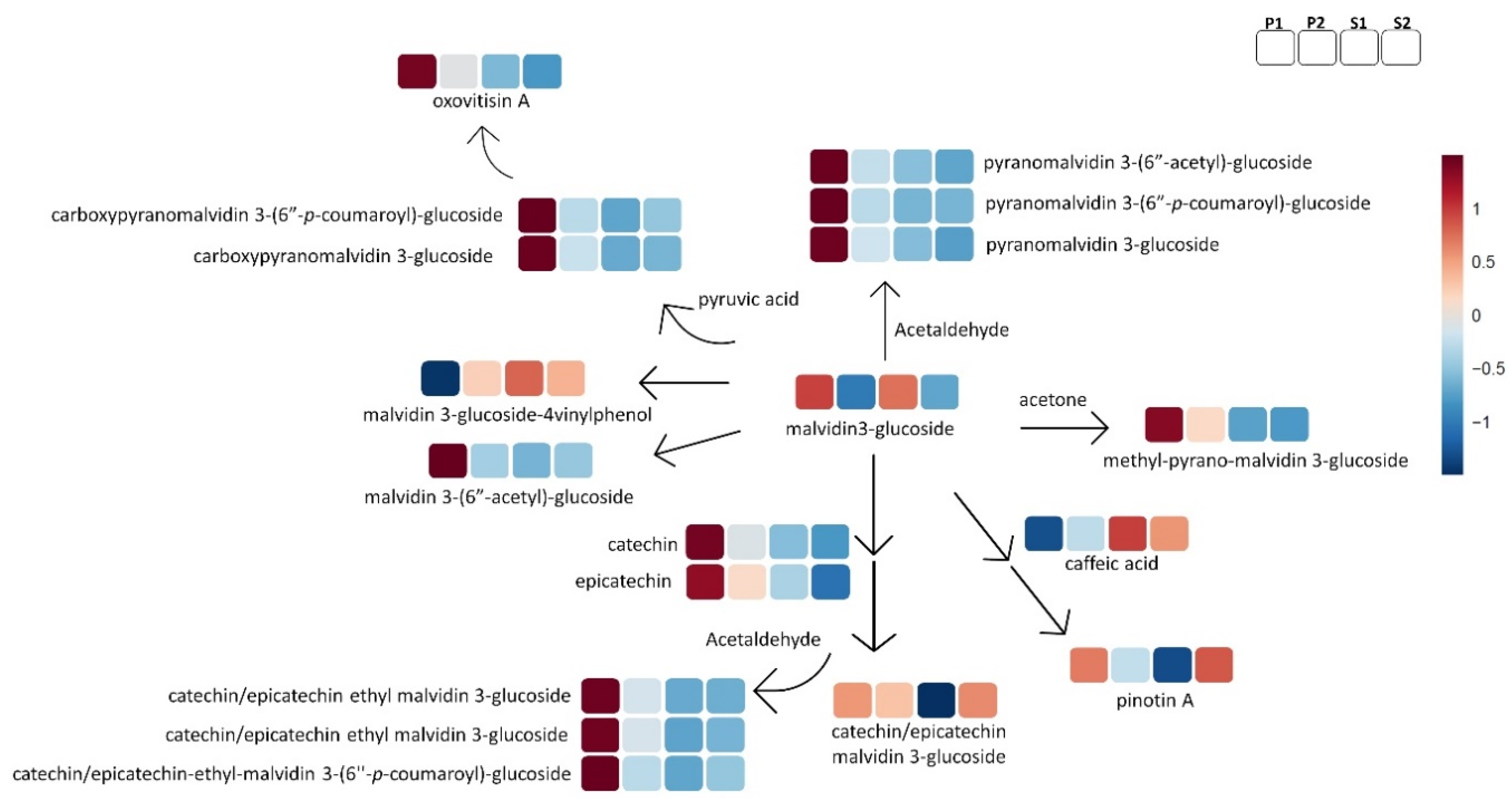
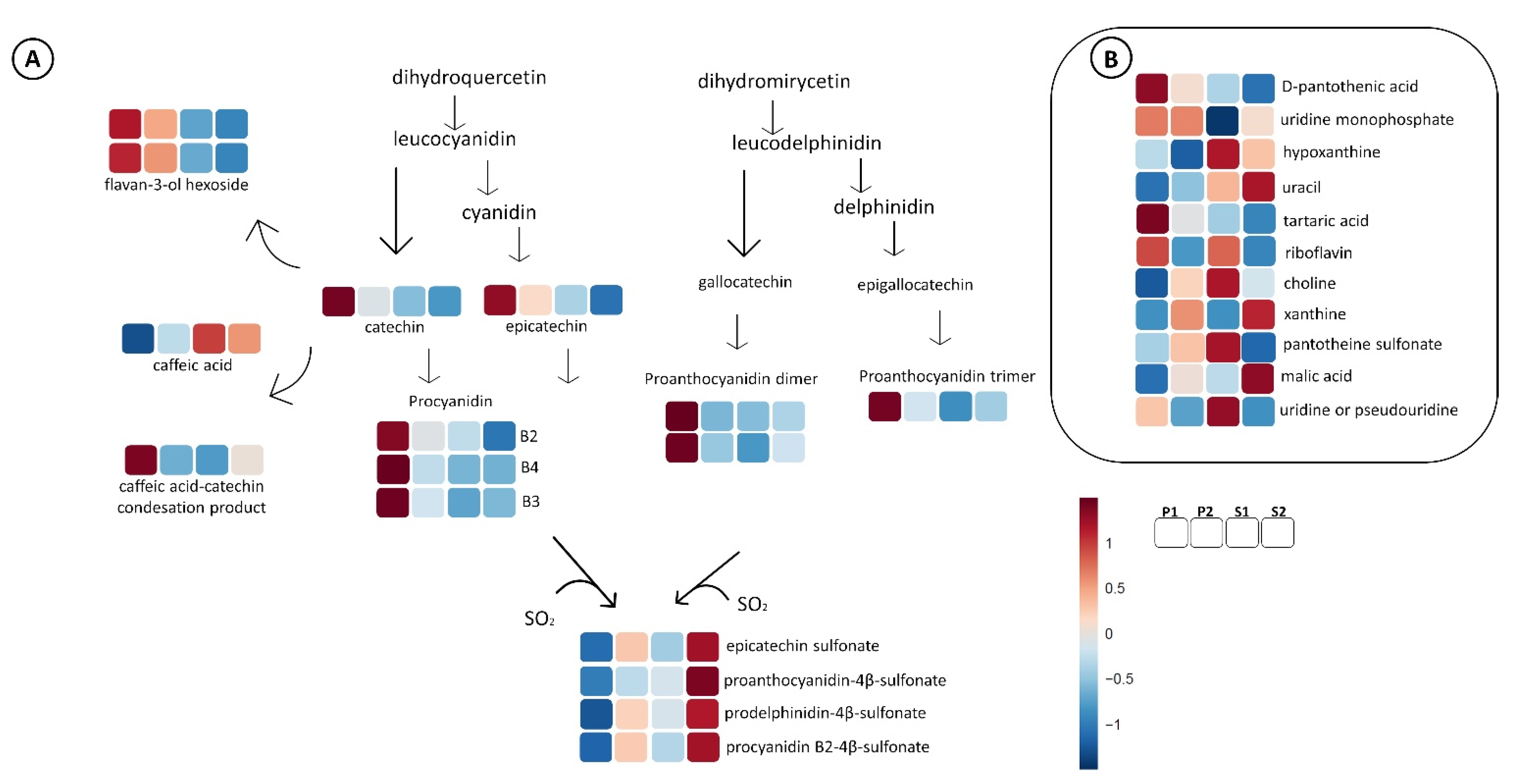
2.2.2. Non-Flavonoid Polyphenols
2.2.3. Flavonoids
2.2.4. Other Compounds
3. Discussion
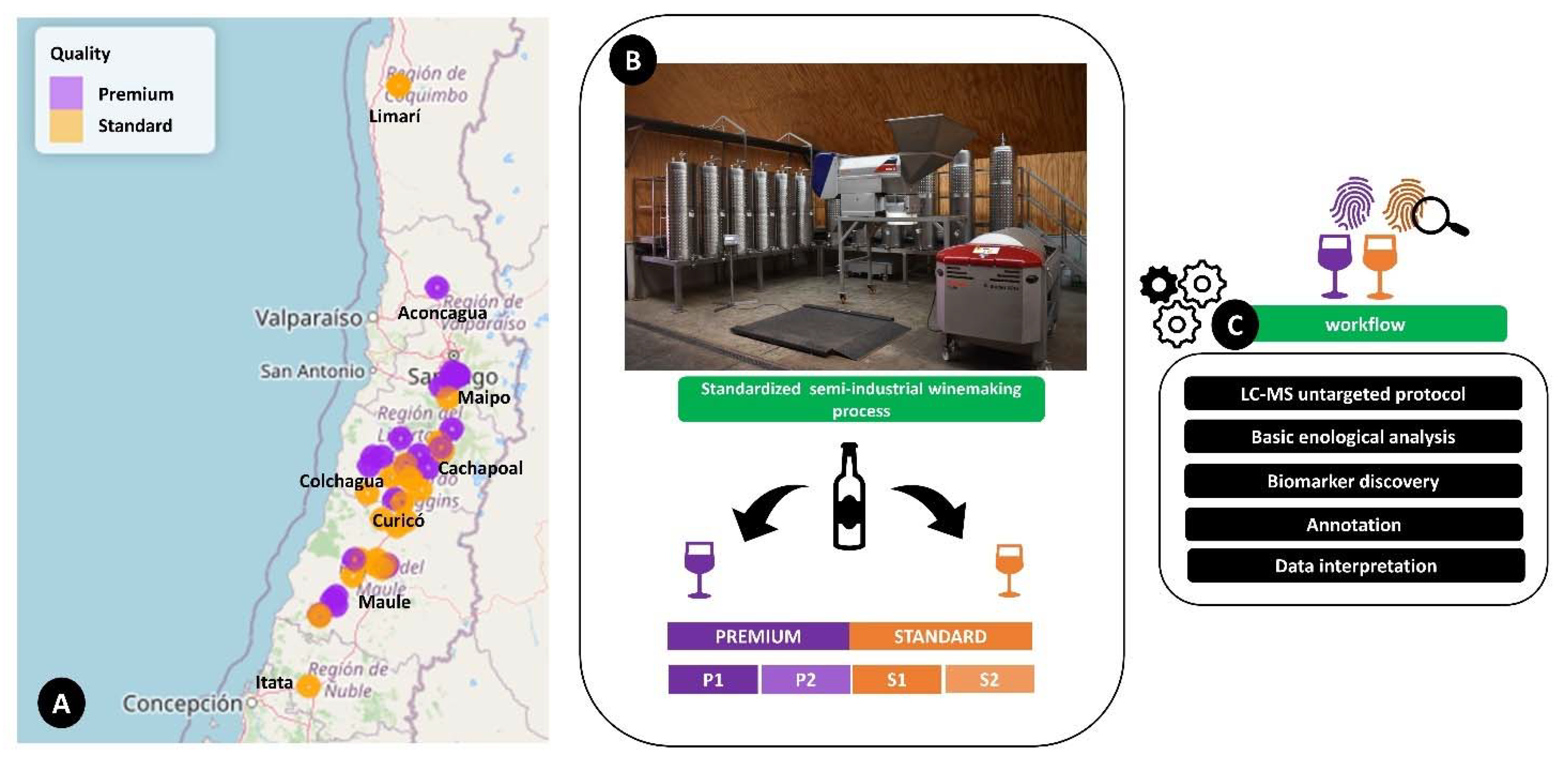
4. Materials and Methods
4.1. Winemaking Procedure
4.2. Chilean cv. Cabernet Sauvignon Wines Samples
4.3. LC–MS-Based Metabolomics Analysis
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Organisation of Vine and Wine (OIV). Distribution of the World’s Grapevine Varieties. Available online: https://www.oiv.int/public/medias/5888/en-distribution-of-the-worlds-grapevine-varieties.pdf (accessed on 31 July 2020).
- Hernández, A. Introducción al Vino de Chile; Ediciones Universidad Católica de Chile: Santiago, Chile, 1997. [Google Scholar]
- Anderson, K.; Nelgen, S. Which Winegrape Varieties Are Grown Where? A Global Empirical Picture; University of Adelaide Press: Adelaide, Australia, 2020; pp. 303–328. [Google Scholar]
- Servicio Agrícola Ganadero (SAG). Catastro Vitícola Nacional. Available online: https://www.sag.gob.cl/noticias/sag-presenta-catastro-viticola-nacional-2019 (accessed on 1 February 2021).
- Sarricolea, P.; Herrera-Ossandon, M.; Meseguer-Ruiz, Ó. Climatic regionalisation of continental Chile. J. Maps 2017, 13, 66–73. [Google Scholar] [CrossRef]
- Montes, C.; Perez-Quezada, J.-F.; Peña-Neira, A.; Tonietto, J. Climatic potential for viticulture in Central Chile. Aust. J. Grape Wine Res. 2012, 18, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Ribereau-Gayon, P.; Glories, Y.; Maujean, A.; Dubourdieu, D. Handbook of Enology, Volume 2: The Chemistry of Wine Stabilization and Treatments, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2021; ISBN 978-1-119-58776-7. [Google Scholar]
- Cuadros-Inostroza, A.; Giavalisco, P.; Hummel, J.; Eckardt, A.; Willmitzer, L.; Peña-Cortés, H. Discrimination of wine attributes by metabolome analysis. Anal. Chem. 2010, 82, 3573–3580. [Google Scholar] [CrossRef]
- Arapitsas, P.; Ugliano, M.; Marangon, M.; Piombino, P.; Rolle, L.; Gerbi, V.; Versari, A.; Mattivi, F. Use of untargeted liquid chromatography–mass spectrometry metabolome to discriminate Italian monovarietal red wines, produced in their different terroirs. J. Agric. Food Chem. 2020, 68, 13353–13366. [Google Scholar] [CrossRef] [PubMed]
- Cuadros-Inostroza, Á.; Verdugo-Alegría, C.; Willmitzer, L.; Moreno-Simunovic, Y.; Vallarino, J.G. Non-targeted metabolite profiles and sensory properties elucidate commonalities and differences of wines made with the same variety but different cultivar clones. Metabolites 2020, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Arapitsas, P.; Scholz, M.; Vrhovsek, U.; Di Blasi, S.; Biondi Bartolini, A.; Masuero, D.; Perenzoni, D.; Rigo, A.; Mattivi, F. A metabolomic approach to the study of wine micro-oxygenation. PLoS ONE 2012, 7, e37783. [Google Scholar] [CrossRef] [PubMed]
- Arapitsas, P.; Ugliano, M.; Perenzoni, D.; Angeli, A.; Pangrazzi, P.; Mattivi, F. Wine metabolomics reveals new sulfonated products in bottled white wines, promoted by small amounts of oxygen. J. Chromatogr. A 2016, 1429, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Arapitsas, P.; Guella, G.; Mattivi, F. The impact of SO2 on wine flavanols and indoles in relation to wine style and age. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Watrelot, A.A.; Addison, B.; Waterhouse, A.L. Condensed tannin reacts with SO2 during wine aging, yielding flavan-3-ol sulfonates. J. Agric. Food Chem. 2018, 66, 9259–9268. [Google Scholar] [CrossRef]
- Roullier-Gall, C.; David, V.; Hemmler, D.; Schmitt-Kopplin, P.; Alexandre, H. Exploring yeast interactions through metabolic profiling. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Arapitsas, P.; Speri, G.; Angeli, A.; Perenzoni, D.; Mattivi, F. The influence of storage on the “chemical age” of red wines. Metabolomics 2014, 10, 816–832. [Google Scholar] [CrossRef]
- Ontañón, I.; Sánchez, D.; Sáez, V.; Mattivi, F.; Ferreira, V.; Arapitsas, P. Liquid chromatography—Mass spectrometry-based metabolomics for understanding the compositional changes induced by oxidative or anoxic storage of red wines. J. Agric. Food Chem. 2020, 68, 13367–13379. [Google Scholar] [CrossRef] [PubMed]
- Sáenz-Navajas, M.P.; Avizcuri, J.M.; Ballester, J.; Fernández-Zurbano, P.; Ferreira, V.; Peyron, D.; Valentin, D. Sensory-active compounds influencing wine experts’ and consumers’ perception of red wine intrinsic quality. LWT—Food Sci. Technol. 2015, 60, 400–411. [Google Scholar] [CrossRef]
- Gambetta, J.M.; Schmidtke, L.M.; Wang, J.; Cozzolino, D.; Bastian, S.E.; Jeffery, D.W. Relating expert quality ratings of australian chardonnay wines to volatile composition and production method. Am. J. Enol. Vitic. 2017, 68, 39–48. [Google Scholar] [CrossRef]
- Sherman, E.; Coe, M.; Grose, C.; Martin, D.; Greenwood, D.R. Metabolomics approach to assess the relative contributions of the volatile and non-volatile composition to expert quality ratings of pinot noir wine quality. J. Agric. Food Chem. 2020, 68, 13380–13396. [Google Scholar] [CrossRef]
- Serni, E.; Pedri, U.; Valls, J.; Sanoll, C.; Dordevic, N.; Überegger, E.; Robatscher, P. Chemical description and organoleptic evaluation of Pinot noir wines from different parts of Italy: A three year investigation. ONEO One 2020, 54, 393–410. [Google Scholar] [CrossRef]
- Becerra-Rodríguez, C.; Marsit, S.; Galeote, V. Diversity of oligopeptide transport in yeast and its impact on adaptation to winemaking conditions. Front. Genet. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Moro, L.; Da Ros, A.; Vieira Da Mota, R.; Purgatto, E.; Mattivi, F.; Arapitsas, P. LC-MS untargeted approach showed that methyl jasmonate application on Vitis labrusca L. grapes increases phenolics at subtropical Brazilian regions. Metabolomics 2020, 16, 1–12. [Google Scholar] [CrossRef]
- Shahaf, N.; Franceschi, P.; Arapitsas, P.; Rogachev, I.; Vrhovsek, U.; Wehrens, R. Constructing a mass measurement error surface to improve automatic annotations in liquid chromatography/mass spectrometry based metabolomics. Rapid Commun. Mass Spectrom. 2013, 27, 2425–2431. [Google Scholar] [CrossRef]
- Fabre, S.; Absalon, C.; Pinaud, N.; Venencie, C.; Teissedre, P.L.; Fouquet, E.; Pianet, I. Isolation, characterization, and determination of a new compound in red wine. Anal. Bioanal. Chem. 2014, 406, 1201–1208. [Google Scholar] [CrossRef]
- Favre, G.; González-Neves, G.; Piccardo, D.; Gómez-Alonso, S.; Pérez-Navarro, J.; Hermosín-Gutiérrez, I. New acylated flavonols identified in Vitis vinifera grapes and wines. Food Res. Int. 2018, 112, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Delcambre, A.; Saucier, C. Identification of new flavan-3-ol monoglycosides by UHPLC-ESI-Q-TOF in grapes and wine. J. Mass Spectrom. 2012, 47, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, F.; Guella, G.; Mattivi, F.; Catorci, D.; Arapitsas, P. Kinetic investigations of sulfite addition to flavanols. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Shiga, K.; Kodama, Y.; Imamura, M.; Uchida, R.; Obata, A.; Bamba, T.; Fukusaki, E. Analysis of the correlation between dipeptides and taste differences among soy sauces by using metabolomics-based component profiling. J. Biosci. Bioeng. 2014, 118, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Bu, T.; Zheng, J.; Liu, L.; Yu, S.; Li, S.; Wu, J. Peptides in brewed wines: Formation, structure, and function. J. Agric. Food Chem. 2021, 69, 2647–2657. [Google Scholar] [CrossRef]
- Sun, X.; Udenigwe, C.C. Chemistry and biofunctional significance of bioactive peptide interactions with food and gut components. J. Agric. Food Chem. 2020, 68, 12972–12977. [Google Scholar] [CrossRef]
- Sáenz-Navajas, M.P.; Fernández-Zurbano, P.; Ferreira, V. Contribution of nonvolatile composition to wine flavor. Food Rev. Int. 2012, 28, 389–411. [Google Scholar] [CrossRef] [Green Version]
- Miyamura, N.; Iida, Y.; Kuroda, M.; Kato, Y.; Yamazaki, J.; Mizukoshi, T.; Miyano, H. Determination and quantification of kokumi peptide, γ-glutamyl-valyl-glycine, in brewed alcoholic beverages. J. Biosci. Bioeng. 2015, 120, 311–314. [Google Scholar] [CrossRef]
- Nikolantonaki, M.; Julien, P.; Coelho, C.; Roullier-Gall, C.; Ballester, J.; Schmitt-Kopplin, P.; Gougeon, R.D. Impact of Glutathione on Wines Oxidative Stability: A Combined Sensory and Metabolomic Study. Front. Chem. 2018, 6, 1–9. [Google Scholar] [CrossRef]
- Schneider, V. Atypical aging defect: Sensory discrimination, viticultural causes, and enological consequences. A review. Am. J. Enol. Vitic. 2014, 65, 277–284. [Google Scholar] [CrossRef] [Green Version]
- Marsit, S.; Sanchez, I.; Galeote, V.; Dequin, S. Horizontally acquired oligopeptide transporters favour adaptation of saccharomyces cerevisiae wine yeast to oenological environment. Environ. Microbiol. 2016, 18, 1148–1161. [Google Scholar] [CrossRef]
- Hellwig, M. Analysis of protein oxidation in food and feed products. J. Agric. Food Chem. 2020, 68, 12870–12885. [Google Scholar] [CrossRef] [PubMed]
- Gawel, R.; Schulkin, A.; McRea, J.; Hack, J.; Pearson, W.; Nandorfy, D.E.; Smith, P. Beyond phenolic bitterness: Tryp-tophol-bisulfites identified as a potential new class of bitter compounds in white wine. In Proceedings of the 17th Australian Wine Industry Technical Conference, Adelaide, Australia, 21–24 July 2019. [Google Scholar]
- Álvarez-Fernández, M.A.; Carafa, I.; Vrhovsek, U.; Arapitsas, P. Modulating wine aromatic amino acid catabolites by using Torulaspora delbrueckii in sequentially inoculated fermentations or Saccharomyces cerevisiae alone. Microorganisms 2020, 8, 1349. [Google Scholar] [CrossRef] [PubMed]
- Piombino, P.; Pittari, E.; Gambuti, A.; Curioni, A.; Giacosa, S.; Mattivi, F.; Parpinello, G.; Rolle, L.; Ugliano, M.; Moio, L. Preliminary sensory characterisation of the diverse astringency of single cultivar Italian red wines and correlation of sub-qualities with chemical composition. Aust. J. Grape Wine Res. 2020, 26, 233–246. [Google Scholar] [CrossRef]
- García-Estévez, I.; Ramos-Pineda, A.; Escribano-Bailón, M.T. Interactions between wine phenolic compounds and human saliva in astringency perception. Food Funct. 2018, 9, 1294–1309. [Google Scholar] [CrossRef]
- Renaud, S.; De Lorgeril, M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet 1992, 339, 1523–1526. [Google Scholar] [CrossRef]
- Flamini, R.; De Rosso, M. High-resolution mass spectrometry and biological properties of grapevine and wine stilbenoids. In Studies in Natural Products Chemistry, 1st ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2019; Volume 61, pp. 175–210. [Google Scholar]
- Gaudette, N.J.; Pickering, G.J. Sensory and chemical characteristics of trans-resveratrol-fortified wine. Aust. J. Grape Wine Res. 2011, 17, 249–257. [Google Scholar] [CrossRef]
- Poklar Ulrih, N.; Opara, R.; Korošec, M.; Wondra, M.; Abram, V. Part II. Influence of Trans-resveratrol addition on the sensory properties of ‘Blaufränkisch’ red wine. Food Chem. Toxicol. 2020, 137, 111124. [Google Scholar] [CrossRef]
- Ferrero-del-Teso, S.; Suárez, A.; Jeffery, D.W.; Ferreira, V.; Fernández-Zurbano, P.; Sáenz-Navajas, M.P. Sensory variability associated with anthocyanic and tannic fractions isolated from red wines. Food Res. Int. 2020, 136, 109340. [Google Scholar] [CrossRef]
- Ferrer-Gallego, R.; Soares, S.; Mateus, N.; Rivas-Gonzalo, J.; Escribano-Bailon, M.T.; Freitas, V. New anthocyanin—Human salivary protein complexes. Langmuir 2015, 31, 8392–8401. [Google Scholar] [CrossRef] [PubMed]
- Schober, D.; Legues, M.; Guidez, H.; Caris Maldonado, J.C.; Vargas, S.; Gonzalez Rojas, A. How geographical origin and vineyard management influence cv. Cabernet-Sauvignon in Chile—Machine learning based quality prediction. In Proceedings of the XIIIth International Terroir Congress, Adelaide, Australia, 17–18 November 2020. [Google Scholar]
- Arapitsas, P.; Mattivi, F. LC-MS untargeted protocol for the analysis of wine. In Metabolic Profiling, 1st ed.; Humana Press: Totowa, NJ, USA, 2018; pp. 225–235. [Google Scholar]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019, 48, 440–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; Anderson, D.; Morais, D.L.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.; Li, S.; et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, 388–396. [Google Scholar] [CrossRef]
- Savoi, S.; Arapitsas, P.; Duchêne, É.; Nikolantonaki, M.; Ontañón, I.; Carlin, S.; Schwander, F.; Gougeon, R.D.; Silva Ferreira, A.; Theodoridis, G.; et al. Grapevine and Wine Metabolomics-Based Guidelines for FAIR Data and Metadata Management. Metabolites 2021, 11, 757. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sáez, V.; Schober, D.; González, Á.; Arapitsas, P. LC–MS-Based Metabolomics Discriminates Premium from Standard Chilean cv. Cabernet Sauvignon Wines from Different Valleys. Metabolites 2021, 11, 829. https://doi.org/10.3390/metabo11120829
Sáez V, Schober D, González Á, Arapitsas P. LC–MS-Based Metabolomics Discriminates Premium from Standard Chilean cv. Cabernet Sauvignon Wines from Different Valleys. Metabolites. 2021; 11(12):829. https://doi.org/10.3390/metabo11120829
Chicago/Turabian StyleSáez, Vania, Doreen Schober, Álvaro González, and Panagiotis Arapitsas. 2021. "LC–MS-Based Metabolomics Discriminates Premium from Standard Chilean cv. Cabernet Sauvignon Wines from Different Valleys" Metabolites 11, no. 12: 829. https://doi.org/10.3390/metabo11120829
APA StyleSáez, V., Schober, D., González, Á., & Arapitsas, P. (2021). LC–MS-Based Metabolomics Discriminates Premium from Standard Chilean cv. Cabernet Sauvignon Wines from Different Valleys. Metabolites, 11(12), 829. https://doi.org/10.3390/metabo11120829