Grape Lipidomics: An Extensive Profiling thorough UHPLC-MS/MS Method
Abstract
1. Introduction
2. Results and Discussion
2.1. Compounds of Interest
| Class | Ionization Mode | Precursor Ion | Product Ion | Reference | Internal Standard | DP (Volts) | EP (Volts) | CE (Volts) | CXP (Volts) |
|---|---|---|---|---|---|---|---|---|---|
| CAR | pos | [M+H]+ | 85.1 | [33,49] | 24:0 (d4) Carnitine | 93 | 10 | 31 | 16 |
| CER | pos | [M+H–18]+ | 264.1 | [33,48] | C15 Ceramide-d7 | 130 | 10 | 55 | 10 |
| DG | pos | [M+Na]+ | [M– (sn2 FA)]+ | [33,44] | 15:0–18:1(d7) DG-Na | 93 | 9 | 42 | 25 |
| DGDG | pos | [M+Na]+ | [M+Na–R2CO2H]+ | [46] | Hydrog DGDG (18:0–18:0) | 80 | 10 | 65 | 20 |
| dhCER | pos | [M+H–18]+ | 266.1 | [33,48] | C15 Ceramide-d7 | 130 | 10 | 55 | 10 |
| FA | neg | [M–H]− | [M–H]− | [33,50] | Stearic acid-d3 | −80 | −10 | −17 | −20 |
| glc-dhCER | pos | [M+H–18]+ | 266.1 | [33] | C15 Ceramide-d7 | 130 | 8 | 45 | 27 |
| glcCER | pos | [M+H–18]+ | 264.1 | [33] | C15 Ceramide-d7 | 130 | 8 | 45 | 27 |
| lac-dhCER | pos | [M+H–18]+ | 266.1 | [33] | C15 Ceramide-d7 | 126 | 10 | 56 | 15 |
| lacCER | pos | [M+H–18]+ | 264.1 | [33] | C15 Ceramide-d7 | 126 | 10 | 56 | 15 |
| LPA | neg | [M–H]− | [sn2 FA]− | [33,41] | 17:0 Lyso PA | −80 | −6 | −45 | −20 |
| LPC | pos | [M+H]+ | 184.1 | [33,43] | 18:1(d7) Lyso PC | 90 | 6 | 35 | 20 |
| LPE | neg | [M–H]− | [sn2 FA]− | [33,40] | 18:1(d7) Lyso PE | −88 | −12 | −42 | −20 |
| LPG | neg | [M–H]− | [sn2 FA]− | [33,42] | 17:1 Lyso PG | −75 | −10 | −38 | −24 |
| LPI | neg | [M–H]− | [sn2 FA]− | [33,36] | 17:1 Lyso PI | −90 | −6 | −40 | −24 |
| LPS | neg | [M–H]− | [sn2 FA]− | [33,37] | 17:1 Lyso PS | −72 | −10 | −53 | −24 |
| MG | pos | [M+H]+ | [M–C3H7O3]+ | [33,44] | 18:1(d7) MG | 140 | 10 | 16 | 10 |
| MGDG | pos | [M+Na]+ | [M+Na–R2CO2H]+ | [46] | Hydrog MGDG (18:0–16:0) | 100 | 10 | 50 | 30 |
| PA | neg | [M–H]− | [sn2 FA]− | [33,40,41] | 15:0–18:1-D7-PA | −80 | −6 | −45 | −20 |
| PC | neg | [M+HCOO]− | [sn2 FA]− | [33,38,39,40] | 15:0–18:1(d7) PC | −90 | −10 | −50 | −20 |
| PE | neg | [M–H]− | [sn2 FA]− | [33,38,40] | 15:0–18:1(d7) PE | −88 | −12 | −42 | −20 |
| PG | neg | [M–H]− | [sn2 FA]− | [33,40,42] | 15:0–18:1(d7) PG | −75 | −10 | −38 | −24 |
| PI | neg | [M–H]− | [sn2 FA]− | [33,36,40] | 15:0–18:1(d7) PI | −50 | −10 | −55 | −10 |
| PS | neg | [M–H]− | [sn2 FA]− | [33,37,40] | 15:0–18:1(d7) PS | −72 | −10 | −53 | −24 |
| SM | pos | [M+H]+ | 184.1 | [33,47] | d18:1–18:1(d9) SM | 124 | 10 | 32.5 | 23 |
| TG | pos | [M+Na]+ | [M– (sn3 FA)]+ | [33,44,45] | 15:0–18:1(d7)-15:0 TG-Na | 90 | 10 | 40 | 10 |
2.2. Chromatographic Optimization
2.3. Method Validation
2.4. Method Application for Grape Maturation Samples
3. Materials and Methods
3.1. Chemicals
3.2. Compounds of Interest and Their Characteristics
3.3. Instrumental Conditions
3.3.1. Optimization of Liquid Chromatography Conditions
3.3.2. Mass Spectrometry Parameters
3.4. Sample Collection and Lipid Extraction
3.5. Method Validation
3.5.1. Recovery
3.5.2. Linearity, Limit of Detection and Limit of Quantification
3.5.3. Repeatability
3.5.4. Intra- and Inter-Day
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACN | acetonitrile |
| BHT | butylated hydroxytoluene |
| CAR | carnitine(s) |
| CE | collision energy |
| CER | ceramide(s) |
| CV | coefficient of variation |
| CXP | collision cell exit potential |
| DG | diacylglycerol(s) |
| DGDG | digalactosyldiacylglycerol(s) |
| dhCER | dihydroceramide(s) |
| DP | declustering potential |
| EP | entrance potential |
| ESI | electrospray ionization |
| FA | free fatty acid(s) |
| GL | glycerolipid(s) |
| glcCER | glucosyl ceramide(s) |
| glc-dhCER | glucosyldihydroceramide(s) |
| GP | glycerophospholipids(s) |
| HPLC | high-performance liquid chromatography |
| IPA | 2-propanol |
| IS | internal standard(s) |
| KMD | Kendrick mass defect |
| lacCER | lactosyl ceramide(s) |
| lac-dhCER | lactosyldihydroceramide(s) |
| LC | liquid chromatography |
| LOD | limit of detection |
| LOQ | limit of quantification |
| LPA | lyso-glycerophosphate(s) |
| LPC | lyso-glycerophosphocholine(s) |
| LPE | lyso-glycerophosphoethanolamine(s) |
| LPI | lyso-glycerophosphoinositol(s) |
| LPG | lyso-glycerophosphoglycerol(s) |
| LPS | lyso-glycerophosphoserine(s) |
| MG | monoacylglycerol(s) |
| MGDG | monogalactosyldiacylglycerol(s) |
| MRM | multiple reaction monitoring |
| MS | mass spectrometry |
| PA | glycerophosphate(s) |
| PC | glycerophosphocholine(s) |
| PCA | principal component analysis |
| PE | glycerophosphoethanolamine(s) |
| PI | glycerophosphoinositol(s) |
| PG | glycerophosphoglycerol(s) |
| PK | polyketide(s) |
| PLS | partial least squares |
| PR | prenol lipid(s) |
| PS | glycerophosphoserine(s) |
| SL | saccharolipids(s) |
| SM | sphingomyelin(s) |
| SP | sphingolipid(s) |
| SQDG | sulfoquinovosyldiacylglycerol(s) |
| ST | sterol(s) |
| TG | triacylglycerol(s) |
| UPLC | ultrahigh performance liquid chromatography |
References
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Astarita, G.; Ahmed, F.; Piomelli, D. Lipidomic analysis of biological samples by liquid chromatography coupled to mass spectrometry. Methods Mol. Biol. 2009, 579, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2011, 1811, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Han, X. Tutorial on lipidomics. Anal. Chim. Acta 2019, 1061, 28–41. [Google Scholar] [CrossRef]
- Li, M.; Yang, L.; Bai, Y.; Liu, H. Analytical methods in lipidomics and their applications. Anal. Chem. 2014, 86, 161–175. [Google Scholar] [CrossRef]
- Wu, B.; Wei, F.; Xu, S.; Xie, Y.; Lv, X.; Chen, H.; Huang, F. Mass spectrometry-based lipidomics as a powerful platform in foodomics research. Trends Food Sci. Technol. 2021, 107, 358–376. [Google Scholar] [CrossRef]
- Blanksby, S.J.; Mitchell, T.W. Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 2010, 3, 433–465. [Google Scholar] [CrossRef]
- Salem, M.A.; Giavalisco, P. Semi-targeted lipidomics of plant acyl lipids using UPLC-HR-MS in combination with a data-independent acquisition mode. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1778, pp. 137–155. [Google Scholar]
- Cajka, T.; Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC—Trends Anal. Chem. 2014, 61, 192–206. [Google Scholar] [CrossRef]
- McDonald, J.G.; Smith, D.D.; Stiles, A.R.; Russell, D.W. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 2012, 53, 1399. [Google Scholar] [CrossRef]
- Wang, M.; Wang, C.; Han, R.H.; Han, X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016, 61, 83–108. [Google Scholar] [CrossRef] [PubMed]
- Han, X. Plant Lipidomics. Lipidomics 2016, 405–426. [Google Scholar] [CrossRef]
- Hölzl, G.; Dörmann, P. Structure and function of glycoglycerolipids in plants and bacteria. Prog. Lipid Res. 2007, 46, 225–243. [Google Scholar] [CrossRef] [PubMed]
- Kehelpannala, C.; Rupasinghe, T.; Pasha, A.; Esteban, E.; Hennessy, T.; Bradley, D.; Ebert, B.; Provart, N.J.; Roessner, U. An Arabidopsis lipid map reveals differences between tissues and dynamic changes throughout development. Plant J. 2021, 107, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Della Corte, A.; Chitarrini, G.; Di Gangi, I.M.; Masuero, D.; Soini, E.; Mattivi, F.; Vrhovsek, U. A rapid LC-MS/MS method for quantitative profiling of fatty acids, sterols, glycerolipids, glycerophospholipids and sphingolipids in grapes. Talanta 2015, 140, 52–61. [Google Scholar] [CrossRef]
- Gallander, J.F.; Peng, A.C. Lipid and fatty acid compositions of different grape types. Am. J. Enol. Vitic. 1980, 31, 24–27. [Google Scholar]
- Deroite, A.; Legras, J.L.; Rigou, P.; Ortiz-Julien, A.; Dequin, S. Lipids modulate acetic acid and thiol final concentrations in wine during fermentation by Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids. AMB Express 2018, 8. [Google Scholar] [CrossRef]
- Podolyan, A.; White, J.; Jordan, B.; Winefield, C.; Podolyan, A.; White, J.; Jordan, B.; Winefield, C. Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct. Plant Biol. 2010, 37, 767–784. [Google Scholar] [CrossRef]
- Šuklje, K.; Carlin, S.; Stanstrup, J.; Antalick, G.; Blackman, J.W.; Meeks, C.; Deloire, A.; Schmidtke, L.M.; Vrhovsek, U. Unravelling wine volatile evolution during Shiraz grape ripening by untargeted HS-SPME-GC × GC-TOFMS. Food Chem. 2019, 277, 753–765. [Google Scholar] [CrossRef]
- Tesnière, C. Importance and role of lipids in wine yeast fermentation. Appl. Microbiol. Biotechnol. 2019, 103, 8293–8300. [Google Scholar] [CrossRef]
- Alexandre, H.; Rousseaux, I.; Charpentier, C. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol. Lett. 1994, 124, 17–22. [Google Scholar] [CrossRef]
- You, K.M.; Rosenfield, C.L.; Knipple, D.C. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl. Environ. Microbiol. 2003, 69, 1499–1503. [Google Scholar] [CrossRef] [PubMed]
- Mbuyane, L.L.; Bauer, F.F.; Divol, B. The metabolism of lipids in yeasts and applications in oenology. Food Res. Int. 2021, 141, 110142. [Google Scholar] [CrossRef] [PubMed]
- Tumanov, S.; Zubenko, Y.; Greven, M.; Greenwood, D.R.; Shmanai, V.; Villas-Boas, S.G. Comprehensive lipidome profiling of Sauvignon blanc grape juice. Food Chem. 2015, 180, 249–256. [Google Scholar] [CrossRef]
- Ruocco, S.; Stefanini, M.; Stanstrup, J.; Perenzoni, D.; Mattivi, F.; Vrhovsek, U. The metabolomic profile of red non-V. vinifera genotypes. Food Res. Int. 2017, 98, 10–19. [Google Scholar] [CrossRef]
- Bauman, J.A.; Gallander, J.F.; Peng, A.C. Effect of maturation on the lipid content of Concord grapes. Am. J. Enol. Vitic. 1977, 28, 241–244. [Google Scholar]
- Le Fur, Y.; Hory, C.; Bard, M.-H.; Olsson, A. Evolution of phytosterols in Chardonnay grape berry skins during last stages of ripening. VITIS—J. Grapevine Res. 1994, 33, 127–131. [Google Scholar] [CrossRef]
- Barron, L.J.R.; Santa-María, G. A relationship between triglycerides and grape-ripening indices. Food Chem. 1990, 37, 37–45. [Google Scholar] [CrossRef]
- Barron, L.J.R.; Celaa, M.V.; Santa-Maria, G. Triacylglycerol changes in grapes in late stages of ripening. Phytochemistry 1989, 28, 3301–3305. [Google Scholar] [CrossRef]
- Arita, K.; Honma, T.; Suzuki, S. Comprehensive and comparative lipidome analysis of Vitis vinifera L. cv. Pinot Noir and Japanese indigenous V. vinifera L. cv. Koshu grape berries. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Pérez-Navarro, J.; Da Ros, A.; Masuero, D.; Izquierdo-Cañas, P.M.; Hermosín-Gutiérrez, I.; Gómez-Alonso, S.; Mattivi, F.; Vrhovsek, U. LC-MS/MS analysis of free fatty acid composition and other lipids in skins and seeds of Vitis vinifera grape cultivars. Food Res. Int. 2019, 125, 108556. [Google Scholar] [CrossRef]
- Han, X. Lipidomics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; ISBN 9781119085263. [Google Scholar]
- Reszczyńska, E.; Hanaka, A. Lipids composition in plant membranes. Cell Biochem. Biophys. 2020, 78, 401–414. [Google Scholar] [CrossRef]
- LIPID MAPS. Available online: https://www.lipidmaps.org/ (accessed on 10 September 2021).
- Hsu, F.F.; Turk, J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: A mechanistic study. J. Am. Soc. Mass Spectrom. 2000, 11, 986–999. [Google Scholar] [CrossRef]
- Hsu, F.-F.; Turk, J. Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: Structural characterization and the fragmentation processes. J. Am. Soc. Mass Spectrom. 2005, 16, 1510–1522. [Google Scholar] [CrossRef]
- Kerwin, J.L.; Tuininga, A.R.; Ericssont, L.H. Identification of molecular species of glycerophospholipids and sphingomyelin using electrospray mass spectrometry. J. Lipid Res. 1994, 35. [Google Scholar] [CrossRef]
- Pulfer, M.; Murphy, R.C. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003, 22, 332–364. [Google Scholar] [CrossRef]
- Hsu, F.F.; Turk, J. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J. Chromatogr. B 2009, 877, 2673–2695. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.F.; Turk, J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000, 11, 797–803. [Google Scholar] [CrossRef]
- Hsu, F.F.; Turk, J. Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: Fragmentation processes and structural characterization. J. Am. Soc. Mass Spectrom. 2001, 12, 1036–1043. [Google Scholar] [CrossRef]
- Khaselev, N.; Murphy, R.C. Electrospray ionization mass spectrometry of lysoglycerophosphocholine lipid subclasses. J. Am. Soc. Mass Spectrom. 2000, 11, 283–291. [Google Scholar] [CrossRef]
- Duffin, K.L.; Henion, J.D.; Shieh, J.J. Electrospray and tandem mass spectrometric characterization of acylglycerol mixtures that are dissolved in nonpolar solvents. Anal. Chem. 2002, 63, 1781–1788. [Google Scholar] [CrossRef] [PubMed]
- Herrera, L.C.; Potvin, M.A.; Melanson, J.E. Quantitative analysis of positional isomers of triacylglycerols via electrospray ionization tandem mass spectrometry of sodiated adducts. Rapid Commun. Mass Spectrom. 2010, 24, 2745–2752. [Google Scholar] [CrossRef] [PubMed]
- Guella, G.; Frassanito, R.; Mancini, I. A new solution for an old problem: The regiochemical distribution of the acyl chains in galactolipids can be established by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 1982–1994. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.F.; Turk, J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2000, 11, 437–449. [Google Scholar] [CrossRef][Green Version]
- Hsu, F.F.; Turk, J.; Stewart, M.E.; Downing, D.T. Structural studies on ceramides as lithiated adducts by low energy collisional-activated dissociation tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2002, 13, 680–695. [Google Scholar] [CrossRef][Green Version]
- Su, X.; Han, X.; Mancuso, D.J.; Abendschein, D.R.; Gross, R.W. Accumulation of long-chain acylcarnitine and 3-hydroxy acylcarnitine molecular species in diabetic myocardium: Identification of alterations in mitochondrial fatty acid processing in diabetic myocardium by shotgun lipidomics. Biochemistry 2005, 44, 5234–5245. [Google Scholar] [CrossRef]
- Kerwin, J.L.; Wiens, A.M.; Ericsson, L.H. Identification of fatty acids by electrospray mass spectrometry and tandem mass spectrometry. J. Mass Spectrom. 1996, 31, 184–192. [Google Scholar] [CrossRef]
- Khan, M.J.; Codreanu, S.G.; Goyal, S.; Wages, P.A.; Gorti, S.K.K.; Pearson, M.J.; Uribe, I.; Sherrod, S.D.; McLean, J.A.; Porter, N.A.; et al. Evaluating a targeted multiple reaction monitoring approach to global untargeted lipidomic analyses of human plasma. Rapid Commun. Mass Spectrom. 2020, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Isaac, G.; Mcdonald, S.; Astarita, G. Lipid Separtion Using UPLC with Charged Surface Hybrid Technology; Waters Corporation: Milford, MA, USA, 2011; pp. 1–8. [Google Scholar]
- Lange, M.; Angelidou, G.; Ni, Z.; Criscuolo, A.; Schiller, J.; Blüher, M.; Fedorova, M. AdipoAtlas: A reference lipidome for human white adipose tissue. Cell Reports Med. 2021, 2, 100407. [Google Scholar] [CrossRef]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2019, 412, 2191–2209. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Bioanalytical Method Validation Guidance for Industry. 2018. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioanalytical-method-validation-guidance-industry (accessed on 10 September 2021).
- Shrestha, P.; Callahan, D.L.; Singh, S.P.; Petrie, J.R.; Zhou, X.-R. Reduced triacylglycerol mobilization during seed germination and early seedling growth in Arabidopsis containing nutritionally important polyunsaturated fatty acids. Front. Plant Sci. 2016, 7, 1402. [Google Scholar] [CrossRef]
- Kim, H.U. Lipid metabolism in plants. Plants 2020, 9, 871. [Google Scholar] [CrossRef]
- Miele, A.; Bouard, J.; Bertrand, A. Fatty acids from lipid fractions of leaves and different tissues of Cabernet Sauvignon grapes. Am. J. Enol. Vitic. 1993, 44, 180–186. [Google Scholar]
- Kobayashi, K. Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 2016, 129, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, L.V.; Napier, J.A.; Molino, D.; Faure, J.D. Plant sphingolipids: Their importance in cellular organization and adaption. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2016, 1861, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.-C.; Liu, Z.; Wu, J.-X.; Liang, H.; Xi, X.-L.; Fang, C.; Sun, T.-J.; Yin, J.; Dai, G.-Y.; Rong, C.; et al. Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 2014, 26, 3449–3467. [Google Scholar] [CrossRef]
- Millán, C.; Vargas, A.; Rubio, A.; Moreno, J.; Ortega, J.M. Fatty acid content of unripe and ripe Pedro Ximénez Vitis vinifera grapes. J. Wine Res. 1992, 3, 235–240. [Google Scholar] [CrossRef]
- Fauconnier, M.L.; Welti, R.; Blée, E.; Marlier, M. Lipid and oxylipin profiles during aging and sprout development in potato tubers (Solanum tuberosum L.). Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2003, 1633, 118–126. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Vrhovsek, U.; Mattivi, F. Quantitative profiling of polar primary metabolites using hydrophilic interaction ultrahigh performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2012, 1259, 121–127. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the siolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
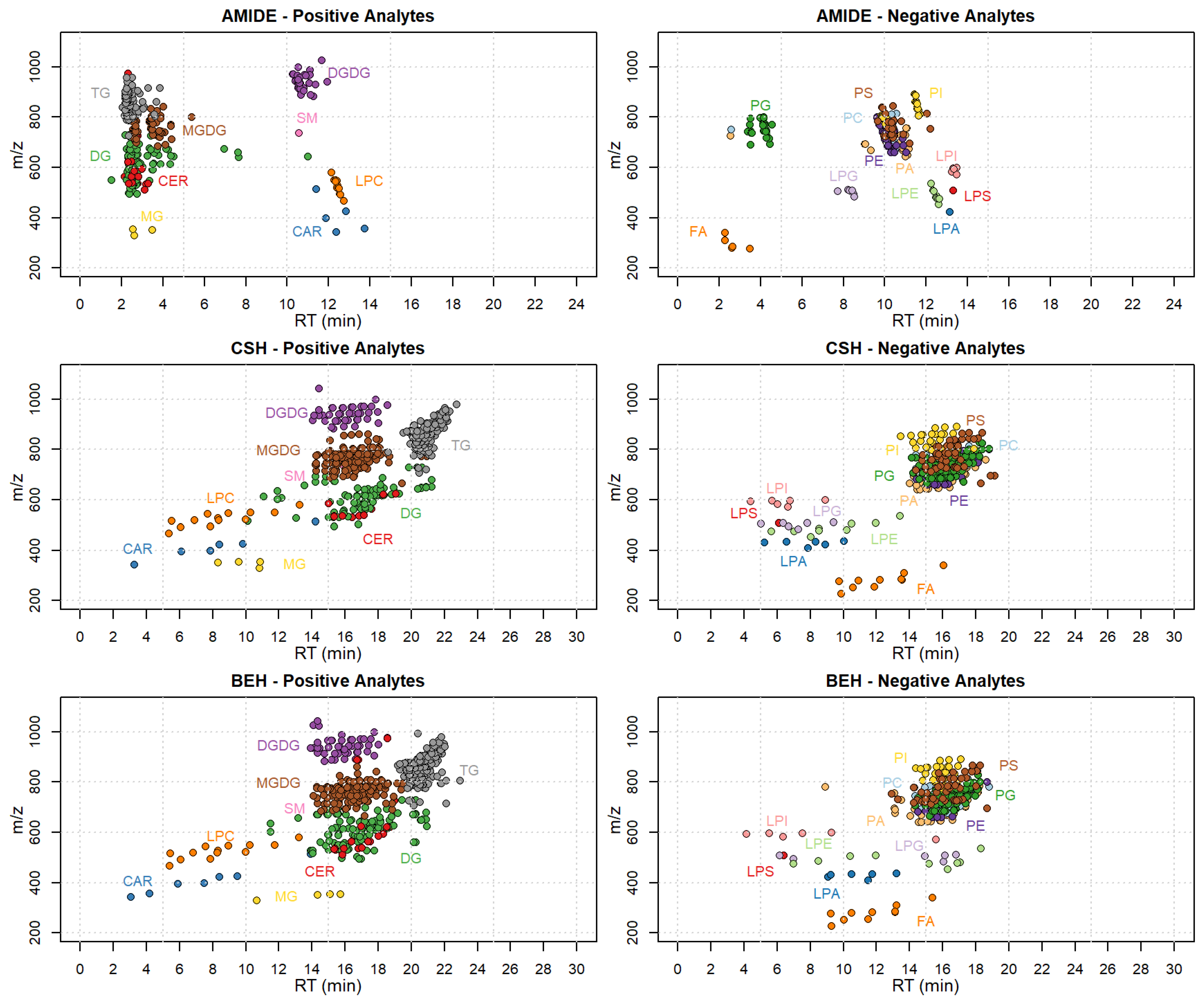
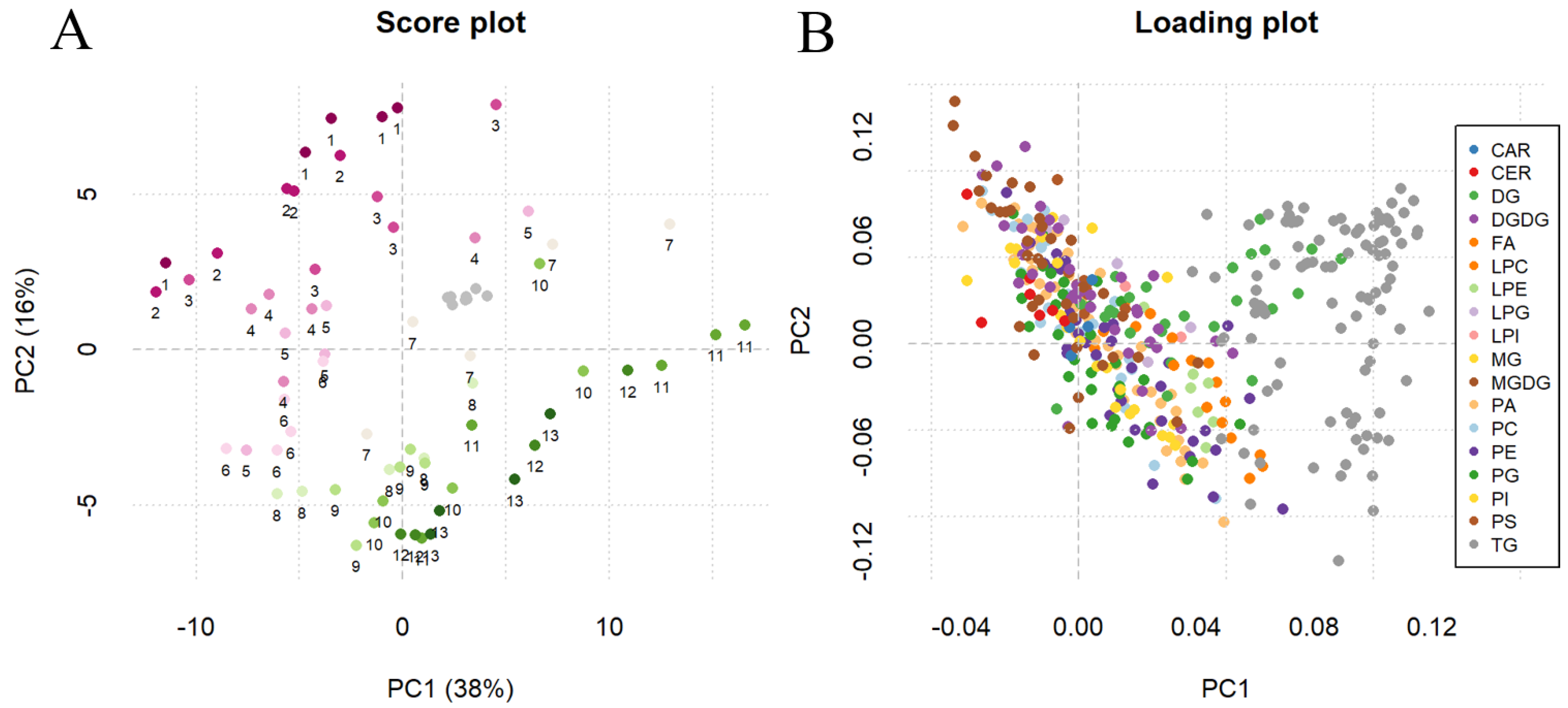
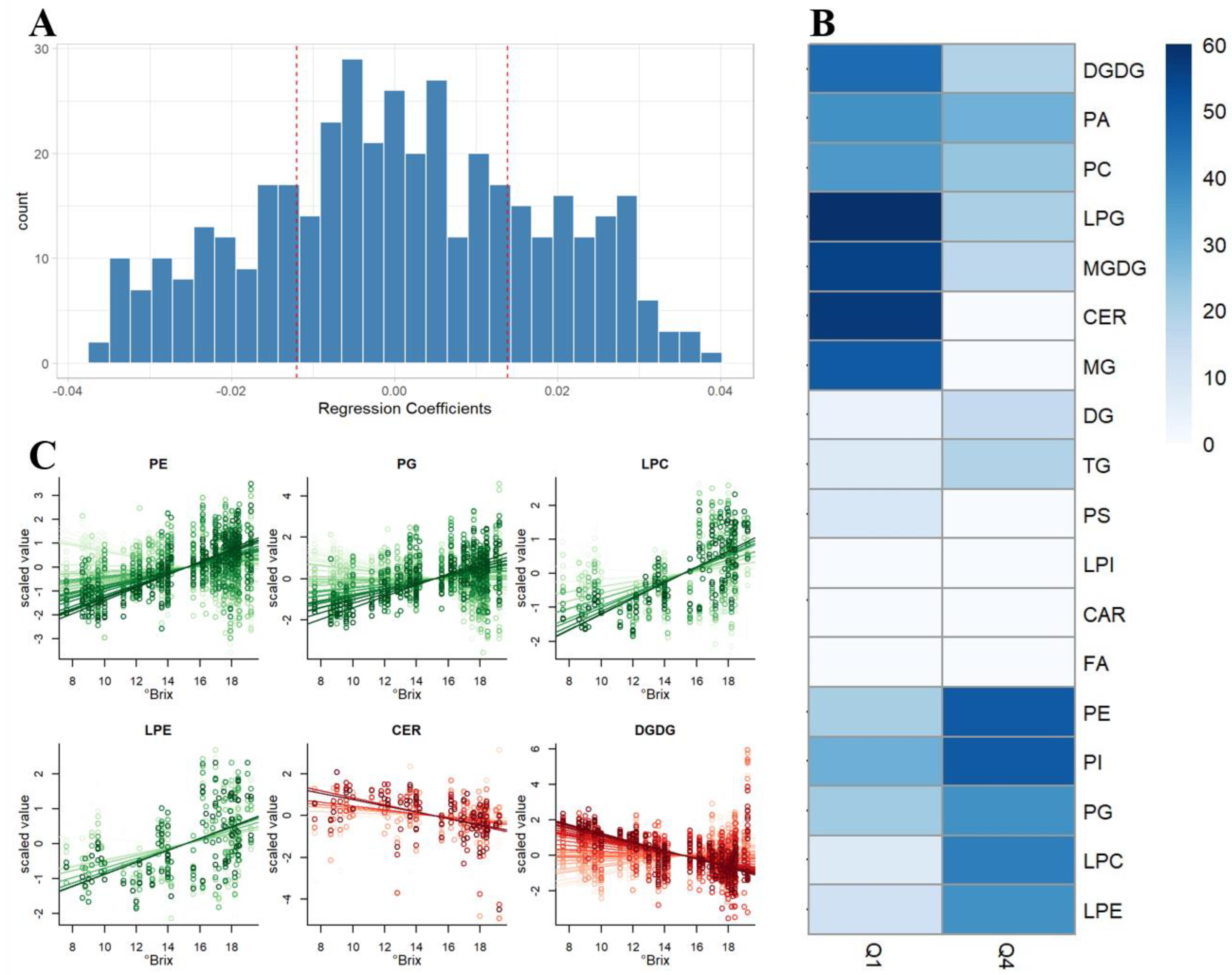
| Class | Compounds in Method | Based on the IS Compounds | Matrix | Validated | Based on the Reference Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|
| Recovery % | LOD (µg/g) | Linearity (µg/g) | Repeatability Range (CV%) | Intra-Day Range (CV%) | Inter-Day Range (CV%) | ||||
| CAR | 48 | 99 | 0.00003 | 0.0003–3 | 5 | 4 | 9–17 | 7–10 | 8–19 |
| CER | 210 | 118 | 0.005 | 0.03–150 | 11 | 7 | 8–15 | 2–15 | 6–21 |
| DG | 630 | 118 | 0.00003 | 0.0015–3 | 132 | 26 | 3–19 | 2–12 | 5–20 |
| DGDG | 630 | 96 | 0.00003 | 0.0003–30 | 43 | 37 | 2–18 | 2–15 | 6–21 |
| FA | 35 | 94 | 0.00003 | 0.003–300 | 8 | 5 | 9–19 | 4–13 | 6–18 |
| LPA | 35 | 76 | 0.05 | 0.15–300 | 0 | 0 | -- | -- | -- |
| LPC | 35 | 100 | 0.00003 | 0.0003–3 | 12 | 12 | 4–9 | 2–7 | 5–7 |
| LPE | 35 | 98 | 0.00003 | 0.0003–150 | 8 | 8 | 2–7 | 2–4 | 4–7 |
| LPG | 35 | 29 | 0.00003 | 0.0003–150 | 5 | 5 | 5–19 | 3–8 | 4–11 |
| LPI | 35 | 4 | 0.00003 | 0.00015–300 | 5 | 2 | 11–14 | 14–15 | 17–19 |
| LPS | 35 | 39 | 0.003 | 0.015–300 | 0 | 0 | -- | -- | -- |
| MG | 35 | 106 | 0.001 | 0.003–150 | 3 | 2 | 10–20 | 5–7 | 8–13 |
| MGDG | 630 | 100 | 0.00003 | 0.00015–3 | 150 | 36 | 2–17 | 1–14 | 4–21 |
| PA | 630 | 101 | 0.001 | 0.003–300 | 53 | 45 | 4–20 | 2–16 | 4–21 |
| PC | 630 | 105 | 0.005 | 0.015–300 | 51 | 25 | 3–20 | 3–16 | 10–21 |
| PE | 630 | 101 | 0.00003 | 0.0003–150 | 60 | 34 | 4–20 | 2–15 | 6–21 |
| PG | 630 | 92 | 0.0001 | 0.0003–30 | 104 | 32 | 4–19 | 2–12 | 5–21 |
| PI | 630 | 68 | 0.00003 | 0.0003–30 | 31 | 20 | 6–21 | 3–16 | 6–21 |
| PS | 630 | 103 | 0.0003 | 0.015–300 | 59 | 11 | 5–18 | 3–14 | 9–20 |
| SM | 35 | 81 | 0.0003 | 0.03–30 | 0 | 0 | -- | -- | -- |
| TG | 1834 | 95 | 0.005 | 0.015–30 | 305 | 101 | 4–21 | 1–16 | 5–21 |
| TOTAL | 8077 | 1045 | 412 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masuero, D.; Škrab, D.; Chitarrini, G.; Garcia-Aloy, M.; Franceschi, P.; Sivilotti, P.; Guella, G.; Vrhovsek, U. Grape Lipidomics: An Extensive Profiling thorough UHPLC-MS/MS Method. Metabolites 2021, 11, 827. https://doi.org/10.3390/metabo11120827
Masuero D, Škrab D, Chitarrini G, Garcia-Aloy M, Franceschi P, Sivilotti P, Guella G, Vrhovsek U. Grape Lipidomics: An Extensive Profiling thorough UHPLC-MS/MS Method. Metabolites. 2021; 11(12):827. https://doi.org/10.3390/metabo11120827
Chicago/Turabian StyleMasuero, Domenico, Domen Škrab, Giulia Chitarrini, Mar Garcia-Aloy, Pietro Franceschi, Paolo Sivilotti, Graziano Guella, and Urska Vrhovsek. 2021. "Grape Lipidomics: An Extensive Profiling thorough UHPLC-MS/MS Method" Metabolites 11, no. 12: 827. https://doi.org/10.3390/metabo11120827
APA StyleMasuero, D., Škrab, D., Chitarrini, G., Garcia-Aloy, M., Franceschi, P., Sivilotti, P., Guella, G., & Vrhovsek, U. (2021). Grape Lipidomics: An Extensive Profiling thorough UHPLC-MS/MS Method. Metabolites, 11(12), 827. https://doi.org/10.3390/metabo11120827








